Abstract
In 2001, Corynebacterium freneyi was described as a new fermentative, α-glucosidase-positive Corynebacterium species related to C. xerosis based on data from three strains. During a review of our extensive culture collection we encountered 18 additional C. freneyi strains and further characterized them in detail. Thirteen of the 18 strains were isolated from female genital tract specimens without any obvious disease association. Phenotypically, C. freneyi can be easily differentiated from C. xerosis by its distinct wrinkled colonies whereas nearly all other routinely applied phenotypic tests do not allow a unanimous separation of C. freneyi from C. xerosis. Restriction length polymorphism analysis using CfoI of the 16S-23S rRNA gene intragenic spacer definitively allows differentiation between the two species. Surprisingly, comparative 16S rRNA gene analysis does not discriminate between C. freneyi and C. xerosis because the designated type strain of C. freneyi is not the most representative strain for this species. The present report also includes detailed data on the antimicrobial susceptibility pattern of C. freneyi presented here for the first time. Based on the large number of additional C. freneyi strains from our culture collection, we provide an extended and emended species description of C. freneyi.
During the last 15 years, clinical microbiologists have witnessed a massive increase in the number of medically relevant species of corynebacteria from 13 in 1993 to 40 in 2007 (5). One of the recently newly described species is Corynebacterium freneyi (13). This taxon was characterized by Renaud et al. in 2001 based on data from three strains (13). Up to now, only one additional C. freneyi strain has appeared in the relevant literature (2). C. freneyi is a fermentative, α-glucosidase-positive Corynebacterium species closely related to C. xerosis (13).
While screening the extensive bacterial species culture collection of one of the authors (G. Funke) we encountered 18 additional C. freneyi strains. As the result of a comprehensive study of these 18 clinical isolates by applying biochemical, chemotaxonomical, and molecular genetic methods, this paper presents an extended and emended description of C. freneyi.
MATERIALS AND METHODS
Strains.
The 18 clinical strains included in the present study (see Table 1) were isolated in our routine clinical laboratory or referred to our reference laboratory during a 3-year period. The strains were definitively not linked epidemiologically.
TABLE 1.
Strains included in the present study
| Species and strain no.a | Origin of strain | Patient age (yr), sexb | 16S rRNA gene GenBank accession no. |
|---|---|---|---|
| C. xerosis | |||
| ATCC 373T | Ear discharge | X84446 | |
| ATCC 7711 | NKc | ||
| C. amycolatum | |||
| CIP 103452T | Human skin | ||
| NCTC 7243 | Infant nose | ||
| C. freneyi | |||
| CIP 106767T | Pus from a toe | AJ292762 | |
| ISPB 16799604 | Subcutaneous abscess fistula | ||
| ISPB 20395347 | Varicose ulcer | ||
| 22 | Vaginal swab | 19, f | EF462397 |
| 2319 | Uterine cervix | 27, f | EF462398 |
| 2484 | Testicle swab | 50, m | EF462399 |
| 2586 | Outer ear canal | 38, m | EF462400 |
| 3008 | Vaginal swab | 31, f | EF462401 |
| 3017 | Vaginal swab | 18, f | EF462402 |
| 3034 | Vaginal swab | 30, f | EF462403 |
| 3077 | Leg | 77, m | EF462404 |
| 3078 | Vaginal swab | 35, f | EF462405 |
| 3171 | Vaginal swab | 70, f | EF462406 |
| 3172 | Vaginal swab | 24, f | EF462407 |
| 3228 | Cervical swab | 71, f | EF462408 |
| 3229 | Duodenal biopsy | 48, f | EF462409 |
| 3296 | Vaginal swab | 33, f | EF462410 |
| 3297 | Urine | 63, m | EF462411 |
| 3437 | Cervical swab | 18, f | EF462412 |
| 3526 | Vaginal swab | 73, f | EF462413 |
| 3527 | Vaginal swab | 24, f | EF462414 |
ATCC, American Type Culture Collection; CIP, Collection de Institut Pasteur; NCTC, National Type Culture Collection; ISPB, Institut des Sciences Pharmaceutiques et Biologiques, Lyon, France.
