Abstract
We recently identified a novel human virus classifiable into a third group in the genus Anellovirus, tentatively designated torque teno midi virus (TTMDV), with a circular DNA genome of 3.2 kb and genomic organization resembling those of torque teno virus (TTV) (3.8 to 3.9 kb) and torque teno mini virus (TTMV) (2.8 to 2.9 kb). TTMDV was characterized by extreme genetic diversity similar to the TTV and TTMV genomes. Taking advantage of universal and virus species-specific primers derived from a highly conserved area located just downstream of the TATA box of the TTV, TTMDV, and TTMV genomes, a PCR method with simultaneous amplification of the genomic DNAs of these three anelloviruses in the first round and subsequent differential amplifications of these viruses in the second round was developed. High prevalence of TTMDV viremia was seen in adults (75/100 [75%]), comparable with the prevalences of TTV viremia (100%) and TTMV viremia (82%). Although none of 10 cord blood samples had detectable TTV, TTMDV, and TTMV DNAs, the prevalences of these three anelloviruses increased with the number of months after birth of the individual and reached 100% for individuals at one year of age. Dual or triple infection of TTV, TTMDV, and/or TTMV was seen in 10 (47.6%) of 21 infants 9 to 180 days of age and more frequently among infants 181 to 364 days of age (20/23 [86.9%]), comparable with the 93.1% (243/261) prevalence among subjects 1 to 81 years of age, indicating early acquisition of dual or triple anellovirus infection during infancy.
Torque teno virus (TTV) was first identified in the serum of a patient with posttransfusion hepatitis of unknown etiology in 1997 (29) and was characterized as a small nonenveloped virus with a circular, single-stranded DNA of 3.8 to 3.9 kb, the first virus of this type known to infect humans (22, 24, 33, 38). In 2000, torque teno mini virus (TTMV) was accidentally discovered in the serum of a blood donor by PCR using TTV-specific primers that partially matched homologous sequences in TTMV but generated a noticeably shorter amplicon than that expected for TTV (48). Although TTV and TTMV have a common presumed genomic organization with four open reading frames (ORF1 to ORF4) and harbor a short region of 80 to 160 nucleotides (nt) with high GC content (approximately 90%) in the noncoding region, they exhibit high dissimilarity in terms of genomic length (3.8 to 3.9 kb and 2.8 to 2.9 kb, respectively) and genetic identity (4, 8, 13, 38, 45, 48, 49). Recently, TTV and TTMV have been classified in a newly designated floating genus, Anellovirus (8).
Based on the wide range of sequence divergence noted among various TTV isolates, TTV has been classified as having at least 39 genotypes, exhibiting >30% nucleotide differences from one another, and five major phylogenetic groups (groups 1 to 5) showing >50% divergence (6, 8, 37, 41). Although the entire nucleotide sequence has been determined for only 13 TTMV isolates, TTMV is also characterized by extreme genetic diversity (4, 8, 48). This situation has led to considerable problems in classification and the development of methods for screening, genetic characterization, and genotype identification. Therefore, the PCR method for sensitive and specific detection of TTV or TTMV DNA has been repeatedly modified and optimized with the accumulation of genomic sequences (2, 4, 25, 34, 47). Improved PCR methods revealed that over 90% of adults are infected with TTV and/or TTMV (16, 25, 38, 42, 49). Frequent infection with multiple genotypes of one or both of these viruses in children and adults has been reported (6, 7, 11, 26, 38, 56). Early acquisition of TTV in infancy has also been reported (20, 41).
