Abstract
In a prospective cohort of 82 renal transplant recipients, we evaluated the capacity of the cytomegalovirus (CMV) load in whole blood (WB) to predict the plasma CMV load, aiming to identify active CMV infections by using WB samples only and to deduce a WB threshold. Using quantitative real-time PCR, a total of 1,474 WB samples were assayed, of which 279 were positive for CMV, and 140 out of the 276 paired plasma samples tested positive. Thirty (36.6%) patients presented with at least one positive plasma PCR result, and 21 infection episodes (19 patients) required curative treatment (median follow-up time, 12 months). When the plasma CMV load was >500 copies/ml (n = 70), more than 94% (95% confidence interval, 86.0%, 98.4%) of WB samples had >500 copies/ml. Two prediction models were built: log10 plasma viral load (VL) was calculated as −0.3777 + 0.9342 × log10 WB VL and as −0.3777 + 0.8563 × log10 WB VL for patients with and without treatment, respectively. In the validation sample (578 routine samples), 77.2% of the observed and expected plasma viral loads were concordant (95% confidence intervals, 73.5 and 80.5%). According to the model, the plasma viral load was >500 copies/ml when the WB load was >3,170 or >4,000 copies/ml in patients with or without treatment, respectively. WB seems to be an appropriate candidate for routine CMV monitoring of transplant recipients by using a single assay.
Cytomegalovirus (CMV) remains a major opportunistic agent among transplant recipients, due to its direct and indirect effects and despite the use of different therapeutic strategies. While detection of CMV pp65 lower matrix protein (pp65 antigen [Ag]) has been widely performed for diagnosis of CMV infection, molecular assays based on quantitative PCR are now routinely used. Indeed, sensitive assays for the early detection of CMV infection are required for monitoring of patients and are essential for timely application of antiviral therapy (6, 12, 16, 24, 25, 28, 37).
Commercial assays were first available for quantification of CMV DNA in plasma and peripheral blood leukocytes (PBL) (23, 24, 28, 29, 36). Then real-time PCR technology became available and represented a considerable improvement, since it was simpler, cheaper, and less time-consuming. In many laboratories, CMV infection diagnosis now relies on real-time PCR assays (7, 8, 10, 11, 14, 16, 17, 19, 21, 26, 28, 31, 34, 40), all the more since multiplex real-time PCR assays enable virological follow-up, including several opportunistic viruses in a single biological sample (33). The question of which type of blood fraction (PBL, plasma, or whole blood [WB]) is best for monitoring CMV DNA in blood is still unresolved and may be context specific. CMV is highly cell associated, and viral loads (VL) have been shown to be higher in PBL and WB than in plasma (1, 7, 9, 18, 29). Plasma viral load monitoring is of modest clinical utility for prediction of CMV disease and delays the detection of CMV DNA, since a negative PCR result for CMV in plasma does not rule out active infection (2, 4, 5, 12, 24, 26). On the other hand, when CMV is detected in the plasma fraction, it reflects active viral replication with virus release into plasma (37) from multiple pools, including endothelial cells and the reticuloendothelial system, in addition to circulating leukocytes.
Some authors have found plasma viral load monitoring of transplant recipients suitable (1, 14, 25, 27). Boeckh et al. (3) concluded that even though the sensitivity of plasma PCR was significantly lower than that of PBL PCR, plasma PCR could be particularly useful when leukocyte counts were inadequate for the performance of cell-based assays. For others, the higher sensitivity of WB and its higher yield of CMV DNA make it an optimal sample for monitoring the CMV DNA load during CMV disease in immunocompromised patients (7, 10, 18, 29). Moreover, the suitability of WB for bone marrow transplant recipients, who are often in aplasia or leukopenic, has been shown (18). However, if the WB assay is chosen as the only test for the monitoring of transplant recipients in routine care management, the information on early active replication provided by the plasma viral load could be missed. Moreover, up to now, no WB CMV threshold that would distinguish a latent from an active CMV infection has ever been defined. Therefore, it is relevant to look for a WB viral load threshold that would indicate active replication which can be defined by a detectable plasma viral load.
We took advantage of the fact that a cohort of patients were routinely monitored for CMV loads after renal transplantation to embark on a prospective study evaluating the capacity of the viral load in the WB compartment to predict the plasma viral load. The objective was to check if the WB CMV load might be able to predict CMV active replication as well as the plasma viral load, to define a threshold if possible, and in the end to monitor CMV infection through the WB CMV load alone.
(This work was presented in part at the Third European Congress of Virology, 2007.)
