Abstract
Hepatitis B virus (HBV) surface antigen (HBsAg) was found in 9.2% of 1,186 pregnant women from Gabon, of whom 10.1% had the HBe antigen and 89.9% had anti-HBe antibodies. Antibodies to the hepatitis delta virus (HDV) were found in 15.6% of the HBsAg-positive women. The HBV strains were of the A3 and E genotypes. The HDV strains belonged to HDV clades 1 and 8. These results provide clear evidence that HDV clade 8 is indigenous to Africa.
Hepatitis B virus (HBV) and hepatitis delta virus (HDV) are highly endemic in Africa. The prevalence of serological markers indicating exposure to HBV in sub-Saharan Africa is very high, up to 90% (3), although the prevalence of HBV carriers varies substantially between regions, from less than 7% to 35% (13).
The molecular characterization of HBV has revealed eight genomic groups, designated genotypes A to H (6, 12). Two major HBV genotypes, genotypes A and E, are predominant in central, south, and west Africa (13). Genotype A has been divided into two subgenotypes, subgenotypes A1 and A2 (8, 20); recently, a new subgenotype, subgenotype A3, was described and characterized in Cameroon and Gabon (9, 11).
HDV is highly endemic in several African countries, the Amazon region, and the Middle East (4). Recent, extensive analyses of the HDV sequences of strains isolated from patients of African origin have shown wide genetic diversity, with seven major clades; their proposed labels are HDV clade 1 (HDV-1) to HDV-7 (16). Recently, a new clade, HDV-8, was described by Le Gal et al. (10).
Perinatal HBV transmission appears to be the most important factor in determining the prevalence of infection in areas where HBV is highly endemic (3). HDV is transmissible only if the recipient is a carrier of HBV.
In Gabon, a central African country, the prevalence of hepatitis B surface antigen (HBsAg) in the general population is often greater than 8 to 10% (1, 2, 17), but nothing is known about the prevalence and genetic diversity of HBV or HDV in pregnant women. There are no data on the prevalence and geographic distributions of the HDV clades in central Africa.
We evaluated the seroprevalence of HBV and HDV in a large population cohort of pregnant women in the five main cities of the country (Table 1), and we characterized the circulating genotypes.
TABLE 1.
Prevalence of HBV and HDV in pregnant women in Gabon, by geographic area and age groupa
| Variable | HBsAg positive
|
HBeAg positive
|
Antibodies to HDV
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. positive/no. tested (%) | OR | 95% CI | No. positive/no. tested (%) | OR | 95% CI | No. positive/no. tested (%) | OR | 95% CI | |
| Sentinel site | |||||||||
| Libreville | 37/394 (9.4) | 1.19 | 0.79-1.78 | 5/37 (13.5) | 2.01 | 0.57-7.06 | 8/37 (21.6) | 2.45 | 0.86-6.95 |
| Port Gentil | 10/171 (5.8) | 0.64 | 0.33-1.25 | 1/10 (10.0) | 1.11 | 0.13-9.68 | 0/10 (0) | ||
| Lambaréné | 29/330 (8.8) | 1.07 | 0.69-1.67 | 3/29 (10.3) | 1.2 | 0.3-4.86 | 0/29 (0) | ||
| Oyem | 6/65 (9.2) | 1.11 | 0.47-2.63 | 0/6 (0) | 3/6 (50.0) | 7.57 | 1.39-41.22 | ||
| Franceville | 27/227 (11.9) | 1.62 | 1.02-2.57 | 2/27 (7.4) | 0.75 | 0.15-3.7 | 6/27 (22.2) | 2.29 | 0.76-6.9 |
| Age range (yr) | |||||||||
| 14-20 | 30/372 (8.1) | 0.94 | 0.61-1.46 | 7/30 (23.3) | 6.54 | 1.76-24.28 | 2/30 (6.7) | 0.39 | 0.08-1.81 |
| 21-25 | 34/334 (10.2) | 1.34 | 0.88-2.05 | 2/34 (5.9) | 0.53 | 0.11-2.59 | 7/34 (20.6) | 2.13 | 0.74-6.14 |
| 26-30 | 23/259 (8.9) | 1.08 | 0.67-1.75 | 1/23 (4.3) | 0.4 | 0.05-3.29 | 6/23 (26.1) | 2.95 | 0.96-9.05 |
| 31-40 | 22/222 (9.9) | 1.25 | 0.76-2.04 | 1/22 (4.5) | 0.38 | 0.05-3.14 | 2/22 (9.1) | 0.59 | 0.12-2.79 |
| All | 109/1187 (9.2) | 11/109 (10.1) | 17/109 (15.6) | ||||||
OR, odds ratio; CI, confidence interval.