Data only for the clinical strains of the present study. f, female; m, male.
NK, not known.
Biochemical profiles.
The methods applied were described in detail before (9). All strains were grown on Columbia base sheep blood agar plates (BD, Heidelberg, Germany) and incubated at 35°C in ambient air. The commercial API Coryne (RAPID Coryne) and API ZYM (both from bioMérieux, Marcy l′Etoile, France) systems were used following the instructions of the manufacturers. The API 50 CH system (bioMérieux) applying CHB medium (bioMérieux) was used for incubation for up to 120 h.
Antimicrobial susceptibility testing.
The recently established CLSI standard for determination and interpretation of antimicrobial MICs for Corynebacterium spp. (3) has been applied. Briefly, by use of a broth microdilution method (prepared in our laboratory), bacterial cells with an inoculum equivalent to a 0.5 McFarland standard were grown in cation-adjusted Mueller-Hinton broth with lysed horse blood and incubated for up to 48 h. Reading of MICs was done by two independent researchers.
Cellular fatty acid analysis.
The methods for preparation of the bacterial cells and analysis of the cellular fatty acid (CFA) profiles have been described before (15).
Molecular genetic investigations.
Analysis of the complete 16S rRNA gene sequences was performed according to a published protocol (6). Full-length (>1,350 bp) 16S rRNA gene sequences were determined for each clinical strain by aligning the resulting sequences by use of a Lasergene 5 package (DNASTAR Inc., Madison, WI). Analysis of the restriction fragment length polymorphism (RFLP) of the 16S-23S rRNA gene intragenic spacer region was performed by use of restriction enzyme CfoI and following the methods outlined by Aubel et al. (1).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the complete 16S rRNA gene sequences of all 18 clinical isolates included in the present study are given in Table 1. Strains 3078, 3172, 3228, 3229, and 3527 mentioned in this report have been deposited in the Culture Collection of the University of Göteborg, Sweden, under accession numbers CCUG 54465, 54466, 54467, 54468, and 54469, respectively.
RESULTS
Table 1 lists the origins of the 18 clinical C. freneyi strains which were further characterized in the present study. The ages of the patients were in a range from 18 to 77 years, and the number of female patients was much higher than the number of the male patients (14 versus 4). Interestingly, 13 of 14 specimens from females came from the genital tract.
The extensive biochemical profiling data obtained from the 18 clinical and 3 reference C. freneyi strains are given in Table 2. C. freneyi strains are consistently positive for α-glucosidase and alkaline phosphatase. Activity of phosphoamidase was stronger in C. freneyi than in C. xerosis strains, whereas weak cystine arylamidase activity was detected in C. xerosis but not in C. freneyi. Only one C. freneyi strain (strain 3171) was negative for leucine arylamidase activity. Nineteen of 21 C. freneyi strains fermented glucose at 42°C, and all strains grew at 20°C.
TABLE 2.
