Abstract
The occurrence of invasive mycoses has progressively increased in recent years. Yeasts of the genus Candida remain the leading etiologic agents of those infections. Early identification of opportunistic yeasts may contribute significantly to improved disease management and the selection of appropriate antifungal therapy. We developed a rapid and reliable molecular identification system for clinically relevant yeasts that makes use of nonspecific primers to amplify a region of the 26S rRNA gene, followed by reverse hybridization of the digoxigenin-labeled products to a panel of species-specific oligonucleotide probes arranged on a nylon membrane macroarray format. DNA amplification was achieved by the recently developed loop-mediated isothermal DNA amplification technology, a promising option for the development of improved laboratory diagnostic kits. The newly developed method was successful in distinguishing among the major clinically relevant yeasts associated with bloodstream infections by using simple, rapid, and cost-effective procedures and equipment.
The occurrence of nosocomial invasive mycoses in immunocompromised patients has increased over the last decade (24). Yeasts of the genus Candida are the main etiologic agents of those infections, with a high prevalence of C. albicans. However, other species (e.g., C. krusei and C. glabrata) have emerged as opportunistic pathogens associated with systemic infections (14, 21), posing difficulties due to the different susceptibilities of these yeasts to antifungal therapy. Sensitive, reliable, and rapid identification of these pathogenic yeasts is of paramount importance to improve disease management and enable the selection of adequate treatment.
Currently, yeast identification in clinical laboratories usually involves the analysis of phenotypic properties, a time-consuming and expensive procedure that often fails to provide clear-cut results. PCR-based methods and other successful molecular diagnostic techniques, such as the peptide nucleic acid-fluorescent in situ hybridization method (1, 39, 47), evaluating the hybridization of specific fluorescent probes to RNA target sites, have been developed, but their implementation for the identification of medically important yeasts in the clinical laboratory has not yet been routinely established, possibly because they are not so easy to perform and require more or less sophisticated equipment.
In order to bypass the PCR step, which until recently was patent protected, several groups have engaged in developing alternative nucleic acid amplification technologies (see, e.g., references 6 and 25). Of particular interest in this context are isothermal amplification processes, which could facilitate their integration in bench molecular diagnostic kits. One such technology is loop-mediated isothermal DNA amplification (LAMP), first described by Notomi et al. (33) and subsequently refined (26-28, 30, 31). This elegant, robust, and very promising isothermal DNA amplification technique relies on autocycling strand displacement DNA synthesis (Fig. 1), using specially designed primer sets that recognize at least six distinct sequences on the target DNA and a DNA polymerase with strand displacement activity. The reaction runs very rapidly in the presence of template DNA and deoxynucleoside triphosphates, usually in less than 90 min at a constant temperature (e.g., 60 to 65°C). The final amplification products present stem-loop DNA structures, encompassing alternate inverted repeats of the target sequence with multiple loops, and appear with a ladder-like pattern in agarose gel electrophoresis. LAMP provides high amplification efficiency, with DNA being amplified 109- to 1010-fold, and shows a detection limit and a specificity comparable to those of standard PCR. Moreover, the sensitivity of LAMP appears not to be affected by the presence of nontarget DNA in samples (33), and the method is also more tolerant of the presence of known PCR inhibitors such as blood, serum, plasma, or heparin (8, 36).
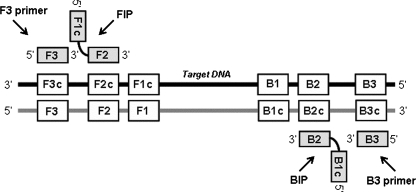
FIG. 1.
General location of the LAMP primer set in relation to previously defined regions of the target DNA. Forward (F3) and backward (B3) outer primers and forward (FIP) and backward (BIP) inner primers are indicated. The specially designed inner primers, FIP and BIP, contain two distinct sequences (F1c plus F2 and B1c plus B2, respectively) corresponding to sense and antisense segments of the target DNA, one for priming in the first stage and the other for self-priming in a subsequent amplification reaction stage (33).
The high potential of LAMP for the development of improved DNA-based diagnostic kits, its simplicity, and the fact that it does not require specific equipment fully justify the increasing number of recent reports on the utilization of this technique for the detection and identification of organisms of clinical and biotechnological importance. There has been a clear emphasis on the diagnosis of viral and bacterial infections (8, 11, 13, 16, 17, 35, 37, 42), but parasites such as Plasmodium falciparum (36) and Trypanosoma spp. (19, 44) and pathogenic fungi such as Paracoccidioides brasiliensis (7) and Ochroconis gallopava (34) have also been addressed. LAMP-based approaches have been applied to a wide range of samples, such as paraffin-embedded tissues (7), whole blood (36), nasopharyngeal swabs (17, 40), dental plaques (23), eggs (13), and potato leaf samples (32).