MATERIALS AND METHODS
Patients and specimen collection.
Renal transplant recipients who gave informed consent were prospectively enrolled in this study from July 2004 to June 2005. As part as the routine follow-up after transplantation, EDTA-blood samples were collected, and 1 ml of WB plus 1 ml of plasma were recovered. CMV quantitative real-time PCR was prospectively performed on WB (7). Whenever a positive result was obtained with WB, the assay was then performed with plasma. CMV DNAemia was assessed weekly for 3 months after transplantation, twice a month from month 3 (M3) to M6, and then monthly from M6 to M12. In case of CMV infection and curative anti-CMV treatment, virological follow-up was strengthened, and PCR was performed weekly until a negative result was obtained.
Treatment protocols.
All the patients received a combination immunosuppressive regimen of cyclosporine (goal, 100 to 200 ng/ml) or tacrolimus (goal, 5 to 15 ng/ml), and mycophenolate mofetil at 2 g/day. They also received intravenous (i.v.) methylprednisolone before transplantation and then high doses of prednisolone that were gradually decreased until 3 months posttransplantation. For immunosuppression induction therapy, all the patients received either anti-CD25 monoclonal antibodies (daclizumab) or anti-lymphocyte polyclonal antibodies (thymoglobulins).
CMV-seronegative patients who received an allograft from a CMV-seropositive donor (D+ R−) and CMV-seropositive recipients (R+) treated with anti-lymphocyte globulins received oral valganciclovir for prevention of CMV infection for 3 months.
Asymptomatic CMV infection was defined as at least one positive plasma PCR result without CMV-related clinical symptoms. Symptomatic CMV infections could be subdivided into CMV syndrome and end-organ disease. In CMV syndrome, positive plasma PCR results were associated with unexplained fever and leukopenia (<3.5 × 109 leukocytes/liter on two consecutive occasions) and/or thrombocytopenia (<5 × 109 platelets/liter on two consecutive occasions) and/or unexplained elevated aminotransferase levels (>2× N). CMV disease was defined as a CMV infection with organ involvement and evidence of localized CMV infection in a biopsy specimen or other appropriate specimen (15, 22).
Asymptomatic CMV infection was treated with a 3-week course of oral ganciclovir (GCV), and symptomatic patients received i.v. GCV for 3 weeks.
Validation samples.
All blood samples routinely tested by CMV PCR on WB and plasma samples between August 2004 and October 2006 were used as validation samples for statistical analysis (see below). Blood samples came from patients with mostly bone marrow or solid organ transplantation but also human immunodeficiency virus infection, pregnancy, and other clinical settings.
CMV assays. (i) DNA extraction.
DNA was extracted from 200 μl WB or 200 μl plasma by using the MagNA Pure instrument (Roche Molecular Biochemicals) with the MagNA Pure LC total nucleic acid isolation kit (Roche Diagnostics) according to the manufacturer's instructions. The purified nucleic acid was eluted in 100 μl low-salt elution buffer, and 10 μl was further used for PCR.
(ii) Quantitative CMV PCR on WB and plasma.
WB and plasma samples were assayed for CMV DNA quantification as previously described (7, 8, 18) with real-time PCR using TaqMan technology on the LightCycler instrument (version 1.0; Roche Diagnostics) with the Fast Start DNA master hybridization probes (Roche Molecular Biochemicals).
A homemade plasmid (pGEM-UL83) containing one copy of the UL83 target sequence was employed for achieving a CMV DNA external quantitative standard curve with dilutions from 5 × 102 (2.70 log10) to 5 × 106 (6.70 log10) copies/ml (7).
A positive control was included from extraction to quantification in each run as well as a distilled water sample to check the absence of contamination. CMV DNA was expressed as copy numbers per milliliter of WB or plasma and as log10 copies per milliliter as well. Accurate quantification was obtained down to 500 copies/ml (2.70 log10 copies/ml). Below 500 copies/ml, samples could have a positive result with unreliable quantification (a result of <500 copies/ml). When no signal was obtained above the noise band, the sample was considered PCR negative. Our laboratory results in 2004, 2005, and 2006 were in agreement with the expected results according to Quality Control for Molecular Diagnostics (Glasgow, Scotland).
Statistical analysis.
Quantitative variables were described by frequency, mean and standard deviation, and/or median and 25th and 75th percentiles. Qualitative variables were described by frequency and proportion. Ninety-five percent confidence intervals (95% CI) were calculated using exact binomial distribution. Viral loads were compared between groups using a nonparametric median score test.