Biochemical properties of C. freneyi and C. xerosis
| Category | % Positive reactionsa
|
|
|---|---|---|
| C. freneyi (n = 21) | C. xerosis (n = 2) | |
| Nitrate reduction | 67 | 50 |
| Urea hydrolysis | 0 | 0 |
| Esculin hydrolysis | 0 | 0 |
| Pyrazinamidase | 100 | 100 |
| Pyrrolidonyl arylamidase | 0 | 0 |
| Gelatinase | 0 | 0 |
| CAMP | 0 | 0 |
| Lipophilia | 0 | 0 |
| Fermentation | ||
| Glucose | 100 | 100 |
| Maltose | 100 | 100 |
| Sucrose | 100 | 100 |
| Mannitol | 10 | 0 |
| Xylose | 0 | 0 |
| Ribose | (100) | 100 |
| Lactose | 48 | 0 |
| Glycogen | (48) | 0 |
| Glycerol | 0 | 50 |
| Erythritol | 0 | 0 |
| d-Arabinose | 0 | 0 |
| l-Arabinose | (5) | 0 |
| Adonitol | (10) | (50) |
| β-Methyl-xyloside | (5) | 0 |
| Galactose | 95 | 50 |
| d-Fructose | 100 | 100 |
| d-Mannose | 100 | 100 |
| l-Sorbose | 10 | 0 |
| Rhamnose | 10 | 0 |
| Dulcitol | (10) | 50 |
| Inositol | (19) | 50 |
| Sorbitol | (14) | 50 |
| α-Methyl-d-mannoside | (10) | 0 |
| α-Methyl-d-glucoside | (10) | 0 |
| N-Acetyl-glucosamine | (10) | 0 |
| Amygdaline | (10) | 0 |
| Arbutine | (24) | 0 |
| Salicine | (10) | 50 |
| Cellobiose | (19) | 0 |
| Melibiose | (10) | 0 |
| Trehalose | 100 | 100 |
| Inuline | (14) | 0 |
| Melezitose | (19) | 0 |
| d-Raffinose | (10) | 0 |
| Amidon | (5) | 0 |
| Xylitol | (29) | 0 |
| β-Gentibiose | (33) | 50 |
| d-Turanose | 100 | 100 |
| d-Lyxose | (14) | 50 |
| d-Tagatose | (100) | (50) |
| d-Fucose | (29) | 50 |
| l-Fucose | (19) | 0 |
| d-Arabitol | (24) | 0 |
| l-Arabitol | (19) | 0 |
| Gluconate | (43) | 0 |
| 2-Keto-Gluconate | 0 | 0 |
| 5-Keto-Gluconate | 100 | 100 |
| Further enzymatic activitiesb | ||
| Alkaline phosphatase | 100 (m) | 100 (w) |
| Esterase (C4) | 100 (m) | 100 (m) |
| Esterase lipase (C8) | 100 (s) | 100 (s) |
| Lipase (C14) | 14 (w) | 50 (w) |
| Leucine arylamidase | 95 (s) | 100 (s) |
| Valine arylamidase | 0 | 0 |
| Cystine arylamidase | 0 | 100 (w) |
| Trypsin | 0 | 0 |
| Chymotrypsin | 0 | 0 |
| Acid phosphatase | 19 (w) | 100 (w) |
| Phosphoamidase | 100 (m) | 100 (w) |
| α-Galactosidase | 0 | 0 |
| β-Galactosidase | ||
| pH 5.4 | 0 | 0 |
| pH 7.4c | 0 | 0 |
| β-Glucuronidase | 0 | 0 |
| α-Glucosidase | ||
| pH 5.4 | 100 (s) | 100 (s) |
| pH 7.4c | 100 (s) | 100 (s) |
| β-Glucosidase | 0 | 0 |
| N-Acetyl-β-glucosaminidase | 0 | 0 |
| α-Mannosidase | 0 | 0 |
| α-Fucosidase | 0 | 0 |
Values in parentheses represent weak acid production after 120 h incubation. (w), hydrolyzed substrate (approximately 5 nmol); (m), hydrolyzed substrate (approximately 20 nmol); (s), hydrolyzed substrate (>40 nmol).
As determined by use of the API ZYM system.
As determined using the API Coryne system.
The following cellular fatty acids were detected at more than 1% of total CFAs: C15:0, 4% (mean); C16:0, 26%; C17:1ω8c, 4%; C17:0, 17%; C18:1ω9c, 21%; and C18:0, 23%. Tuberculostearic acid was not detected.
Table 3 outlines the antimicrobial susceptibility pattern of C. freneyi. All 21 strains tested were susceptible to doxycycline, gentamicin, linezolid, meropenem, rifampin, and vancomycin, with gentamicin and rifampin exhibiting extremely low MICs for all C. freneyi strains.