Previous reports on the application of “isothermal” nucleic acid amplification techniques to yeast identification (3, 4, 22, 46) are all based on nucleic acid sequence-based amplification (6), but this method is rather unspecific due to the need to use a relatively low temperature (40°C) for amplification (33). We are interested in the development of a simple and user-friendly bench DNA-based diagnostic kit for the identification of clinically relevant yeasts. To the best of our knowledge, this is the first report on the utilization of LAMP to amplify digoxigenin (DIG)-labeled yeast DNA amplicons. Our concept is different from that used in all LAMP-based methods published so far in that they involve the utilization of species-specific LAMP primer sets for the detection and identification of a single organism. In contrast, our system progresses in two steps. The first involves the utilization of a relatively conserved panfungal LAMP primer set that leads to the amplification of a common DIG-labeled DNA fragment from a broad range of yeast species. A specific species, either alone or in a mixed yeast population, can be identified subsequently by reverse hybridization to an array of membrane-bound species-specific oligonucleotide probes.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used in this study are listed in Table 1 and are maintained at the Portuguese Yeast Culture Collection (PYCC), Caparica, Portugal. Eight yeast species were selected on the basis of their clinical importance in relation to invasive mycosis: Candida albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. lusitaniae, C. krusei, Pichia anomala, and Saccharomyces cerevisiae. All the other species were used as negative controls for the reverse hybridization assays. We took the option to conserve common yeast names recognized in the clinical set, despite changes in nomenclature for some of the species (see Table 1 footnotes).
TABLE 1.
List of yeasts used in this study
| Yeast | Strain | Origin |
|---|---|---|
| Candida albicans | PYCC 3436T | Skin of man with interdigital mycosis |
| PYCC 2411 | Sputum (Portugal) | |
| PYCC 2746 | Sputum of asthma patient (Norway) | |
| PYCC 4079 | Unknown | |
| Candida glabrata | PYCC 2418T | Human feces |
| PYCC 3109 | Digestive tract of domestic animal (Germany) | |
| PYCC 2716 | Unknown | |
| Candida kruseia | PYCC 3341T | Sputum of bronchitic patient (Sri Lanka) |
| PYCC 2631 | Digestive tract of a horse (Portugal) | |
| PYCC 4740 | Seawater (Portugal) | |
| Candida lusitaniaeb | PYCC 2705T | Cecum of pig (Portugal) |
| PYCC 4093c | Citrus essence (Israel) | |
| PYCC 4175 | Unknown | |
| Candida maltosa | PYCC 3860T | Neutralizing tank for monosodium glutamate (Japan) |
| Candida oleophila | PYCC 4296 | Unknown origin |
| Candida parapsilosis | PYCC 2545T | Case of sprue (Puerto Rico) |
| PYCC 5124 | Unknown | |
| Candida tropicalis | PYCC 3097T | Bronchitic patient |
| PYCC 4672 | Soil near a polluted river (Portugal) | |
| PYCC 2508 | Liver of an elephant (Portugal) | |
| Candida viswanathii | PYCC 2811 | Penaeus braziliensis (shrimp) (Gulf of Mexico) |
| Kluyveromyces polysporusd | PYCC 3887T | Soil (South Africa) |
| Lodderomyces elongisporus | PYCC 4136T | Concentrated orange juice (United States) |
| Pichia anomala | PYCC 4121T | Unknown |
| PYCC 3294 | Pus from lung of a dead tuberculosis patient (Italy) | |
| PYCC 5618 | Unknown | |
| Saccharomyces bayanus | PYCC 4456T | Turbid beer |
| Saccharomyces cerevisiae | PYCC 4455T | Brewer's top yeast (The Netherlands) |
| Saccharomyces exiguuse | PYCC 2543T | Japan |
| Saccharomyces paradoxus | PYCC 4570T | Tree exudate |
| Stephanoascus ciferrii | PYCC 3818 | Neck of a cow (The Netherlands) |
Anamorph of Issatchenkia orientalis.
Anamorph of Clavispora lusitaniae.
Type strain of Clavispora lusitaniae.
Current synonym of Vanderwaltozyma polyspora.
Current synonym of Kazachstania exigua.
DNA extraction.
For DNA extraction, two loopfuls of cultures grown on MYP agar (0.05% [wt/vol] yeast extract, 0.7% malt extract, 0.25% Soytone, and 1.5% agar) for 2 to 5 days at 25°C were suspended in 500 μl lysing buffer (50 mM Tris, 250 mM NaCl, 50 mM EDTA, 0.3% sodium dodecyl sulfate [SDS] [pH 8]) plus the equivalent of a 200-μl volume of 425- to 600-μm-diameter glass beads (Sigma). After being vortexed for 2 min, the tubes were incubated for 1 h at 65°C and vortexed again for another 2 min. The suspensions were centrifuged for 15 min at 14,000 rpm, and the supernatant (DNA concentration, 10 to 30 ng/μl) was diluted in sterilized double-distilled water (1:750) and used directly for DNA amplification. The concentration of genomic DNA was estimated by visual comparison to serial dilutions of reference standards (GeneRuler DNA ladder mix; Fermentas) in an ethidium bromide-stained agarose gel. DNA solutions could be kept for several months at −20°C without noticeable degradation.
LAMP primers.