Recently, in the process of searching for small anellovirus (SAV) types 1 and 2 (SAV-1 and SAV-2) that were cloned from plasma samples of individuals at high risk for human immunodeficiency virus infection (17), we identified a novel DNA virus classifiable into a third group in the genus Anellovirus, with a circular DNA genome of 3.2 kb and with a genomic organization resembling those of TTV and TTMV (27), and provisionally designated this new virus torque teno midi virus (TTMDV), since its genomic length was intermediate between those of TTV and TTMV. Our findings indicated that the originally published SAV-1 and SAV-2 sequences may have been incomplete. Determination and comparative analysis of the full-length genomic sequence of 18 TTMDV isolates in our previous studies indicated the extremely wide range of sequence divergence among TTMDV isolates (27, 28). However, when the TTMDV sequences were aligned with those of TTV and TTMV over the entire genome so as to obtain maximal homology, a specific region of approximately 130 nt located just downstream of a common internal promoter, i.e., the TATA box (ATATAA), in each anellovirus that contains highly conserved areas at both ends and TTV-, TTMDV-, or TTMV-specific sequences in the central area, was recognized. This finding encouraged us to develop a specific PCR method for simultaneous amplification of the genomic DNAs of three anelloviruses in the first-round PCR and subsequent differential amplifications of these viruses in the three second-round PCRs. In the present study, the newly developed PCR method was applied to investigate the prevalence of TTMDV DNA in the general population, the possibility of early acquisition of TTMDV during infancy similar to TTV acquisition, and the prevalence of dual or triple anellovirus infection in children and adults in Japan.
MATERIALS AND METHODS
Serum samples.
Three serum samples obtained from healthy adults (subjects D1 to D3) who had the genomic DNAs of all three anelloviruses detectable by the untranslated region PCR method for TTV (52), the 3.2-kb long-distance PCR method for TTMDV (28), and a previously reported TTMV-specific PCR method (56) were used in the present study. In addition, serum samples that had been collected between 2001 and 2003 from a total of 305 apparently healthy Japanese subjects, including 100 adults (54.9 ± 11.9 years of age [mean ± standard deviation]; 57 males and 43 females) and 205 children (6.1 ± 5.7 years; 99 males and 106 females), including 44 infants of 9 to 364 days of age, were used in the present study. Aliquots of the same serum samples obtained from the 100 adults had been used for detection of TTMDV DNA by the 3.2-kb long-distance PCR method described in a previous study (28). Ten umbilical cord blood samples were also used in the present study. The sera were stored at −80°C until testing.
All of the serum samples were negative for hepatitis B virus (HBV) surface antigen (by the commercial MyCell kit; Institute of Immunology Co. Ltd., Tokyo, Japan), antibodies to hepatitis C virus (hepatitis C virus phytohemagglutinin kit; Abbott Japan, Tokyo, Japan), and antibodies to human immunodeficiency virus type 1 (SERODIA-HIV; Fujirebio, Tokyo, Japan). The study conforms to the ethical guidelines and was approved by the ethics committees of the institutions.
Detection of TTV, TTMDV, and TTMV DNAs in serum.
Nucleic acids were extracted from 100 μl of serum using the High Pure viral nucleic acid kit (Roche Molecular Biochemicals, Mannheim, Germany) and dissolved in 40 μl of nuclease-free distilled water. A 20-μl aliquot of extracted nucleic acids, equivalent to 50 μl of the sample, was tested for the presence of TTV, TTMDV, and TTMV DNAs by the PCR method described below. In the presence of Ex Taq polymerase (TaKaRa Bio Inc., Shiga, Japan) in a reaction volume of 50 μl, PCR for the universal amplification of the genomic DNAs of the three anelloviruses TTV, TTMDV, and TTMV was performed with the mixed primers NG779/NG780 (sense) and NG781/NG782 (antisense) for 35 cycles (94°C for 30 s with an additional 2 min in the first cycle, 55°C for 30 s, and 72°C for 30 s, with an additional 7 min in the last cycle) in the first round (Table 1). By use of a 2-μl portion of each of the viral amplification products of the first round, the TTV, TTMDV, and TTMV genomes were separately amplified by PCRs with seminested or nested primers which were made to be specific for each anellovirus species. In more detail, in the presence of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), the second-round PCR was carried out with primers NG779/NG780 (sense) and NG785 (antisense) for the detection of TTV DNA, with NG795 (sense) and NG796 (antisense) for TTMDV DNA, and with NG792/NG793/NG794 (sense) and NG791 (antisense) for TTMV DNA (Table 1), for 25 cycles (94°C for 30 s with an additional 2 min in the first cycle, 55°C for 30 s, and 72°C for 30 s, with an additional 7 min in the last cycle). The sizes of the amplification products of TTV DNA were 112 to 117 bp, those of TTMDV DNA were 88 bp and those of TTMV DNA were 70 to 72 bp. The amplification products were electrophoresed on a 3.5% (wt/vol) NuSieve 3:1 agarose gel (Cambrex, Rockland, ME), stained with ethidium bromide, and photographed under UV light to detect a band specific for each anellovirus species.