TABLE 3.
Antimicrobial susceptibility pattern of C. freneyi strainsa
| Antimicrobial agent | MIC (μg/ml)
|
No. (%) of isolates in indicated category
|
||||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | Susceptible | Intermediate | Resistant | |
| Cefotaxime | 0.12-8 | 0.5 | 1 | 20 (95) | 0 (0) | 1 (5) |
| Ciprofloxacin | 0.06-2 | 0.12 | 0.25 | 20 (95) | 1 (5) | 0 (0) |
| Clindamycin | 0.015->32 | 0.06 | >32 | 18 (86) | 0 (0) | 3 (14) |
| Doxycycline | 0.12-4 | 0.25 | 1 | 21 (100) | 0 (0) | 0 (0) |
| Erythromycin | ≤0.015->32 | 0.12 | >32 | 13 (62) | 1 (5) | 7 (33) |
| Gentamicin | ≤0.06 | ≤0.06 | ≤0.06 | 21 (100) | 0 (0) | 0 (0) |
| Linezolid | 0.5-2 | 2 | 2 | 21 (100) | 0 (0) | 0 (0) |
| Meropenem | ≤0.03-1 | 0.06 | 1 | 21 (100) | 0 (0) | 0 (0) |
| Penicillin | 0.06-32 | 0.25 | 8 | 18 (86) | 0 (0) | 3 (14) |
| Rifampin | ≤0.015-0.03 | ≤0.015 | 0.03 | 21 (100) | 0 (0) | 0 (0) |
| Vancomycin | 0.5-1 | 1 | 1 | 21 (100) | 0 (0) | 0 (0) |
The 21 strains listed in Table 1 were tested.
Table 4 and Table 5 show the overall numbers and exact details of 16S rRNA gene base pair mismatches between all 18 clinical C. freneyi strains and the comparative sequence AJ 292762 of the C. freneyi CIP 106767T type strain and sequence X84446 of the of C. xerosis ATCC 373T type strain. In general, the 18 clinical strains had similar numbers of mismatches with either C. freneyi or C. xerosis. The 16S rRNA gene homology of the 18 clinical strains was in a range from 98.9 to 99.3% (mean, 99.0%) for C. freneyi and in a range from 98.7 to 99.1% (mean, 98.8%) for C. xerosis.
TABLE 4.
16S rRNA gene sequence comparison between the C. freneyi ISPB 6695110 type strain and the 18 clinical strains
| Alignment position | Position in C. freneyi | Base in C. freneyi | Event in comparison to C. freneyi result | No. of bases in the majority of the 18 clinical strains | No. of bases in the minority of the 18 clinical strainsa |
|---|---|---|---|---|---|
| 19 | 19 | T | Point mutation | 16 C | 2 T (2586, 3526) |
| 72 | Between 71 and 72 | Insertion | 18 C | ||
| 94 | Between 92 and 93 | Insertion | 18 C | ||
| 138 | 141 | G | Point mutation | 15 G | 3 A (3077, 3297, 3437) |
| 180 | 178 | T | Point mutation | 17 T | 1 A (3296) |
| 304 | Between 301 and 302 | Insertion | 18 A | ||
| 926 | 923 | C | Point mutation | 17 T | 1 Y (22) |
| 929 | 926 | G | Point mutation | 14 C | 3 S (22, 3008, 3034), 1 G (3297) |
| 930 | 927 | G | Point mutation | 14 C | 2 G (22, 3297), 2 S (3008, 3034) |
| 931 | 928 | C | Point mutation | 14 G | 3 C (3008, 3034, 3297), 1 S (22) |
| 940 | 937 | K | Point mutation | 14 T | 3 G (3008, 3034, 3297), 1 K (22) |
| 941 | 938 | T | Point mutation | 14 G | 3 K (22, 3008, 3034), 1 T (3297) |
| 942 | 939 | C | Point mutation | 14 G | 2 C (3034, 3297), 2 S (22, 3008) |
| 943 | 940 | C | Point mutation | 18 T | |
| 1055 | 1052 | G | Point mutation | 13 G | 3 T (3077, 3297, 3437), 2 K (22, 3296) |
| 1341 | 1338 | T | Deletion |
Numbers in parentheses represent strain designations.