Between January and March 2005, a total of 1,186 samples were obtained from pregnant women. The mean age of the women was 25.0 ± 6.4 years (range, 14 to 40 years). Plasma samples were assessed for the presence of HBsAg as described previously (11). HBsAg was detected in 109 plasma samples (9.2%). The prevalence of HBsAg did not differ significantly by age. However, the prevalence of HBe antigen positivity was significantly higher (23.3%; P = 0.045) in the 14- to 20-year-old age group than in the other age groups (Table 1).
The presence of HDV antibodies was determined by the Murex anti-delta assay (Abbott, Wiesbaden, Germany). Of the 109 HBsAg-positive samples, 17 (15.6%) had antibodies to HDV. Antibodies were detected in women in all age groups. The percentage of HDV antibodies was lower in women aged 14 to 20 years (6.7%) than in the women in the other age groups (Table 1).
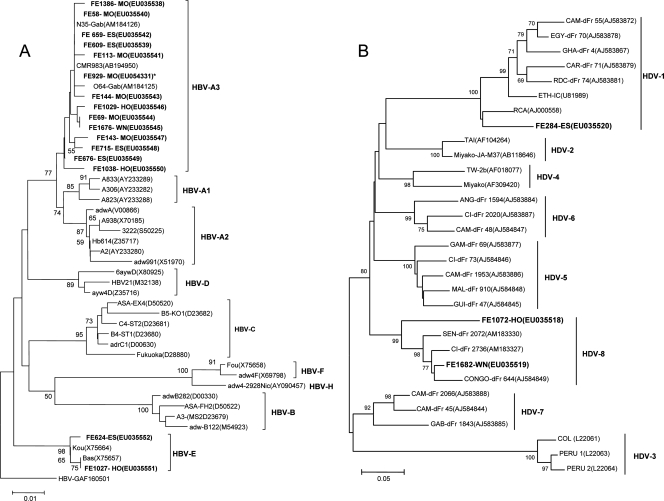
A 315-bp fragment of the HBV-S gene was amplified and sequenced for 16 HBsAg-positive isolates, as described previously (11). After alignment with reference isolates (HBV genotypes A to H), 14 of the new HBV isolates were shown to belong to subgenotype A3 and 2 were found to be closely related to HBV genotype E (99.4% and 99.6% similarities, respectively). Phylogenetic analysis showed that the strains from Gabon belonged to HBV subgenotype A3 or genotype E (Fig. 1A). In the subgenotype A3 cluster, our new strains clustered with strains from Cameroon and with other strains from Gabon described previously (11). The strains in genotype E from Gabon were closely related to other strains of African origin (bootstrap value, 98%).
FIG. 1.
Phylogenetic analysis of HBV and HDV strains obtained from pregnant women in Gabon, central Africa. (A) Phylogenetic analysis of a 315-bp fragment of the HBV-S gene from different HBV isolates by the neighbor-joining method with HBV-G (GenBank accession number AF160501) as the outgroup. Fourteen HBV-A3-FE sequences and 2 HBV-E sequences (highlighted in boldface) were compared with 31 HBV sequences from GenBank. *, a complete genome sequence was obtained for the FE-929-MO sample (GenBank accession number EU054331). This HBV subgenotype A3 clustering was confirmed by a separate phylogenetic analysis of the partially overlapping open reading frames coding for the polymerase-reverse transcriptase protein Pol (bootstrap value, 99%); envelope proteins S, M, and L (bootstrap value, 91%); the core protein (bootstrap value, 65%); and the transcriptional trans-activator protein (bootstrap value, 67%). No mutations in cis-acting elements were observed in the FE-929-MO isolate. pFDW294 (GenBank accession number M57663) was used as the reference strain (data not shown). (B) Phylogenetic analysis of a 326-bp fragment of the sHD gene from various HDV isolates by the neighbor-joining method, with HDV-3 used as the outgroup. One HDV-1 sequence (strain FE284-ES) and two HDV-8 sequences (strains FE1072-HO and FE1682-WN) (in boldface) were analyzed with 28 HDV sequences from GenBank.