A set of LAMP primers targeting relatively conserved sequences within the D1/D2 domains of the fungal 26S ribosomal DNA (rDNA) was designed in order to amplify a 190-bp DNA fragment from a variety of yeast species. The primers were as follows: F3, forward outer primer (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′); B3, backward outer primer (5′-CCT TCC CTT TCA ACA ATT TCA C-3′); FIP, forward inner primer (5′-CTG CAT TCC CAA ACA ACT CGA CTC ACA GAG GGT GAG AAT CCC G-3′); BIP, backward inner primer (5′-TAT TGG CGA GAG ACC GAT AGC GTT TCA CTC TCT TTT CAA AGT TC-3′). The primer set is fully complementary to segments inside the 26S rDNAs of three of the species under study, C. albicans, C. parapsilosis, and C. tropicalis. For the other species, primer FIP had the maximum number of nucleotide substitutions: five nucleotide substitutions in comparison to the sequences of C. krusei and C. lusitaniae. Primers were designed according to the instructions of Notomi et al. (33) and those found at the LoopAmp Eiken Genome website (http://loopamp.eiken.co.jp/e/lamp/index.html). All 26S rDNA sequences used for primer design were retrieved from GenBank, with special emphasis on the sequences made available earlier by comprehensive yeast systematics studies (9, 20).
Species-specific oligonucleotide probes.
A universal probe for fungi, U210, was designed. Species-specific oligonucleotide probes were designed for selected clinically relevant yeasts based on the comparative analysis of 26S rDNA sequences retrieved from GenBank (Table 2). The targets for the specific probes are located inside the FIP/BIP LAMP-amplified fragment (Fig. 1). All probes were synthesized with an additional 3′ tail of six thymine bases to ensure efficient binding to nylon membranes and capture of the target amplicons (5). All primers and probes were synthesized by STAB Vida Lda. (Oeiras, Portugal).
TABLE 2.
DNA probes developed in this study
| Probe | Sequence (5′→3′) | % GCa | Tm (°C)a | Target species |
|---|---|---|---|---|
| U210 | TCG AGT TGT TTG GGA ATG CAG CTC | 50.0 | 65 | Panfungal |
| Ca170 | TGA GAT GAC CCG GGT CTG TGT AAA | 50.0 | 65 | Candida albicans |
| Ca176 | GAC CCG GGT CTG TGT AAA GTT CC | 56.5 | 66 | C. albicans |
| Cg175 | GTG TCA GTT CTT TGT AAA GGG TGC TCG | 48.0 | 68 | C. glabrata |
| Cd176 | GGC CCG GGT CTA TGT AAA GTT CC | 56.5 | 66 | C. dubliniensis |
| Cl180 | GAC TCT TTG CAC CGC GGC TCC | 66.7 | 67 | C. lusitaniae |
| Ct171 | GAG ATG ATC CAG GCC TAT GTA AAG T | 44.0 | 64 | C. tropicalis |
| Cp171 | GAG ATG TCC CAG ACC TAT GTA AAG TTC | 44.4 | 67 | C. parapsilosis |
| Ck175 | GGC GGA AGC AGT GAG GCC CTT CT | 65.2 | 70 | C. krusei |
| Pa176 | GCC CAT TCC TAT GTA AGG TGC TAT C | 48.0 | 66 | Pichia anomala |
| Sc176 | GTG CGG TTC TTT GTA AAG TGC CTT CG | 50.0 | 68 | Saccharomyces cerevisiae |
Data obtained with Oligo Analyzer, version 1.0.2 (Teemu Kuulasmaa, University of Kuopio, Finland).
LAMP reaction.
The LAMP reaction mixture was optimized for our identification system and contained 1.6 μM (each) FIP and BIP, 0.2 μM (each) F3 and B3, 900 μM each deoxynucleoside triphosphate, 1.4 μl of the template DNA solution, 0.8 M betaine (Sigma), 3 mM MgCl2, 3.2 U Bst polymerase, and the respective 1× buffer from New England Biolabs, for a final volume of 10 μl. When the amplicons were labeled, 1/40 of the dTTP was in the form of DIG-labeled dUTP (Roche Diagnostics). The template DNA was denatured (at 94°C for 4 min; then it was kept on ice) prior to the amplification reaction. The LAMP mixture was incubated at 64°C for 90 min in a heater block, followed by a final step of 80°C for 5 min to inactivate the enzyme. Amplicons were separated by subjecting the amplification mixture to electrophoresis in a 1.4% agarose gel and were detected with ethidium bromide. LAMP reactions were also performed by the addition of whole-yeast-cell suspensions directly to the reaction mixture (cells grown for 2 to 5 days on MYP agar at 25°C were suspended in water [at a McFarland standard of 5] and heated to 99°C for 5 min before amplification).
Reverse hybridization.
DIG-labeled LAMP amplicons were hybridized to a panel of species-specific oligonucleotide probes in a nylon membrane macroarray format. The 11 oligonucleotide probes (Table 2) were first immobilized on nylon strips (1 by 2 cm; Hybond-N; Amersham Pharmacia Biotech): 0.3 μl of each 50 pM probe aqueous solution was spotted onto a specific location on the nylon membrane, followed by irradiation with short-wave UV light for 2.5 min to cross-link the oligonucleotides to the membranes. Membranes were washed once in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 2 min at 37°C to remove any unbound probes. The strips were dried and stored at room temperature. For the hybridization, each strip was transferred to a 2-ml microcentrifuge tube containing 1 ml of prewarmed hybridization buffer (Dig Easy Hyb; Roche Diagnostics) and incubated with gentle agitation for 10 min at 55°C. After denaturation of the DIG-labeled LAMP amplicons (at 95°C for 5 min), after which they were kept on ice, 4 μl of the reaction mixture was added to the microcentrifuge tube containing the strip and hybridization buffer. Hybridization was performed for 3 h at 55°C, with inversion of the tubes. The strip was then removed from the tube and washed once in 0.25× SSC-0.1% SDS (40 ml for each batch of 30 strips, in a Falcon tube) at 55°C for 10 min. Positive hybridization was detected by using an alkaline phosphatase-labeled anti-DIG antibody and a color substrate detection system according to the manufacturer's instructions (Dig labeling and detection kit; Roche Diagnostics). Color developed 5 to 30 min after the start of the reaction. The universal probe U210 was used as a positive control on each strip, and water was used as a negative control.