TABLE 5.
16S rRNA gene sequence comparison between the C. xerosis ATCC 373 type strain and the 18 clinical strains
| Alignment position | Position in C. xerosis | Base in C. xerosis | Event in comparison to C. xerosis result | No. of bases in the majority of the 18 clinical strains | No. of bases in the minority of the 18 clinical strainsa |
|---|---|---|---|---|---|
| 4 | 4 | N | Point mutation | 15 G | |
| 17 | 17 | N | Point mutation | 17 G | |
| 19 | 19 | C | Point mutation | 15 C | 2 T (2586, 3526) |
| 27 | 27 | C | Point mutation | 17 G | |
| 130 | 130 | C | Point mutation | 18 T | |
| 138 | 138 | G | Point mutation | 16 G | 2 A (3077, 3437) |
| 155 | Between 154 and 155 | Insertion | 18 T | ||
| 180 | 179 | T | Point mutation | 17 T | 1 A (3296) |
| 217 | 216 | N | Point mutation | 18 G | |
| 926 | 925 | C | Point mutation | 17 T | 1 Y (22) |
| 929 | 928 | G | Point mutation | 14 C | 3 S (22, 3008, 3034) |
| 930 | 929 | G | Point mutation | 14 C | 2 S (3008, 3034) |
| 931 | 930 | C | Point mutation | 14 G | 1 S (22) |
| 940 | 939 | G | Point mutation | 14 T | 1 K (22) |
| 941 | 940 | T | Point mutation | 14 G | 3 K (22, 3008, 3034) |
| 942 | 941 | C | Point mutation | 14 G | 2 S (22, 3008) |
| 943 | 942 | C | Point mutation | 18 T | |
| 1055 | 1054 | A | Point mutation | 15 G | 2 T (3007, 3437), 2 K (22, 3296) |
| 1182 | 1181 | G | Point mutation | 18 A | |
| 1205 | 1204 | G | Point mutation | 18 T |
Numbers in parentheses represent strain designations.
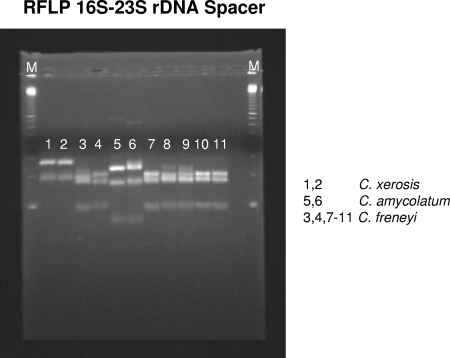
In order to clarify the exact genetic status (i.e., species identity) of the 18 clinical strains, RFLP analysis of the amplified 16S-23S rRNA gene intragenic spacer region was performed. Figure 1 shows the RFLP patterns for five clinical strains which undoubtedly represented C. freneyi strains. The RFLP patterns for the remaining 13 clinical strains were identical to those of the other five clinical strains (data not shown).
FIG. 1.
Lane 1, C. xerosis ATCC 373T; lane 2, C. xerosis ATCC 7711; lane 3, C. freneyi CIP 106767T; lane 4, C. freneyi ISPB 16799604; lane 5, C. amycolatum CIP 103452T; lane 6, C. amycolatum NCTC 7243; lanes 7 to 11, clinical strains 3078, 3172, 3228, 3229, and 3527, respectively; lanes M, 100-bp ladder (Amersham/GE Healthcare, Buckinghamshire, United Kingdom).