The sensitivity of the WB threshold of 500 copies/ml for detection of a plasma viral load of >500 copies/ml was defined by the proportion of samples with a WB viral load of >500 copies/ml among all samples with a plasma viral load of >500 copies/ml.
The prediction of the plasma viral load according to the quantified WB viral load of >500 copies/ml was estimated using a linear mixed model taking the quantification limit and repeated data into account (35). The base-10 logarithm of viral load was fitted in accordance with model assumptions. The proportion of variability explained by the regression model (R2) was also estimated (39). The model predictive capacity was validated by using external data collected from August 2004 to October 2006 in routine practice in the same ward. Concordance between observed and predicted plasma viral loads was first estimated using the proportion of concordant pairs in terms of quantified or nonquantified results. Then, when both plasma viral loads were quantified as >500 copies/ml, the mean difference between these two loads was compared to null. All analyses were performed with SAS software, version 9.2 (SAS Institute, Cary, NC).
RESULTS
Patients.
Eighty-six patients (62 males and 24 females; mean age ± standard deviation, 48.7 ± 13.3 years) among whom 72 had received their first renal transplant, 13 their second graft, and 1 the third graft were consecutively enrolled from July 2004 to June 2005. Four patients had their kidney allograft rapidly removed because of thrombosis. Therefore, statistical analysis was performed on 82 patients. Two patients died during the follow-up: one with multiple infections including CMV disease and one with bacterial septicemia.
The characteristics of the 82 patients according to their donor/recipient serostatus are presented in Table 1. The median (25th to 75th percentile) duration of follow-up after renal transplantation was 12 (11.4 to 12.7) months, with 18 (16 to 20) follow-up visits. Thirty (36.6%) patients presented with CMV infections (positive qualitative plasma PCR), 20 of whom had plasma CMV loads of >500 copies/ml. Nineteen patients (all but the D− R+ patient) received the first curative treatment in a median (25th to 75th percentile) time of 3.6 (1.9 to 6.1) months after engraftment (calculated from the data obtained for 18 of the 19 patients, since the start-of-treatment dates were available for those 18 patients): 7 patients received i.v. GCV, 5 received oral GCV, and 7 received i.v. GCV followed by oral GCV.
TABLE 1.
Characteristics of the 82 patients according to their donor/recipient serostatus
| Patient characteristic | No. (%) of patients with the following CMV donor/recipient serostatus and the indicated characteristic:
|
|||
|---|---|---|---|---|
| D+ R− | D+ R+ | D− R+ | D− R− | |
| Total | 24 (29.3) | 25 (30.5) | 24 (29.3) | 9 (11) |
| 3-mo anti-CMV prophylaxy | 22 (92) | 1 (4) | 2 (8.3) | 1 (11) |
| Plasma CMV load of >500 copies/ml | ||||
| Total | 11 (45.8) | 8 (32) | 1 (4.2) | 0 (0) |
| With CMV-related symptomsa | 8 (72.7) | 1 (12.5) | 0 (0) | 0 (0) |
| Anti-CMV curative treatment | 11 (45.8) | 8 (32) | 0 (0) | 0 (0) |
Percentages for patients with CMV-related symptoms are relative to the number of all patients of the same serostatus with plasma CMV loads of >500 copies/ml.
As a whole, 21 episodes of infection requiring curative treatment occurred in 19 patients during follow-up. At the first visit under treatment, one symptomatic patient (D+ R−) and one asymptomatic patient (D− R+) tested negative for CMV in WB. The plasma viral load was missing for one asymptomatic patient (D+ R+), negative for three asymptomatic patients (one D− R+ and two D+ R+), and below the quantification limit for two symptomatic D+ R− patients.
Among 21 episodes, viral loads tended to be higher during symptomatic episodes than during asymptomatic episodes: median (25th to 75th percentile) WB viral loads were 4.45 (4.03 to 5.46) (n = 9) and 3.74 (3.36 to 4.44) (n = 12) log10 copies/ml, respectively (P = 0.14), and median (25th to 75th percentile) plasma viral loads were 4.25 (3.25 to 4.42) (n = 9) and 3.18 (<2.70 to 3.78) (n = 11) log10 copies/ml, respectively (P = 0.19). In D+ R− (11 episodes) compared to D+ R+ (9 episodes) patients, viral loads tended to be higher in WB (4.66 [3.79 to 5.46] versus 3.68 [3.46 to 4.10] log10 copies/ml, respectively [P = 0.19]) and significantly higher in plasma (4.25 [3.57 to 4.43] [n = 11] versus 3.07 [<2.70 to 3.26] [n = 8] log10 copies/ml, respectively [P = 0.01]).