A fragment of 326 bp in the sHD gene of HDV from the three regions in which HDV was detected was amplified and sequenced as described previously (16). After alignment of the sequences with those of HDV isolates representing HDV-1 to HDV-8, one strain belonged to HDV-1 with an 84.4% similarity. The other two strains showed strong similarity with the newly described HDV-8 (84.9% and 95.7%). Phylogenetic analysis showed that these two strains clustered with HDV-8 with a bootstrap value of 99% (Fig. 1B).
In our study, the prevalence of HBsAg in pregnant women was as high as that in other African countries (5, 14, 18, 19). However, the hepatitis B envelope antigen (HBeAg) level was higher in pregnant women in Gabon than in pregnant women in other African countries, suggesting the earlier exposure and transmission of HBV in pregnant women in central Africa than in other regions of Africa.
We also showed that the HBV genotypes circulating in pregnant women belong to HBV subgenotype A3 and genotype E. Subgenotype A3 predominates in five widely separated geographical regions of Gabon. HBV genotype E has previously been reported mainly in West Africa (7, 13, 15), although it was also recently described in Cameroon (9). The E genotype was first suspected of circulating in Gabon when we found an HBV genotype A-E recombinant strain in a rural population (11). In the present study, we clearly demonstrated the occurrence of HBV genotype E in pregnant women in at least two regions of the country.
Few data on the prevalence of HDV in the general population in central Africa are available, although one study (17) showed a prevalence of 8.5% in three villages in a rural area of Gabon. In the present study, the overall prevalence of HDV among pregnant women was high (15.6%).
Up to September 2006, seven major clades, HDV-1 to HDV-7 (16), had been identified, with strong phylogenetic support. Recently, Le Gal (10) described a new clade, HDV- 8, the strains of which were isolated from patients of African origin living in France. These strains clustered with another strain (strain dFr-644), previously closely related to HDV-7, isolated from a patient originating from Brazzaville, Congo, but living in France (16). Therefore, Le Gal and coauthors proposed the identification of a new clade with a probable African origin (10). To date, few sequences have been identified directly from indigenous African individuals, and no HDV-8 sequences have been identified in Africa. In this study, we showed that the new HDV strains circulating in various regions of Gabon belong to the new clade, HDV-8, and are indigenous to Africa.
This study was restricted to pregnant women, and more extensive studies of the HDV clades in central Africa are needed to better characterize the circulation of HDV in autochthonous African populations.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the new HBV subgenotype A3 and HBV genotype E strains from Gabon are EU035538 to EU035552. The GenBank accession numbers of the new HDV strains are EU035518, EU035519, and EU035520.
Acknowledgments
We thank Marie-Thérèse Bedjabaga and Patricia Keba for technical help.
The CIRMF is funded by the Gabonese Government, Total-Fina-Elf Gabon, and the French Foreign Ministry. This work was supported by funds from the Service de Coopération et d'Action Culturelle, French Embassy, Libreville, Gabon.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Bertherat, E., R. Nabias, M. C. Georges-Courbot, and A. Renaut. 1999. Seroprevalence of HIV, hepatitis B, and syphilis in an urban population and isolated villages in Gabon. Sex. Transm. Infect. 75271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupont, A., E. Delaporte, J.-M. Jego, D. Schrijvers, M. Merlin, and R. Josse. 1988. Prevalence of hepatitis B antigen among randomized representative urban and rural populations in Gabon. Ann. Soc. Belg. Med. Trop. 68157-158. [PubMed] [Google Scholar]
- 3.Hou, J., Z. Liu, and F. Gu. 2005. Epidemiology and prevention of hepatitis B virus infection. Int. J. Med. Sci. 250-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husa, P., A. Linhartova, V. Nemecek, and L. Husova. 2005. Hepatitis D. Acta Virol. 49219-225. [PubMed] [Google Scholar]