DISCUSSION
None of the 18 clinical Corynebacterium strains included in the present study that exhibited dry colonies, fermentative metabolism, and α-glucosidase positivity turned out to be C. xerosis, indicating that this species is extremely rarely encountered in clinical specimens, as reported previously (4, 7).
Of the 4 C. freneyi strains for which studies have appeared in the literature (2, 13), none came from urogenital specimens, whereas 13 of the 18 strains described in the present report came from the female genital tract. This finding certainly cannot be explained by increased awareness on the part of the plate-reading personnel, since 12 of the 18 strains were referred to our laboratory from other independent laboratories. The limited patient clinical data available did not indicate a clear disease association with C. freneyi or any pathogenic potential. The prevalence of C. freneyi in the female genital tract is presently not known.
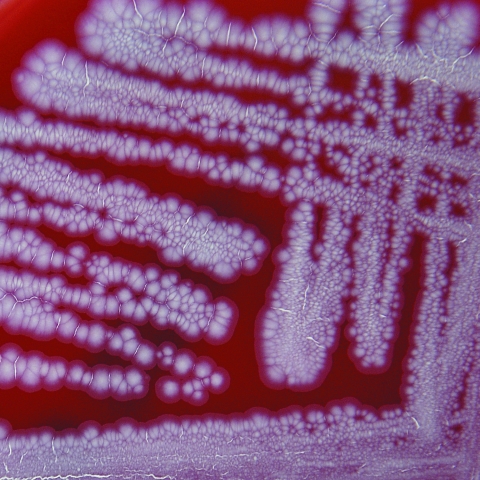
The most striking feature of C. freneyi strains is their distinctive macroscopic morphology (Fig. 2), showing dry, buff, and rough colonies with a wrinkled surface which strongly reminds the clinical microbiologist of a Mycobacterium tuberculosis culture on Middlebrook 7H10 agar or of certain Tsukamurella species. Some of the plate-reading personnel in our laboratory often described C. freneyi colonies as “folded-up colonies.” The majority of C. freneyi strains had a whitish-grayish color, but two strains (strains 3077 and 3437) showed a yellowish pigment (which had not been reported previously), as constantly reported from examinations of the few true published C. xerosis strains (7), making the distinction between these two species even more complicated. However, strongly wrinkled or folded colonies are seen neither in C. xerosis nor in the morphologically related dryish C. amycolatum strains (when applying the above-mentioned incubation conditions), making C. freneyi easily recognizable in the routine clinical laboratory.
FIG. 2.
Colonies of C. freneyi on sheep blood agar plates after 48 h incubation at 35°C. Note the wrinkled colonies.
Because of the larger number of C. freneyi strains in the present study (n = 18) compared to the initial C. freneyi study (n = 3), we were able to detect the following biochemical features not described previously. C. freneyi strains are variable for fermentation of lactose and glycogen. A minority of C. freneyi strains (2 of 21) are able to produce acid from mannitol. In addition, the present report outlines API ZYM data on C. freneyi for the first time.
C. freneyi strains can be differentiated from the recently described, phylogenetically related C. hansenii species (14) by the fact that it gives positive results for α-glucosidase and alkaline phosphatase activity whereas C. hansenii is negative for these reactions. Furthermore, the majority of C. freneyi strains ferment glucose at 42°C whereas C. hansenii does not.
For CFA analysis, the MIDI system (MIDI Inc., Newark, DE) tentatively identified some fatty acids as pentadecanoic acid (C15:0), margaric acid (C17:0), and cis-heptadec-8-enoic acid (C17:1ω8c) but these components may have been degradation products of mycolic acids cleaved at the temperature produced in the injection port of the system (300°C). Normally, cleaved mycolic acids account for only 2% to 10% of all CFAs in Corynebacterium spp. A similar observation of an even higher percentage of cleaved mycolic acids has been reported for another Corynebacterium species, C. auris (8).