Description of PCR results in WB and plasma.
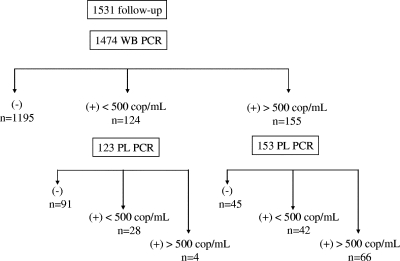
Figure 1 shows the results of PCR for WB and plasma. A total of 1,474 WB samples were assayed, of which 1,195 were negative and 279 were positive for CMV. Among patients with PCR-positive WB samples, 276 plasma samples were tested: 108/153 (70.6%) plasma samples were positive when the WB CMV load was >500 copies/ml, whereas 32/123 (26%) were positive when the WB CMV load was <500 copies/ml.
FIG. 1.
Follow-up of the 82 patients, with distribution of real-time quantitative PCR results for WB and plasma (PL). cop, copies.
As can be noticed, in four pairs of samples, the WB sample had <500 copies/ml by PCR and the plasma samples had >500 copies/ml by PCR, but all plasma loads were low (<900 copies/ml): three pairs of samples were from two patients receiving anti-CMV treatment and were preceded and/or followed by a positive result at least for the WB sample and by a negative result or a result of <500 copies/ml for the plasma sample, and the last pair of samples was from a patient receiving no anti-CMV treatment, with <500 copies/ml in WB by PCR and a negative result for the plasma sample by PCR before and after this sample. These four pairs of samples were not used for the analysis because only WB CMV quantifications of >500 copies/ml were used in the prediction analysis, as discussed below.
Prediction of the plasma CMV load from the WB CMV load.
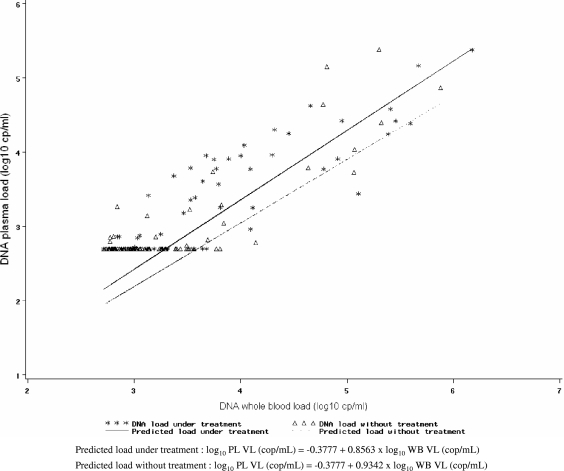
Because the CMV load in WB is expected to be higher than that in plasma, we expected the former to be positive when the plasma CMV load was >500 copies/ml. Actually, when the plasma viral load was >500 copies/ml (n = 70), more than 94% (95% CI, 86.0%, 98.4%) of WB samples had >500 copies/ml (Table 2). Discrepancies came from 4/70 samples with a WB viral load of <500 copies/ml; these 4 samples have been described above. With the idea of using the WB viral load rather than the plasma viral load because the former is more sensitive, we were interested in predicting what the level of the plasma viral load would be according to the WB viral load when the latter was >500 copies/ml. For this purpose, we used 147 samples for which WB and plasma viral loads were available. Because the prediction differed according to the presence or absence of curative treatment, we did two separate models (Fig. 2). For patients with no treatment, log10 plasma VL (in copies per milliliter) was calculated as −0.3777 + 0.8563 × log10 WB VL (in copies per milliliter). For patients receiving treatment, log10 plasma VL (in copies per milliliter) was calculated as −0.3777 + 0.9342 × log10 WB VL (in copies per milliliter). The percentage of variation of the plasma viral load explained by the regression model (R2) was rather good but not perfect (78%). In addition to their use for prediction, these equations confirmed the trend toward a lower viral load in plasma than in WB, since both slope coefficients (0.8563 and 0.9342) were below 1.
TABLE 2.
Results for WB and plasma CMV loads according to the threshold of 500 copies/ml (n = 276)
| WB CMV load | No. (%) of patients with the indicated WB CMV load and a plasma CMV load of:
|
|
|---|---|---|
| >500 copies/ml | <500 copies/ml | |
| >500 copies/ml | 66 (94.3) | 87 (42.2) |
| <500 copies/ml | 4 (5.7) | 119 (57.8) |
| Total | 70 (100) | 206 (100) |
FIG. 2.