RESULTS
LAMP primers.
The first and perhaps the most important step in LAMP optimization is the primer set design. We corroborate the experience of others (2, 8) in that it may be necessary to design several primer sets before finding one that works efficiently in the LAMP reaction. The LAMP primers developed in this investigation targeted conserved sequences inside the fungal D1/D2 domains of the 26S rDNA. This rDNA region was chosen not only because it is much used in yeast systematics and identification studies but also because of the availability of its sequences in public databases and its earlier utilization for designing fungal LAMP primers (34). Unfortunately, some of the available sequences were of low quality and/or were incorrectly labeled. Regions of the rDNA unit have been used before (34, 36) to develop LAMP primers, but the majority of LAMP-based identification systems targeted species- or group-specific genes, such as the gp43 gene of Paracoccidioides brasiliensis (7), which encodes the major glycoprotein antigen of this fungus, the pertussis toxin gene promoter region of Bordetella pertussis (17), or the amoA gene in ammonia-oxidizing bacteria (2).
Species-specific oligonucleotide probes.
The probes we designed are listed in Table 2. The probes designed for C. albicans, Ca170 and Ca176, have identical sequences in the closely related and recently described species Candida africana (45). The C. tropicalis probe, Ct171, has an identical sequence in C. sojae and one mismatch in C. maltosa. The C. parapsilosis probe, Cp171, has an identical sequence in C. orthopsilosis and one mismatch in C. metapsilosis, both recently described (43). Probe Pa176, for Pichia anomala, has an identical sequence in Pichia subpelliculosa, and probe Sc176, for S. cerevisiae, has one mismatch relative to Saccharomyces bayanus and Saccharomyces paradoxus.
LAMP amplification.
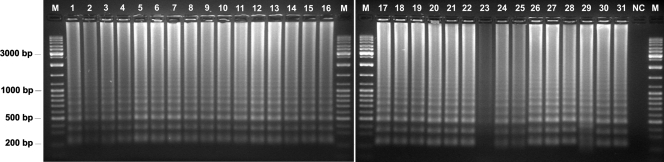
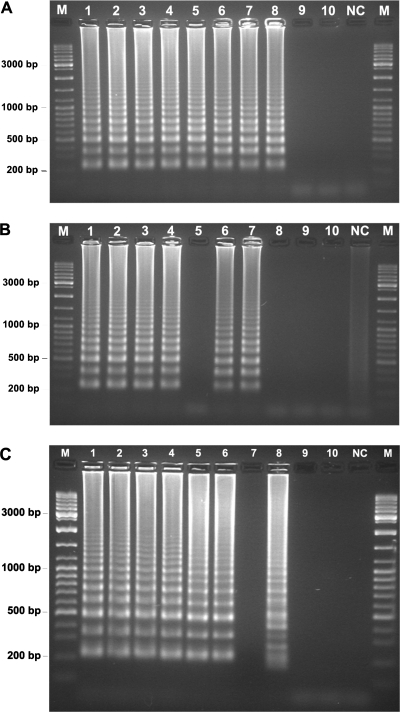
The LAMP primer set designed in this work successfully amplified genomic DNA from all yeasts tested, producing the expected ladder-like patterns on the agarose gel electrophoregram (Fig. 2). Occasionally, with a very low frequency (10−2), LAMP produced a minor amount of amplified DNA (e.g., C. krusei PYCC 3341T [Fig. 2, lane 23]). To determine the sensitivity of the LAMP reaction, the extracted genomic DNAs from C. albicans PYCC 3436T and C. krusei PYCC 3341T were quantified, serially diluted, denatured, and used as templates in LAMP reactions (Fig. 3A). The detection limit assessed with denatured genomic DNA of C. albicans PYCC 3436T, which shows a 26S rDNA segment fully complementary to the designed primer set, was around 50 fg (Fig. 3A). This sensitivity is comparable to that of standard PCR and similar to values (10 to 100 fg) mentioned by other authors for their LAMP-based systems (7, 17, 34). However, the LAMP detection limit was only 1 pg when C. krusei PYCC 3341T genomic DNA was used (Fig. 3A), which may be ascribed to the five mismatches between the FIP primer and the respective C. krusei target site. An alternative to circumvent this lower sensitivity could be to utilize degenerated primers or a mixture of primers, which has proved successful in other LAMP assays (10, 38). We confirmed that the LAMP reaction proceeds without a previous thermal denaturation of the template DNA (30), making this technique really isothermal (Fig. 3B). However, as other authors found (17), the detection limit was 5 to 10 times less sensitive when a nondenatured DNA template was used (Fig. 3B). There was no difference in band intensity over the genomic DNA concentration range tested, which has also been observed by other authors (17, 19, 41). It is possible that the incubation time used (90 min) was sufficient to complete the reaction, even when the lowest amount of template DNA was used.