TABLE 1.
Positions and nucleotide sequences of oligonucleotide primers used for amplification of TTV, TTMDV, and TTMV
| Primer | PCR round | Polarity | Nucleotide position | Nucleotide sequence (5′ to 3′)a |
|---|---|---|---|---|
| Universal | ||||
| NG779 | First | Sense | 99-118b | ACWKMCGAATGGCTGAGTTT |
| NG780 | First | Sense | 99-118b | RGTGRCGAATGGYWGAGTTT |
| NG781 | First | Antisense | 208-227b | CCCKWGCCCGARTTGCCCCT |
| NG782 | First | Antisense | 208-227b | AYCTWGCCCGAATTGCCCCT |
| TTV-specific | ||||
| NG785 | Second | Antisense | 192-212b | CCCCTTGACTBCGGTGTGTAA |
| TTMDV-specific | ||||
| NG795 | Second | Sense | 78-97c | SGABCGAGCGCAGCGAGGAG |
| NG796 | Second | Antisense | 146-165c | GCCCGARTTGCCCCTAGACC |
| TTMV-specific | ||||
| NG792 | Second | Sense | 195-214d | TTTATGCYGCYAGACGRAGA |
| NG793 | Second | Sense | 195-214d | TTTAYCMYGCCAGACGGAGA |
| NG794 | Second | Sense | 195-214d | TTTATGCCGCCAGACGRAGG |
| NG791 | Second | Antisense | 247-266d | CTCACCTYSGGCWCCCGCCC |
W denotes A or T, K denotes G or T, M denotes A or C, R denotes A or G, Y denotes C or T, S denotes C or G, and B denotes C, G, or T.
Nucleotides were numbered in accordance with the prototype TTV isolate of TA278 (AB017610).
Nucleotides were numbered in accordance with the prototype TTMDV isolate of MD1-073 (AB290918).
Nucleotides were numbered in accordance with the prototype TTMV isolate of CBD231 (AB026930).
For convenience, the newly developed PCR assays for detecting TTV DNA, TTMDV DNA, and TTMV DNA in the second round, are designated Anello-TTV PCR, Anello-TTMDV PCR, and Anello-TTMV PCR, respectively, in this paper.
For comparison, previously reported PCR methods for detection of TTV DNA (52) and TTMV DNA (6, 56) were also used.
Molecular cloning of partial TTV, TTMDV, and TTMV sequences.
To assess the reliability of the newly developed PCR assays for detecting TTV DNA, TTMDV DNA, and TTMV DNA, respectively, recombinant DNA clones containing a DNA fragment of TTV, TTMDV, or TTMV were prepared. Briefly, nucleic acids extracted from serum samples from three healthy subjects (D1 to D3) who each tested positive for TTV, TTMDV, and TTMV DNAs were subjected to PCR with the above-mentioned mixed primers NG779/NG780 and NG781/NG782, and the amplicons of approximately 130 bp were ligated into pT7 Blue T-Vector (Novagen Inc., Madison, WI). Independent clones thus obtained were purified by use of the NucleoSpin Plasmid kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany) and subjected to the TTV-, TTMDV-, or TTMV-specific PCR for 25 cycles as described above, followed by sequence analysis.
Evaluation of sensitivities and specificities of the newly developed PCR assays.
The sensitivities of the newly developed PCR assays (Anello-TTV PCR, Anello-TTMDV PCR, and Anello-TTMV PCR) were assessed by using as templates serial dilutions of cloned DNAs (1.5, 5, 7.5, 15, and 150 copies per test) of each anellovirus species (TTV [D3-03, D3-08, and D3-93 clones], TTMDV [D3-01, D3-02, and D3-15 clones], and TTMV [D3-06, D3-07, and D3-57 clones]) isolated from subject D3 (see Fig. S1C in the supplemental material), with or without the coexistence of the other two anellovirus species (1.5 × 103 copies each per test). The lower limit of detection was calculated using Probit analysis.