Prediction of plasma (PL) VL from WB VL (n = 147). cp or cop, copies. Of note, some of the viral DNA loads in plasma were undetectable (i.e., left-censored) and are plotted at the detection limit (2.70 log10 copies/ml); however, they were taken into account as left-censored values in the model (see Materials and Methods).
Using the validation sample (n = 578), 77.2% of the observed and expected plasma viral loads were concordant (95% confidence intervals, 73.5 and 80.5%). When both observed and predicted plasma viral loads were quantified as >500 copies/ml, the mean difference was statistically but not clinically significant: 2.3 copies/ml (0.36 log10 copies/ml). Information about treatment was not available for the validation sample, so we used the model estimated without interaction with treatment. However, since separate models were better specified, validation of these models would have given results as good as those presented above.
Correspondences between plasma and WB CMV loads, predicted from WB CMV loads of >500 copies/ml (2.70 log10 copies/ml) and according to anti-CMV curative therapy, are presented in Table 3. During active CMV infection (with plasma viral loads of >500 copies/ml), WB viral loads were >4,000 copies/ml (>3,170 copies/ml for patients receiving anti-CMV treatment).
TABLE 3.
Mean predicted plasma viral loads according to WB viral loads from 500 copies/mla
| Patient group | WB CMV load
|
Plasma CMV load (log10 copies/ml)b (95% CI) | |
|---|---|---|---|
| Log10 copies/ml | Copies/ml | ||
| Without curative treatment | 2.70 | 500 | 1.93* (1.68-2.17) |
| 3 | 1,000 | 2.19* (1.97-2.41) | |
| 3.6 | 4,000 | 2.70 (2.49-2.91) | |
| 4 | 10,000 | 3.05 (2.82-3.27) | |
| 5 | 100,000 | 3.90 (3.59-4.22) | |
| 6 | 1,000,000 | 4.76 (4.32-5.20) | |
| During curative treatment | 2.70 | 500 | 2.00* (1.77-2.24) |
| 3 | 1,000 | 2.27* (2.06-2.48) | |
| 3.5 | 3,170 | 2.70 (2.51-2.89) | |
| 4 | 10,000 | 3.13 (2.93-3.32) | |
| 5 | 100,000 | 3.98 (3.70-4.27) | |
| 6 | 1,000,000 | 4.84 (4.43-5.25) | |
Equivalent to 2.70 log10 copies/ml, the quantification limit.
Asterisked values are given for information only, since they are below the quantification threshold.
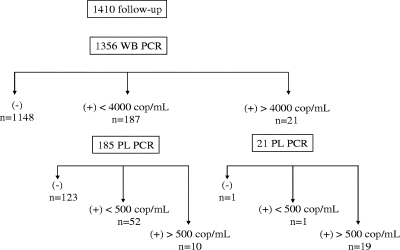
When we considered samples collected in the absence of treatment, 187 WB samples had viral loads of <4,000 copies/ml and 21 had >4,000 copies/ml (of the corresponding plasma samples, 19 had viral loads of >500 copies/ml, 1 had <500 copies/ml, and 1 tested negative) (Fig. 3).
FIG. 3.
Distribution of WB and plasma (PL) real-time quantitative PCR results for the 82 patients without treatment. cop, copies.
DISCUSSION
Many molecular techniques have been assessed for the diagnosis and follow-up of CMV infection in immunosuppressed patients: in-house or commercially available, traditional or real-time PCR, qualitative or quantitative, with detection of DNA or late mRNA, in different body or blood compartments. In previous studies, PBL or WB assays had demonstrated higher sensitivity than plasma assays (3, 4, 16, 18, 24, 26, 38). WB specimens offer the advantage of easier processing with no centrifugation or cell separation step. Our results argue for the preferential use of WB. On the other hand, the clinical impact of low-level CMV DNA in WB is unresolved, but our study was not designed for answering such a question, which requires a much larger cohort. This early detection has the advantage of warning physicians to keep track of the evolution of infection and to carefully follow up viral load kinetics (6).
As previously reported, higher viral loads were detected in WB than in plasma (1, 7, 18, 29). Since CMV replication starts in cells and is followed by the release of viral particles into plasma, CMV DNA in plasma can represent a valuable indicator for viral replication providing that specimens are prepared without excessive delay so as to avoid positive plasma results due to cell lysis (20, 32). We aimed at predicting plasma CMV loads from WB CMV loads in order to identify active CMV infections. To our knowledge, this is the first work aiming at such a prediction. In our prospective renal transplant cohort of 82 patients, prediction of the plasma CMV load from a low-level WB viral load (500 copies/ml) was possible through two equations, for patients with and without curative anti-CMV treatment, respectively. Indeed, the implementation of treatment modifies the natural kinetics of CMV; this impact of therapy on viral kinetics merits further studies.