FIG. 2.
Agarose gel electrophoresis of LAMP products from clinically relevant yeasts obtained by using the primer set designed in this work. Lanes: 1 to 3, Candida lusitaniae PYCC 2705T, PYCC 4093, and PYCC 4175; 4, Saccharomyces cerevisiae PYCC 4455T; 5, S. bayanus PYCC 4456T; 6, S. paradoxus PYCC 4570T; 7, Saccharomyces exiguus PYCC 2543T; 8 to 10, C. glabrata PYCC 2418T, PYCC 3109, and PYCC 2716; 11 to 14, C. albicans PYCC 3436T, PYCC 2411, PYCC 2746, and PYCC 4079; 15 to 17, C. tropicalis PYCC 3097T, PYCC 4672, and PYCC 2508; 18 and 19, C. parapsilosis PYCC 2545T and PYCC 5124; 20 to 22, Pichia anomala PYCC 4121T, PYCC 3294, and PYCC 5618; 23 to 25, C. krusei PYCC 3341T, PYCC 2631, and PYCC 4740; 26, C. viswanathii PYCC 2811; 27, C. maltosa PYCC 3860T; 28, C. oleophila PYCC 4296; 29, Lodderomyces elongisporus PYCC 4136T; 30, Kluyveromyces polysporus PYCC 3887T; 31, Stephanoascus ciferrii PYCC 3818; NC, negative control; M, molecular weight marker (GeneRuler DNA ladder mix; Fermentas).
FIG. 3.
LAMP sensitivity. (A and B) Different amounts of genomic DNA from C. albicans PYCC 3436T (lanes 1 to 5) and C. krusei PYCC 3341T (lanes 6 to 10), subjected (A) or not (B) to a previous thermal denaturation step, were used in the reaction mixture: 500 pg (lanes 1 and 6), 5 pg (lanes 2 and 7), 1 pg (lanes 3 and 8), 0.5 pg (lanes 4 and 9). and 0.05 pg (lanes 5 and 10). (C) LAMP sensitivity determined with heat-treated whole cells of C. albicans PYCC 3436T placed directly in the reaction mixture. Estimated numbers of cells in 10 μl of the reaction mixture are as follows: lane 1, 7 × 103; lane 2, 3.5 × 103; lane 3, 103; lane 4, 700; lane 5, 70; lane 6, 7; lane 7, 1; lanes 8 to 10, <1. Lane NC, negative control; lane M, molecular weight marker (GeneRuler DNA ladder mix; Fermentas).
To shorten the identification response time by avoiding the DNA extraction step, the direct utilization of heat-treated whole yeast cells in the LAMP assay was tested. Heat-treated C. albicans cell suspensions prepared directly from growth on plates showed a detection limit for the LAMP reaction consistently below 10 cells in the reaction mixture (Fig. 3C), in good agreement with the results of other authors who followed the same experimental approach using whole bacterial cells (13, 15). A different electrophoretic banding pattern was occasionally (<1%) detected in LAMP assays (Fig. 2, lane 29, and Fig. 3C, lane 8). This unusual banding pattern was not observed consistently for a given species or strain. Since it occurred with a negative control, with the Lodderomyces elongisporus DNA template (Fig. 2, lane 29), and in a sensitivity assay of the isothermal amplification step (Fig. 3C, lane 8), we were not able to determine whether the respective LAMP products can hybridize with the species-specific probes. This banding pattern was observed only once in reactions using DIG-modified nucleotides, with L. elongisporus. In this case, the LAMP product hybridized only with the panfungal U210 probe, as expected. We have no clear explanation for the formation of these odd banding patterns, but it appears to be the result of specific linear target isothermal multimerization and amplification of the template DNA (19), a property of the Bst DNA polymerase that has been demonstrated by Hafner et al. (12).
Reverse hybridization.
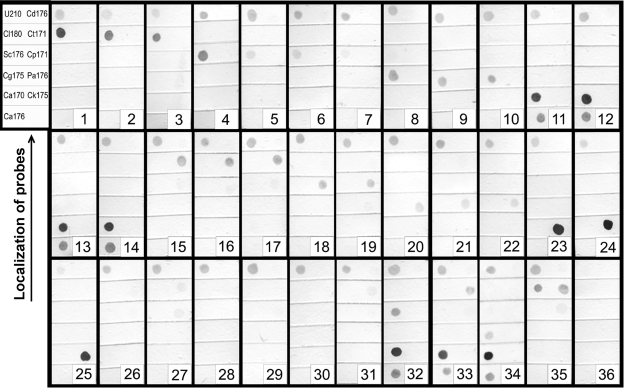
The reverse hybridization of the DIG-labeled LAMP amplicons to a panel of species-specific oligonucleotide probes (Table 2) on nylon membrane strips correctly identified the respective yeast species. Each amplicon yielded a clearly visible hybridization signal with the respective species-specific probe and the U210 universal probe for fungi, albeit with different intensities depending on the probe (Fig. 4). In a future improvement of the method, it should be possible to standardize signal intensities for all probes, e.g., by modifying their concentrations and/or the size of the 3′ thymine tail (5). Control yeast species, e.g., Candida viswanathii PYCC 2811 and Kluyveromyces polysporus PYCC 3887T, for which no specific probe was available, hybridized intensely only to the universal probe. Weak, cross-reacting hybridization signals were observed for S. bayanus PYCC 4456T and S. paradoxus PYCC 4570T DNA with the S. cerevisiae probe, Sc176 (Fig. 4, strips 5 and 6, respectively). This is most likely the result of the fact that the S. bayanus and S. paradoxus sequences have only one internal mismatch with the Sc176 probe sequence. This weak cross-reactivity contrasts with the strong, distinct signal observed for S. cerevisiae PYCC 4455T DNA with the Sc176 probe (Fig. 4, strip 4). Similar weak cross-reactivity between C. maltosa PYCC 3860T DNA and the Ct171 and Cp171 probes was observed; C. maltosa DNA has only one and two internal mismatches with the Ct171 and Cp171 probe sequences, respectively. The signal produced can be well differentiated from those obtained with the same probes and DNAs from C. tropicalis (Fig. 4, strips 15 to 17) and C. parapsilosis (Fig. 4, strips 18 and 19), respectively. When DNA mixtures from two yeast species were used for the LAMP reaction (Fig. 4, strips 32 to 35), hybridization signals were obtained only with the corresponding species-specific probes. The reverse hybridization system also allowed the detection of LAMP amplicons that could hardly be visualized after gel electrophoresis, as in the case of C. krusei PYCC 3341T (Fig. 2, lane 23, versus Fig. 4, strip 23), which demonstrates the high sensitivity of the method developed.