To assess the specificity of the new PCR assays developed in the present study, per each test, 105 copies of each of the cloned DNAs from the nine representative TTV isolates classifiable into the five major groups (TA278, Pt-TTV6, Kt-08F, TUS01, TJN01, JT03F, CT23F, JT33F, and CT39F), the 18 TTMDV isolates (MD1-032, MD1-073, MD2-013, MDJHem2, MDJHem3-1, MDJHem3-2, MDJHem5, MDJHem8-1, MDJHem8-2, MDJN1, MDJN2, MDJN14, MDJN47, MDJN51, MDJN52, MDJN69, MDJN91, and MDJN97), and the two representative TTMV isolates (TGP96 and Pt-TTV8-II), for which entire nucleotide sequences were determined in our previous studies (23, 27, 28, 35, 36, 41, 55), were used as templates.
Sequence analysis of molecular clones.
Molecular clones obtained in the present study were sequenced using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 3100 genetic analyzer (Applied Biosystems). For comparison, nucleotide sequence data of 49 TTV isolates, 20 TTMDV isolates, including SAV-1 and SAV-2, and 13 TTMV isolates (see Fig. 1 for accession numbers) were retrieved from the DDBJ/GenBank/EMBL nucleotide databases. Sequence analysis was performed using Genetyx version 8 (Genetyx Corp., Tokyo, Japan) and ODEN (version 1.1.1) programs from the DNA Data Bank of Japan (National Institute of Genetics, Mishima, Japan) (14). Sequence alignments were generated by CLUSTAL W (version 1.8) (51), and the nucleotide sequence alignment was rearranged manually so as to obtain maximal homology. Phylogenetic trees were constructed by the neighbor-joining method (43). Bootstrap values were determined on 1,000 resamplings of the data sets (10). The final tree was obtained with the TreeView program (version 1.6.6) (39).
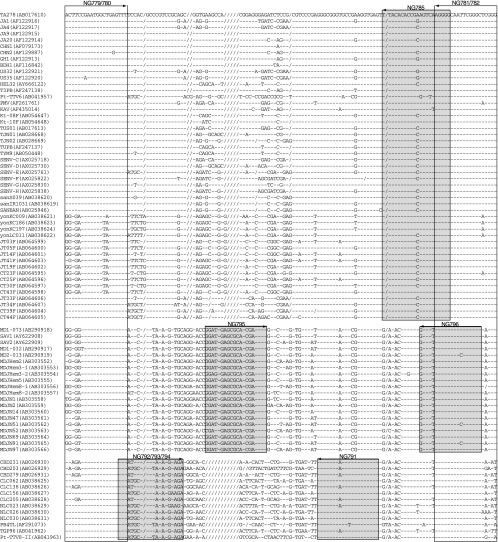
FIG. 1.
Alignment of partial sequences located just downstream of the TATA box of 82 TTV, TTMDV, and TTMV isolates whose entire sequence is known or for whom nearly the entire sequence is known. Open boxes indicate the positions of the primers common to TTV, TTMDV, and TTMV, and shaded boxes represent the positions of the primers specific to each of the three anelloviruses. Arrows denote the polarities (sense and antisense) of the primers designed in the present study. Dashes indicate nucleotides that were identical to the top sequence, and slashes denote deletions of nucleotides.
Nucleotide sequence accession numbers.
The sequences determined in the present study have been deposited in the DDBJ/GenBank/EMBL nucleotide databases under accession numbers AB355152 to AB355408.
RESULTS
Designing primers.