Since 2006, a CLSI standard has been available for performance and interpretation of antimicrobial susceptibility testing of Corynebacterium species by use of a microdilution method. Therefore, the antimicrobial susceptibility data of the present study are more significant than the data of Renaud et al. obtained using an unvalidated commercial system (API Staph gallery) or a disk diffusion method (13). In addition, in the publication by Renaud et al., the interpretation standard used remained unclear. The present study outlines MIC data for C. freneyi for the first time. As observed for many other Corynebacterium spp. (5, 10, 11), β-lactams show good activity against C. freneyi strains. In contrast, in the related C. amycolatum strains the activity of β-lactams is often limited (11). As with many other Corynebacterium spp. (5, 10, 11), resistance to erythromycin is the most frequently encountered form of single-substance resistance. Both linezolid and vancomycin are active against C. freneyi strains, as seen with all other true corynebacteria (10). A peculiar feature of C. freneyi is the consistently very low MICs for gentamicin, which have not been detected in studies of many other true corynebacteria.
Unfortunately, Renaud et al. (13) did not give any precise 16S rRNA data with respect to gene homology between C. freneyi and C. xerosis strains. Our 16S rRNA gene data obtained using a very reliable double-stranded DNA reading approach (with multiple overlapping partial sequences) indicate that our 18 C. freneyi strains share on average only 99.0% 16S rRNA gene homology with the C. freneyi type strain. However, the 16S rRNA gene homology within our 18 C. freneyi strains often reached 100% (i.e., within 8 of 18 strains, not a single 16S rRNA gene base pair mismatch was seen), indicating that the C. freneyi type strain might not be the most representative C. freneyi strain. Our sequencing data confirm that C. freneyi and C. xerosis are the most closest related phylogenetic neighbors presently known, although they are distinct species with a relatively high level (13% to 23%) of DNA-DNA homology (13).
Another molecular genetic method for discerning closely related Corynebacterium species is sequencing of the rpoB gene (12). Khamis et al. (12) demonstrated that for either the complete or a partial rpoB gene sequence the similarity of C. xerosis and C. freneyi was greatest (>95.0%) among all closely related Corynebacterium species, indicating again that sequencing approaches directed at identification of C. freneyi may not lead to unanimous identification.
Using another molecular genetic technique, RFLP analysis of the 16S-23S rRNA gene intragenic spacer, we definitively assigned all 18 of our clinical isolates to the species C. freneyi. However, for the routine clinical laboratory the simple morphological and biochemical features outlined above are certainly sufficient to conclusively identify C. freneyi strains.
Based on the large number of strains characterized in the present study, we provide an extended and emended description of C. freneyi.
Extended and emended description of Corynebacterium freneyi Renaud et al. 2001, corr. Funke & Frodl 2008.
Corynebacterium freneyi (fre'ney.i. N.L. gen. n. freneyi of Freney, to honor Jean Freney, a contemporary French microbiologist).
The description given below is based on the results of studying 21 strains. Cells are gram positive, non-spore-forming, and nonmotile. They are typical club-shaped rods. Colonies are whitish-grayish (19 of 21 strains) or yellowish (2 of 21 strains), dry, and rough, with a distinct wrinkled morphology (see Fig. 2). Colonies are 1 to 2 mm in diameter after 48 h of incubation on a blood-enriched medium. The edges are irregular. Growth is not enhanced in a medium containing lipids. The organism is catalase positive. Reduction of nitrates is variable. The strains express α-glucosidase, pyrazinamidase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase (20 of 21 strains), and phosphoamidase. They do not produce pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, N-acetyl-β-glucosaminidase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, α-galactosidase, α-mannosidase, or α-fucosidase. They do not hydrolyze esculin, gelatin, or urea. The CAMP reaction is negative. They produce acid from glucose, maltose, sucrose, galactose (20 of 21 strains), d-fructose, d-mannose, trehalose, d-turanose, and 5-keto-gluconate. Ribose and d-tagatose acidifications are slow and weak. The results of fermentation of lactose and glycogen are variable, and very few strains (2 of 21) ferment mannitol. Xylose, glycerol, erythritol, d-arabinose, and 2-keto-gluconate are not fermented. The cell wall contains meso-diaminopimelic acid, arabinose, galactose, and mycolic acids.