FIG. 4.
Hybridization of DIG-labeled LAMP amplification products to species-specific probes. (Upper left panel) Spatial distribution of the DNA probes immobilized onto each nylon membrane strip (U210, universal panfungal probe; Cl180, C. lusitaniae probe; Sc176, S. cerevisiae probe; Cg175, C. glabrata probe; Ca170 and Ca176, C. albicans probes; Cd176, C. dubliniensis probe; Ct171, C. tropicalis probe; Cp171, C. parapsilosis probe; Pa176, P. anomala probe; Ck175, C. krusei probe). The species in strips 1 to 31 correspond to those in Fig. 2, lanes 1 to 31; strips 32 to 35, DNA mixtures from C. albicans PYCC 3436T plus S. cerevisiae PYCC 4455T, C. albicans PYCC 3436T plus C. tropicalis PYCC 3097T, C. albicans PYCC 3436T plus C. glabrata PYCC 2418T, and C. lusitaniae PYCC 2705 plus C. tropicalis PYCC 3097T, respectively; strip 36, negative control (DNA replaced with water).
Our LAMP amplicons obtained with the panfungal probe produced clearly more-intense specific hybridization signals than standard PCR for 26S rDNA-based amplification products under similar experimental conditions (data not shown). The production of higher-molecular-weight DIG-labeled amplicons containing several inverted repeats of the target DNA certainly contributes to that better performance. The oligonucleotide probe panel can easily be extended to accommodate additional species and/or variants.
DISCUSSION
This report describes the development of a DNA-based identification system for clinically relevant yeasts that provides accurate identification of an isolate in less than 6 h. Our concept comprises the amplification of a 26S rDNA fragment relatively conserved in a wide range of yeast species, followed by reverse hybridization to a panel of species-specific oligonucleotide probes for identification at the species level. The present work provides the proof of a principle that ultimately may be applied to the development of an improved qualitative yeast identification molecular diagnostic kit. Validation with clinical samples will be required before application to a clinical setting. Routine methods in clinical diagnostics must be reliable, sensitive, simple to execute, and cost-effective. PCR-based diagnostics combine some of these characteristics but involve the use of expensive equipment and/or reagents. Simple diagnostic kits are in high demand, because they can be used wherever a shortage of resources exists. In order to overcome the limitations, we adapted and optimized a recently described isothermal DNA amplification technology, known as LAMP, for use with yeasts. Compared to a standard PCR protocol, LAMP requires a single bath at a constant temperature instead of a thermal cycler.
The performances of LAMP- and PCR-based diagnostic systems, including real-time technologies (23, 40), have been extensively compared. In general, LAMP was found to be either similar or superior to PCR, and more specific (e.g., 8, 13, 15), but a few studies proved otherwise, such as those reported by Kato et al. (18), who showed that although LAMP was 10-fold more sensitive than standard PCR for the detection of the Clostridium difficile toxin B gene (tcdB), an optimized nested-PCR assay performed much better than LAMP. In our experience, an optimization step can be critical for improving the sensitivity of both LAMP- and PCR-based methods. For instance, we obtained better results in the LAMP reaction with a 90-min incubation at 64°C than with incubation for a standard period of 60 min or less. Some authors would corroborate this result (7, 8, 36), while others would disagree (29). Another possibility for increasing LAMP sensitivity and accelerating the response time would be the additional utilization of loop primers in the reaction mixture (31). These hybridize to the stem-loops in amplified template DNA and initiate new strand displacement DNA synthesis.
Occasionally, we observed false-positive LAMP reactions in negative controls. Kuboki et al. (19) also mentioned the occurrence of false positives in work with Trypanosoma spp., probably due to cross-contamination. To avoid this, they recommended a few precautions and careful manipulation in preparing the samples and reaction mixtures. We stress the need to guarantee a clean environment by sterilizing all the labware utilized in the LAMP reaction and using a UV-sterilized laminar flow chamber. The utilization of a lower MgCl2 concentration in the reaction mixtures (see Materials and Methods) also helped to eliminate the occurrence of false-positive results in negative controls. A rough estimate of costs involved in a single identification by the LAMP-based system reported here provides a value of around 3.8 euros, approximately half the amount spent in clinical mycology laboratories for current identification systems (e.g., API 20 C AUX and API Candida galleries).