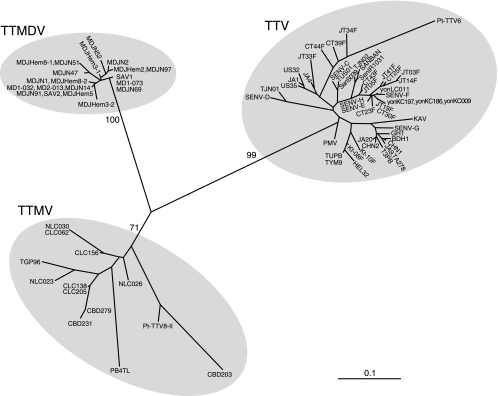
The nucleotide sequences of TTV, TTMDV, and TTMV isolates whose entire sequence is known or for whom nearly the entire sequence is known (as of August 2007) were retrieved from the DDBJ/GenBank/EMBL databases and aligned so as to obtain maximal homology. As illustrated in Fig. 1, a highly conserved genomic area located just downstream of the TATA box, corresponding to nt 99 to 227 in the prototype TTV isolate (TA278), nt 34 to 170 in the prototype TTMDV isolate (MD1-073), and nt 178 to 303 in the prototype TTMV isolate (CBD231), was noted. Universal primers suitable for simultaneous amplification of TTV, TTMDV, and TTMV DNAs in the first round, a mixture of NG779 and NG780 as sense primers and another mixture of NG781 and NG782 as antisense primers (Table 1), were designed. For differential detection of TTV, TTMDV, and TTMV DNAs, seminested or nested primers specific to each anellovirus species were designed (Fig. 1). The amplification products of TTV-specific PCR (Anello-TTV PCR) were 112 to 117 bp (primer sequences included) in size, those of TTMDV-specific PCR (Anello-TTMDV PCR) were 88 bp long, and those of the TTMV-specific PCR (Anello-TTMV PCR) were 70 to 72 bp long. Figure 2 depicts a phylogenetic tree constructed based on the 84- to 97-nt sequence, corresponding to the first-round PCR products amplifiable by universal primers (primer sequences at both ends excluded), of three human anelloviruses, including 49 TTV isolates, 20 TTMDV isolates, and 13 TTMV isolates (Fig. 1 indicates the accession numbers), indicating that these anellovirus isolates are phylogenetically classifiable as the respective virus even within this very limited area of genomic sequence.
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence of the noncoding region (84 to 97 nt) of 82 TTV, TTMDV, and TTMV isolates whose entire sequence is known or for whom nearly the entire sequence is known. The accession number of each isolate is shown in Fig. 1. Bootstrap values are indicated for the major nodes as a percentage obtained from 1,000 resamplings of the data. Bar, 0.1 nucleotide substitutions per site.
Reliability of the newly developed PCR assays for the detection of TTV, TTMDV, and TTMV DNAs.
A total of 81 anellovirus DNA clones amplified by the universal primers NG779/NG780 and NG781/NG782 were isolated from subject D1, who was known to be coinfected with TTV, TTMDV, and TTMV, and were classified as 34 TTV clones, 43 TTMDV clones, and 4 TTMV clones by the newly developed differential PCR assays. The phylogenetic tree constructed based on the 86- to 97-nt sequence of the 81 clones (primer sequences at both ends excluded) confirmed the distribution of TTV, TTMDV, and TTMV in subject D1 (see Fig. S1A in the supplemental material). Similarly, 91 DNA clones obtained from subject D2 were grouped as TTV (n = 44), TTMDV (n = 31), and TTMV (n = 16), and 85 clones from subject D3 as TTV (n = 20), TTMDV (n = 28), and TTMV (n = 37) by the differential PCR assays developed in the present study. These results were consistent with those obtained by phylogenetic analyses (see Fig. S1B and C in the supplemental material).
Sensitivity of the newly developed PCR assays.
Probit analysis determined that the 95% detection levels of TTV DNA by the Anello-TTV PCR assay were 5.1 and 5.4 copies per test with or without the presence of TTMDV and TTMV clones, respectively. The sensitivity of Anello-TTMDV PCR and Anello-TTMV PCR were almost equal (5.3 to 5.4 copies per test) to that of the Anello-TTV PCR assay regardless of the coexistence of the other two anellovirus species.
Specificity of the newly developed PCR assays.
When cloned DNAs from the 18 TTMDV isolates, whose full-length genomic sequences are known (27, 28), were used as templates, no amplification signals were detectable by both the newly developed TTV PCR (Anello-TTV PCR) assay and a reported TTV PCR assay (52) and, although two reported TTMV PCR assays (6, 56) yielded nonspecific bands of 121 bp and 443 to 444 bp, respectively, due to misannealing of the used primers to TTMDV DNA, no amplification products were obtained by the Anello-TTMV PCR assay. When serum samples from 100 adults 20 to 81 years of age were used as a panel, TTV DNA was detectable in all 100 samples by the Anello-TTV PCR assay, comparable with 99 samples (99%) with detectable virus by the reported TTV PCR assay (52). Specificity of the Anello-TTV PCR for the TTV DNA in one sample that tested negative for TTV DNA by the previously reported TTV PCR assay (52) was confirmed by sequence analysis (data not shown).