The four discrepancies highlighted in Fig. 1 deserve a short comment: since three samples were from two patients receiving anti-CMV treatment, it may be hypothesized that due to CMV kinetics, a weak plasma viral load (<900 copies/ml) may appear undetectable in WB. On the other hand, since the main goal of this study was to monitor CMV infection through the WB CMV load alone, these results would have had no clinical impact. Moreover, these four samples did not have to be taken into account in the prediction analysis (based on the WB samples with >500 copies/ml).
Despite the heterogeneity of the validation sample, the results were satisfying and could have been even more conclusive with a homogeneous renal transplant population for validation. Nevertheless, this prediction model cannot be used in clinical settings other than renal transplantation without previous evaluation.
A recent article by Ruell et al. (30) indicated that active CMV disease does not always correlate with viral load detection: in their population of bone marrow recipients, CMV end-organ disease could occur in the absence of detectable WB CMV DNAemia throughout the course of the disease and in spite of the use of sensitive real-time PCR detection. This could underline the possible compartmentalization of viral replication occurring during CMV disease. In our group of renal transplant recipients, CMV-related symptoms were always associated with a positive WB PCR before treatment.
As in other areas (13), in the routine practice of our renal transplantation center, a threshold of 2,000 copies/ml in WB has recently been chosen for the initiation of preemptive therapy; by following the prediction equation of the present study, this WB viral load (2,000 copies/ml, or 3.3 log10 copies/ml) corresponds to 2.53 log10 copies/ml in plasma, i.e., a positive plasma load below the quantification threshold of our real-time PCR (2.70 log10 copies/ml). However, detection of a few viral copies in plasma confirms viral replication. In a previous work (7), this threshold of 2,000 copies/ml in WB has also been correlated with a pp65 Ag result of 10 positive cells/200,000 polynuclear cells, which was previously the pp65 Ag threshold used to implement anti-CMV treatment for our renal transplant recipients.
The resulting WB viral load threshold of 4,000 copies/ml (corresponding to a plasma viral load of >500 copies/ml) is not to be used as a clinical threshold and needs first to be clinically evaluated. Moreover, in our study, a WB viral load greater than 4,000 copies/ml permits one to ascertain the presence of an active infection, but an active infection can occur below this threshold, which is still in accordance with the current threshold of 2,000 copies/ml.
It is well recognized that D+ R− patients have a higher risk of developing CMV disease. In our study, 55% of patients with at least one plasma viral load of >500 copies/ml during their follow-up were D+ R− (these patients constituted 45.8% of the D+ R− group, and 72.7% of them developed symptomatic CMV infections). Viral loads showed a trend to be higher for symptomatic than for asymptomatic patients by both assays. Detection of CMV in clinical samples may represent asymptomatic viral shedding and does not necessarily indicate the presence of current or impending symptomatic disease. To identify patients at high risk of CMV disease among those who are infected, viral load thresholds and kinetics still need to be determined.
For routine performance of a single test to monitor CMV infection in transplant patients, WB real-time quantitative PCR seems to be an appropriate candidate. Besides its ease of processing and its sensitivity, we have shown here that WB CMV load results could be used to predict plasma CMV load results and thus to evidence an active infection: the plasma CMV load was found to be greater than 500 copies/ml when the WB CMV load was greater than 4,000 copies/ml (3.6 log10 copies/ml) for patients without treatment. However, the WB viral load thresholds for initiation of anti-CMV therapy should be determined in further specific studies, taking into account the baseline risk of the patients for developing symptomatic CMV infections.
Acknowledgments
This work was supported by a grant from Bordeaux University Hospital (Appel d'Offres Interne, CHU de Bordeaux, 2003).