Overall, our results indicate that robust and simple “PCR-free” isothermal DNA amplification methodologies could greatly contribute to the development of rapid and reliable molecular diagnostic kits to be used in clinical laboratories worldwide for the identification of pathogens in general and infectious yeasts in particular.
Acknowledgments
J.I. holds a postdoctoral fellowship (SFRH/BPD/18453/2004) from the Fundação para a Ciência e a Tecnologia, Portugal.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Alexander, B. D., E. D. Ashley, L. B. Reller, and S. D. Reed. 2006. Cost savings with implementation of PNA FISH testing for identification of Candida albicans in blood cultures. Diagn. Microbiol. Infect. Dis. 54277-282. [DOI] [PubMed] [Google Scholar]
- 2.Aoi, Y., M. Hosogai, and S. Tsuneda. 2006. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J. Biotechnol. 125484-491. [DOI] [PubMed] [Google Scholar]
- 3.Borst, A., J. Verhoef, E. Boel, and A. C. Fluit. 2002. Clinical evaluation of a NASBA-based assay for detection of Candida spp. in blood and blood cultures. Clin. Lab. 48487-492. [PubMed] [Google Scholar]
- 4.Borst, A., M. A. Leverstein-Van Hall, J. Verhoef, and A. C. Fluit. 2001. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39155-160. [DOI] [PubMed] [Google Scholar]
- 5.Brown, T. J., and R. M. Anthony. 2000. The addition of low numbers of 3′ thymine bases can be used to improve the hybridization signal of oligonucleotides for use within arrays on nylon supports. J. Microbiol. Methods 42203-207. [DOI] [PubMed] [Google Scholar]
- 6.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 35091-92. [DOI] [PubMed] [Google Scholar]
- 7.Endo, S., T. Komori, G. Ricci, A. Sano, K. Yokoyama, A. Ohori, K. Kamei, M. Franco, M. Miyaji, and K. Nishimura. 2004. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol. Lett. 23493-97. [DOI] [PubMed] [Google Scholar]
- 8.Enosawa, M., S. Kageyama, K. Sawai, K. Watanabe, T. Notomi, S. Onoe, Y. Mori, and Y. Yokomizo. 2003. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 414359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 501351-1371. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, S., S. Takao, M. Kuwayama, Y. Shimazu, and K. Miyazaki. 2006. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 441376-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuhata, K., T. Annaka, M. Ikedo, M. Fukuyama, and S.-I. Yoshida. 2005. Comparison of loop-mediated isothermal amplification (LAMP) and conventional culture for the detection of Legionella species in hot spring water samples in Japan. Biocontrol Sci. 10117-120. [Google Scholar]
- 12.Hafner, G. J., I. C. Yang, L. C. Wolter, M. R. Stafford, and P. M. Giffard. 2001. Isothermal amplification and multimerization of DNA by Bst DNA polymerase. BioTechniques 30852-867. [DOI] [PubMed] [Google Scholar]
- 13.Hara-Kudo, Y., M. Yoshino, T. Kojima, and M. Ikedo. 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253155-161. [DOI] [PubMed] [Google Scholar]
- 14.Hobson, R. P. 2003. The global epidemiology of invasive Candida infections—is the tide turning? J. Hosp. Infect. 55159-168. [DOI] [PubMed] [Google Scholar]
- 15.Horisaka, T., K. Fujita, T. Iwata, A. Nakadai, A. T. Okatani, T. Horikita, T. Taniguchi, E. Honda, Y. Yokomizo, and H. Hayashidani. 2004. Sensitive and specific detection of Yersinia pseudotuberculosis by loop-mediated isothermal amplification. J. Clin. Microbiol. 425349-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, M., A. Ninomiya, H. Minekawa, T. Notomi, T. Ishizaki, M. Tashiro, and T. Odagiri. 2006. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 246679-6682. [DOI] [PubMed] [Google Scholar]
- 17.Kamachi, K., H. Toyoizumi-Ajisaka, K. Toda, S. C. Soeung, S. Sarath, Y. Nareth, Y. Horiuchi, K. Kojima, M. Takahashi, and Y. Arakawa. 2006. Development and evaluation of a loop-mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection. J. Clin. Microbiol. 441899-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, H., T. Yokoyama, H. Kato, and Y. Arakawa. 2005. Rapid and simple method for detecting the toxin B gene of Clostridium difficile in stool specimens by loop-mediated isothermal amplification. J. Clin. Microbiol. 436108-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuboki, N., N. Inoue, T. Sakurai, F. Di Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and I. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 415517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73331-371. [DOI] [PubMed] [Google Scholar]
- 21.Linton, C. L., A. M. Borman, G. Cheung, A. D. Holmes, A. Szekely, M. D. Palmer, P. D. Bridge, C. K. Campbell, and E. M. Johnson. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 451152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeffler, J., C. Dorn, H. Hebart, P. Cox, S. Magga, and H. Einsele. 2003. Development and evaluation of the Nuclisens Basic Kit NASBA for the detection of RNA from Candida species frequently resistant to antifungal drugs. Diagn. Microbiol. Infect. Dis. 45217-220. [DOI] [PubMed] [Google Scholar]