- 5.Itoua-Ngaporro, A., M. V. Sapoulou, J. R. Ibarra, L. H. Iloki, and F. Denis. 1995. Prevalence of hepatitis B viral markers in a population of pregnant women in Brazzaville (Congo). J. Gynecol. Obstet. Biol. Reprod. (Paris) 24534-536. [PubMed] [Google Scholar]
- 6.Kramvis, A., M. Kew, and F. Guido. 2005. Hepatitis B virus genotypes. Vaccine 232409-2423. [DOI] [PubMed] [Google Scholar]
- 7.Kramvis, A., K. Restorp, H. Norder, J. F. Botha, L. O. Magnius, and M. C. Kew. 2005. Full genome analysis of hepatitis B virus genotype E strains from south-western Africa and Madagascar reveals low genetic variability. J. Med. Virol. 7747-52. [DOI] [PubMed] [Google Scholar]
- 8.Kramvis, A., L. Weitzmann, W. K. Owiredu, and M. C. Kew. 2002. Analysis of the complete genome of subgroup A′ hepatitis B virus isolates from South Africa. J. Gen. Virol. 83835-839. [DOI] [PubMed] [Google Scholar]
- 9.Kurbanov, F., Y. Tanaka, K. Fujiwara, F. Sugauchi, D. Mbanya, L. Zekeng, N. Ndembi, C. Ngansop, L. Kaptue, T. Miura, E. Ido, M. Hayami, H. Ichimura, and M. Mizokami. 2005. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J. Gen. Virol. 862047-2056. [DOI] [PubMed] [Google Scholar]
- 10.Le Gal, F., E. Gault, M. P. Ripault, J. Serpaggi, J. C. Trinchet, E. Gordien, and P. Deny. 2006. Eight major clade for hepatitis Delta virus. Emerg. Infect. Dis. 91447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makuwa, M., S. Souquiere, P. Telfer, C. Apetrei, M. Vray, I. Bedjabaga, A. Mouinga-Ondeme, R. Onanga, P. A. Marx, M. Kazanji, P. Roques, and F. Simon. 2006. Identification of hepatitis B virus subgenotype A3 in rural Gabon. J. Med. Virol. 781175-1184. [DOI] [PubMed] [Google Scholar]
- 12.Miyakawa, Y., and M. Mizokami. 2003. Classifying hepatitis B virus genotypes. Intervirology 46329-338. [DOI] [PubMed] [Google Scholar]
- 13.Mulders, M. N., V. Venard, M. Njayou, A. P. Edorh, A. O. Bola Oyefolu, M. O. Kehinde, J. J. Muyembe Tamfum, Y. K. Nebie, I. Maiga, W. Ammerlaan, F. Fack, S. A. Omilabu, A. Le Faou, and C. P. Muller. 2004. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J. Infect. Dis. 190400-408. [DOI] [PubMed] [Google Scholar]
- 14.Ndumbe, P. M., J. Skalsky, and H. Joller-Jemelka. 1994. Seroprevalence of hepatitis and HIV infection among rural pregnant women in Cameroon. APMIS 102662-666. [DOI] [PubMed] [Google Scholar]
- 15.Olinger, C. M., V. Venard, M. Njayou, A. O. Bola Oyefolu, I. Maiga, A. J. Kemps, S. A. Omilabu, A. le Faou, and C. P. Muller. 2006. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J. Gen. Virol. 871163-1173. [DOI] [PubMed] [Google Scholar]
- 16.Radjef, N., E. Gordien, V. Ivaniushina, E. Gault, P. Anais, T. Drugan, J. C. Trinchet, D. Roulot, M. Tamby, M. C. Milinkovitch, and P. Deny. 2004. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J. Virol. 782537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard-Lenoble, D., O. Traore, M. Y. Kombila, P. Roingeard, F. Dubois, and A. Goudeau. 1995. Hepatitis B, C, D, and E markers in rural equatorial African villages (Gabon). Am. J. Trop. Med. Hyg. 53338-341. [DOI] [PubMed] [Google Scholar]
- 18.Roingeard, P., A. Diouf, J. L. Sankale, C. Boye, S. Mboup, F. Diadhirou, and M. Essex. 1993. Perinatal transmission of hepatitis B virus in Senegal, west Africa. Viral Immunol. 665-73. [DOI] [PubMed] [Google Scholar]
- 19.Simpore, J., A. Savadogo, D. Ilboudo, M. C. Nadambega, M. Esposito, J. Yara, S. Pignatelli, V. Pietra, and S. Musumeci. 2006. Toxoplasma gondii, HCV, and HBV seroprevalence and co-infection among HIV-positive and - negative pregnant women in Burkina Faso. J. Med. Virol. 78730-733. [DOI] [PubMed] [Google Scholar]
- 20.Sugauchi, F., E. Orito, H. Kato, S. Suzuki, S. Kawakita, Y. Sakamoto, K. Fukushima, T. Akiba, N. Yoshihara, R. Ueda, and M. Mizokami. 2003. Genotype, serotype, and phylogenetic characterization of the complete genome sequence of hepatitis B virus isolates from Malawian chronic carriers of the virus. J. Med. Virol. 6933-40. [DOI] [PubMed] [Google Scholar]