We thank the patients who participated in this study and the following individuals for their contributions to this work: Anne Caumont, Marie-Hélène Schrive, Marie-José Defrance, Delphine Bachellerie, Carole Arnaud, and all the laboratory technicians.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Barrett-Muir, W., J. Breuer, C. Millar, J. Thomas, D. Jeffries, M. Yaqoob, and C. Aitken. 2000. CMV viral load measurements in whole blood and plasma—which is best following renal transplantation? Transplantation 70116-119. [PubMed] [Google Scholar]
- 2.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation 64108-113. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 384356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliendo, A. M., K. St George, S. Y. Kao, J. Allega, B. H. Tan, R. LaFontaine, L. Bui, and C. R. Rinaldo. 2000. Comparison of quantitative cytomegalovirus (CMV) PCR in plasma and CMV antigenemia assay: clinical utility of the prototype AMPLICOR CMV MONITOR test in transplant recipients. J. Clin. Microbiol. 382122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 3552032-2036. [DOI] [PubMed] [Google Scholar]
- 7.Garrigue, I., S. Boucher, L. Couzi, A. Caumont, C. Dromer, M. Neau-Cransac, R. Tabrizi, M. H. Schrive, H. Fleury, and M. E. Lafon. 2006. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J. Clin. Virol. 3672-75. [DOI] [PubMed] [Google Scholar]
- 8.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerna, G., M. Zavattoni, F. Baldanti, A. Sarasini, L. Chezzi, P. Grossi, and M. G. Revello. 1998. Human cytomegalovirus (HCMV) leukoDNAemia correlates more closely with clinical symptoms than antigenemia and viremia in heart and heart-lung transplant recipients with primary HCMV infection. Transplantation 651378-1385. [DOI] [PubMed] [Google Scholar]
- 10.Gouarin, S., A. Vabret, E. Gault, J. Petitjean, A. Regeasse, B. Hurault de Ligny, and F. Freymuth. 2004. Quantitative analysis of HCMV DNA load in whole blood of renal transplant patients using real-time PCR assay. J. Clin. Virol. 29194-201. [DOI] [PubMed] [Google Scholar]
- 11.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J. H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 394362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humar, A., C. Paya, M. D. Pescovitz, E. Dominguez, K. Washburn, E. Blumberg, B. Alexander, R. Freeman, N. Heaton, and B. Mueller. 2004. Clinical utility of cytomegalovirus viral load testing for predicting CMV disease in D+/R− solid organ transplant recipients. Am. J. Transplant. 4644-649. [DOI] [PubMed] [Google Scholar]
- 13.Khoury, J. A., G. A. Storch, D. L. Bohl, R. M. Schuessler, S. M. Torrence, M. Lockwood, M. Gaudreault-Keener, M. J. Koch, B. W. Miller, K. L. Hardinger, M. A. Schnitzler, and D. C. Brennan. 2006. Prophylactic versus preemptive oral valganciclovir for the management of cytomegalovirus infection in adult renal transplant recipients. Am. J. Transplant. 62134-2143. [DOI] [PubMed] [Google Scholar]
- 14.Leruez-Ville, M., M. Ouachee, R. Delarue, A. S. Sauget, S. Blanche, A. Buzyn, and C. Rouzioux. 2003. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J. Clin. Microbiol. 412040-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ljungman, P., P. Griffiths, and C. Paya. 2002. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 341094-1097. [DOI] [PubMed] [Google Scholar]
- 16.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 382536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mengelle, C., C. Pasquier, L. Rostaing, K. Sandres-Saune, J. Puel, L. Berges, L. Righi, C. Bouquies, and J. Izopet. 2003. Quantitation of human cytomegalovirus in recipients of solid organ transplants by real-time quantitative PCR and pp65 antigenemia. J. Med. Virol. 69225-231. [DOI] [PubMed] [Google Scholar]
- 18.Mengelle, C., K. Sandres-Saune, C. Pasquier, L. Rostaing, J. M. Mansuy, M. Marty, I. Da Silva, M. Attal, P. Massip, and J. Izopet. 2003. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J. Clin. Microbiol. 413840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, T., S. Okamoto, S. Matsuoka, T. Yajima, M. Wakui, R. Watanabe, A. Ishida, Y. Iwao, M. Mukai, T. Hibi, and Y. Ikeda. 2000. Risk-adapted pre-emptive therapy for cytomegalovirus disease in patients undergoing allogeneic bone marrow transplantation. Bone Marrow Transplant. 25765-769. [DOI] [PubMed] [Google Scholar]