The type strain, ISPB 6695110T (= CIP 106767T = DSM 44506T), was isolated from pus from a toe.
Acknowledgments
We thank F. N. R. Renaud for kindly providing strains CIP 106767T, ISPB 16799604, and ISPB 20395347.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Aubel, D., F. N. R. Renaud, and J. Freney. 1997. Genomic diversity of several Corynebacterium species identified by amplification of the 16S-23S rRNA gene spacer regions. Int. J. Syst. Bacteriol. 47767-772. [Google Scholar]
- 2.Auzias, A., C. Bollet, R. Ayari, M. Drancourt, and D. Raoult. 2003. Corynebacterium freneyi bacteremia. J. Clin. Microbiol. 412777-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Document M45-A. CLSI, Wayne, PA. [DOI] [PubMed]
- 4.Coyle, M. B., R. B. Leonard, D. J. Nowowiejski, A. Malekniazi, and D. J. Finn. 1993. Evidence of multiple taxa within commercially available reference strains of Corynebacterium xerosis. J. Clin. Microbiol. 311788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funke, G., and K. A. Bernard. 2007. Coryneform gram-positive rods, p. 485-514. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 6.Funke, G., R. Frodl, and H. Sommer. 2004. First comprehensively documented case of Paracoccus yeei infection in a human. J. Clin. Microbiol. 423366-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke, G., P. A. Lawson, K.A. Bernard, and M. D. Collins. 1996. Most Corynebacterium xerosis strains identified in the routine clinical laboratory correspond to Corynebacterium amycolatum. J. Clin. Microbiol. 341124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funke, G., P. A. Lawson, and M. D. Collins. 1995. Heterogeneity within human-derived Centers for Disease Control and Prevention (CDC) coryneform group ANF-1-like bacteria and description of Corynebacterium auris sp. nov. Int. J. Syst. Bacteriol. 45735-739. [DOI] [PubMed] [Google Scholar]
- 9.Funke, G., G. Martinetti Lucchini, G. E. Pfyffer, M. Marchiani, and A. von Graevenitz. 1993. Characteristics of CDC group 1 and group 1-like coryneform bacteria isolated from clinical specimens. J. Clin. Microbiol. 312907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funke, G., and C. Nietznik. 2005. Minimal inhibitory concentrations of linezolid against clinical isolates of coryneform bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 24612-614. [DOI] [PubMed] [Google Scholar]
- 11.Funke, G., V. Pünter, and A. von Graevenitz. 1996. Antimicrobial susceptibility patterns of some recently defined coryneform bacteria. Antimicrob. Agents Chemother. 402874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khamis, A., D. Raoult, and B. La Scola. 2004. rpoB gene sequencing for identification of Corynebacterium species. J. Clin. Microbiol. 423925-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renaud, F. N. R., D. Aubel, P. Riegel, H. Meugnier, and C. Bollet. 2001. Corynebacterium freneyi sp. nov., α-glucosidase-positive strains related to Corynebacterium xerosis. Int. J. Syst. Evol. Microbiol. 511723-1728. [DOI] [PubMed] [Google Scholar]
- 14.Renaud, F. N. R., A. Le Coustumier, N. Wilhem, D. Aubel, P. Riegel, C. Bollet, and J. Freney. 2007. Corynebacterium hansenii sp. nov., an α-glucosidase-negative bacterium related to Corynebacterium xerosis. Int. J. Syst. Evol. Microbiol. 571113-1116. [DOI] [PubMed] [Google Scholar]
- 15.von Graevenitz, A., G. Osterhout, and J. Dick. 1991. Grouping of some clinically relevant gram-positive rods by automated fatty acid analysis. APMIS 99147-154. [DOI] [PubMed] [Google Scholar]