- 23.Maeda, H., S. Kokeguchi, C. Fujimoto, I. Tanimoto, W. Yoshizumi, F. Nishimura, and S. Takashiba. 2005. Detection of periodontal pathogen Porphyromonas gingivalis by loop-mediated isothermal amplification method. FEMS Immunol. Med. Microbiol. 43233-239. [DOI] [PubMed] [Google Scholar]
- 24.Martin, G. S., D. M. Mannino, S. Eaton, and M. Moss. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 3481546-1554. [DOI] [PubMed] [Google Scholar]
- 25.Monis, P. T., and S. Giglio. 2006. Nucleic acid amplification-based techniques for pathogen detection and identification. Infect. Genet. Evol. 62-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289150-154. [DOI] [PubMed] [Google Scholar]
- 27.Mori, Y., M. Kitao, N. Tomita, and T. Notomi. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59145-157. [DOI] [PubMed] [Google Scholar]
- 28.Mori, Y., T. Hirano, and T. Notomi. 2006. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukai, T., Y. Miyamoto, T. Yamazaki, and M. Makino. 2006. Identification of Mycobacterium species by comparative analysis of the dnaA gene. FEMS Microbiol. Lett. 254232-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 471742-1743. [PubMed] [Google Scholar]
- 31.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16223-229. [DOI] [PubMed] [Google Scholar]
- 32.Nie, X. 2005. Reverse transcription loop-mediated isothermal amplification of DNA for detection of Potato virus Y. Plant Dis. 89605-610. [DOI] [PubMed] [Google Scholar]
- 33.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop mediated isothermal amplification of DNA. Nucleic Acids Res. 28E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohori, A., S. Endo, A. Sano, K. Yokoyama, K. Yarita, M. Yamaguchi, K. Kamei, M. Miyaji, and K. Nishimura. 2006. Rapid identification of Ochroconis gallopava by a loop-mediated isothermal amplification (LAMP) method. Vet. Microbiol. 114359-365. [DOI] [PubMed] [Google Scholar]
- 35.Parida, M., K. Horioke, H. Ishida, P. K. Dash, P. Saxena, A. M. Jana, M. A. Islam, S. Inoue, N. Hosaka, and K. Morita. 2005. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 432895-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poon, L. L. M., B. W. Y. Wong, E. H. T. Ma, K. H. Chan, L. M. C. Chow, W. Abeyewickreme, N. Tangpukdee, K. Y. Yuen, Y. Guan, S. Looareesuwan, and J. S. M. Peiris. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 52303-306. [DOI] [PubMed] [Google Scholar]
- 37.Poon, L. L. M., B. W. Y. Wong, K. H. Chan, S. S. F. Ng, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J. Clin. Microbiol. 433457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon, L. L. M., C. S. W. Leung, K. H. Chan, J. H. C. Lee, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 43427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigby, S., G. W. Procop, G. Haase, D. Wilson, G. Hall, C. Kurtzman, K. Oliveira, S. V. Oy, J. J. Hyldig-Nielsen, J. Coull, and H. Stender. 2002. Fluorescence in situ hybridization with peptide nucleic acid probes for rapid identification of Candida albicans directly from blood culture bottles. J. Clin. Microbiol. 402182-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, R., Y. Misawa, K. Moriya, K. Koike, K. Ubukata, and N. Okamura. 2005. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Mycoplasma pneumoniae. J. Med. Microbiol. 541037-1041. [DOI] [PubMed] [Google Scholar]
- 41.Seki, M., Y. Yamashita, H. Torigoe, H. Tsuda, S. Sato, and M. Maeno. 2005. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J. Clin. Microbiol. 431581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song, T., C. Toma, N. Nakasone, and M. Iwanaga. 2005. Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243259-263. [DOI] [PubMed] [Google Scholar]
- 43.Tavanti, A., A. D. Davidson, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thekisoe, O. M. M., N. Inoue, N. Kuboki, D. Tuntasuvan, W. Bunnoy, S. Borisutsuwan, I. Igarashi, and C. Sugimoto. 2005. Evaluation of loop-mediated isothermal amplification (LAMP), PCR and parasitological tests for detection of Trypanosoma evansi in experimentally infected pigs. Vet. Parasitol. 130327-330. [DOI] [PubMed] [Google Scholar]
- 45.Tietz, H.-J., M. Hopp, A. Schmalreck, W. Sterry, and V. Czaika. 2001. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses 44437-445. [DOI] [PubMed] [Google Scholar]
- 46.Widjojoatmodjo, M. N., A. Borst, R. A. F. Schukkink, A. T. A. Box, N. M. M. Tacken, B. V. Gemen, J. Verhoef, B. Top, and A. C. Fluit. 1999. Nucleic acid sequence-based amplification (NASBA) detection of medically important Candida species. J. Microbiol. Methods 3881-90. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, D. A., M. J. Joyce, L. S. Hall, L. B. Reller, G. D. Roberts, G. S. Hall, B. D. Alexander, and G. W. Procop. 2005. Multicenter evaluation of a Candida albicans peptide nucleic acid fluorescent in situ hybridization probe for characterization of yeast isolates from blood cultures. J. Clin. Microbiol. 432909-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]