- 20.Nesbitt, S. E., L. Cook, and K. R. Jerome. 2004. Cytomegalovirus quantitation by real-time PCR is unaffected by delayed separation of plasma from whole blood. J. Clin. Microbiol. 421296-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 382734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paya, C., A. Humar, E. Dominguez, K. Washburn, E. Blumberg, B. Alexander, R. Freeman, N. Heaton, and M. D. Pescovitz. 2004. Efficacy and safety of valganciclovir vs. oral ganciclovir for prevention of cytomegalovirus disease in solid organ transplant recipients. Am. J. Transplant. 4611-620. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrin, I., I. Garrigue, C. Binquet, G. Chene, D. Neau, P. Bonot, F. Bonnet, H. Fleury, and J. L. Pellegrin. 1999. Evaluation of new quantitative assays for diagnosis and monitoring of cytomegalovirus disease in human immunodeficiency virus-positive patients. J. Clin. Microbiol. 373124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellegrin, I., I. Garrigue, D. Ekouevi, L. Couzi, P. Merville, P. Merel, G. Chene, M. H. Schrive, P. Trimoulet, M. E. Lafon, and H. Fleury. 2000. New molecular assays to predict occurrence of cytomegalovirus disease in renal transplant recipients. J. Infect. Dis. 18236-42. [DOI] [PubMed] [Google Scholar]
- 25.Piiparinen, H., I. Helantera, M. Lappalainen, J. Suni, P. Koskinen, C. Gronhagen-Riska, and I. Lautenschlager. 2005. Quantitative PCR in the diagnosis of CMV infection and in the monitoring of viral load during the antiviral treatment in renal transplant patients. J. Med. Virol. 76367-372. [DOI] [PubMed] [Google Scholar]
- 26.Piiparinen, H., K. Hockerstedt, C. Gronhagen-Riska, M. Lappalainen, J. Suni, and I. Lautenschlager. 2001. Comparison of plasma polymerase chain reaction and pp65-antigenemia assay in the quantification of cytomegalovirus in liver and kidney transplant patients. J. Clin. Virol. 22111-116. [DOI] [PubMed] [Google Scholar]
- 27.Piiparinen, H., K. Hockerstedt, C. Gronhagen-Riska, and I. Lautenschlager. 2004. Comparison of two quantitative CMV PCR tests, Cobas Amplicor CMV Monitor and TaqMan assay, and pp65-antigenemia assay in the determination of viral loads from peripheral blood of organ transplant patients. J. Clin. Virol. 30258-266. [DOI] [PubMed] [Google Scholar]
- 28.Razonable, R. R., R. A. Brown, M. J. Espy, A. Rivero, W. Kremers, J. Wilson, C. Groettum, T. F. Smith, and C. V. Paya. 2001. Comparative quantitation of cytomegalovirus (CMV) DNA in solid organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 394472-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razonable, R. R., R. A. Brown, J. Wilson, C. Groettum, W. Kremers, M. Espy, T. F. Smith, and C. V. Paya. 2002. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation 73968-973. [DOI] [PubMed] [Google Scholar]
- 30.Ruell, J., C. Barnes, K. Mutton, B. Foulkes, J. Chang, J. Cavet, M. Guiver, L. Menasce, M. Dougal, and R. Chopra. 2007. Active CMV disease does not always correlate with viral load detection. Bone Marrow Transplant. 4055-61. [DOI] [PubMed] [Google Scholar]
- 31.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol. 384006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schäfer, P., W. Tenschert, M. Schroter, K. Gutensohn, and R. Laufs. 2000. False-positive results of plasma PCR for cytomegalovirus DNA due to delayed sample preparation. J. Clin. Microbiol. 383249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stöcher, M., G. Holzl, H. Stekel, and J. Berg. 2004. Automated detection of five human herpes virus DNAs by a set of LightCycler PCRs complemented with a single multiple internal control. J. Clin. Virol. 29171-178. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60455-462. [DOI] [PubMed] [Google Scholar]
- 35.Thiébaut, R., and H. Jacqmin-Gadda. 2004. Mixed models for longitudinal left-censored repeated measures. Comput. Methods Programs Biomed. 74255-260. [DOI] [PubMed] [Google Scholar]
- 36.von Müller, L., J. Hinz, M. Bommer, W. Hampl, S. Kluwick, M. Wiedmann, D. Bunjes, and T. Mertens. 2007. CMV monitoring using blood cells and plasma: a comparison of apples with oranges? Bone Marrow Transplant. 39353-357. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, D. G., and S. A. Spector. 1993. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation 56330-334. [DOI] [PubMed] [Google Scholar]
- 38.Woo, P. C., S. K. Lo, K. Y. Yuen, J. S. Peiris, H. Siau, E. K. Chiu, R. H. Liang, and T. K. Chan. 1997. Detection of CMV DNA in bone marrow transplant recipients: plasma versus leucocyte polymerase chain reaction. J. Clin. Pathol. 50231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, R. 2003. Measuring explained variation in linear mixed effects models. Stat. Med. 223527-3541. [DOI] [PubMed] [Google Scholar]
- 40.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, and A. Vahlne. 2000. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation 691733-1736. [DOI] [PubMed] [Google Scholar]