When cloned DNAs from the nine representative TTV isolates classifiable into five major groups were used as templates, no amplification signals were detectable by both the Anello-TTMV PCR assay and the Anello-TTMDV PCR assay. In addition, when cloned DNAs from the two representative TTMV isolates were used as templates, no specific bands were amplified by both the Anello-TTV PCR assay and the Anello-TTMDV PCR assay.
Detection of TTV, TTMDV, and TTMV DNAs in serum samples.
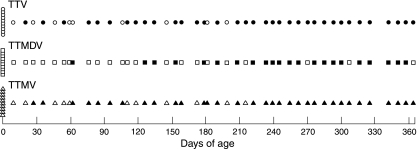
TTMDV DNA was detected in serum samples from 75 (75%) of the 100 adults 20 to 81 years of age, including 35 adults who tested negative for TTMDV DNA by the 3.2-kb long-distance PCR method (28), indicating the higher sensitivity of the present PCR assay for TTMDV. In order to investigate the prevalence of anelloviruses according to age and virus species (TTV, TTMDV, and TTMV), the newly developed PCR method was applied to 10 umbilical cord blood samples and serum samples collected from 305 apparently healthy children and adults in Japan, including the 100 adults mentioned above. Overall, the prevalence of TTMDV viremia was found to be high (75.2%), comparable with those of TTV viremia (93.3%) and TTMV viremia (84.8%) (Table 2). Although none of the 10 cord blood samples had detectable TTV, TTMDV, and TTMV DNAs, the prevalence of these three anelloviruses increased rapidly with the number of months after birth, with the first appearance of TTV at 20 days of age, the first appearance of TTMV at 27 days of age, and the first appearance of TTMDV at 62 days of age, and viral prevalence reached 100% at the age of one year, indicating early acquisition of TTV, TTMDV, and TTMV during infancy (Fig. 3). Children 1 to 4 years of age showed extremely high prevalences (98.2 to 100%) of TTV, TTMDV, and TTMV viremias. Although the prevalence of TTV DNA was almost 100% in all groups of individuals older than one year of age (Table 2), TTMDV DNA was detectable in only 68.8 to 85.7% of the age groups of 5 to 9, 10 to 19, 20 to 49, and 50 to 81 years, and TTMV DNA was detectable in 75.0 to 88.7% of the individuals in age groups of 10 to 19, 20 to 49, and 50 to 81 years. The prevalences of TTV DNA, TTMDV DNA, and TTMV DNA among subjects ≥1 year of age were 99.2%, 82.4%, and 89.7%, respectively.
TABLE 2.
Age-specific prevalence of TTV, TTMDV, and TTMV DNAs
| Age (yrs) and sample type | No. of samples tested | No. (%) of samples with:
|
||
|---|---|---|---|---|
| TTV DNA | TTMDV DNA | TTMV DNA | ||
| <1 | ||||
| Cord blood | 10 | 0 | 0 | 0 |
| Seruma | 44 | 35 (79.5) | 22 (50.0) | 33 (75.0) |
| Subtotal | 54 | 35 (64.8) | 22 (40.7) | 33 (61.1) |
| ≥1 (serum) | ||||
| 1 | 28 | 28 (100) | 28 (100) | 28 (100) |
| 2-4 | 29 | 29 (100) | 28 (96.6) | 28 (96.6) |
| 5-9 | 42 | 41 (97.6) | 36 (85.7) | 41 (97.6) |
| 10-19 | 62 | 61 (98.4) | 48 (77.4) | 55 (88.7) |
| 20-49 | 32 | 32 (100) | 22 (68.8) | 24 (75.0) |
| 50-81 | 68 | 68 (100) | 53 (77.9) | 58 (85.3) |
| Subtotal | 261 | 259 (99.2) | 215 (82.4) | 234 (89.7) |
| Total | 315 | 294 (93.3) | 237 (75.2) | 267 (84.8) |
The serum samples were from infants <1 year of age (9 to 364 days).
FIG. 3.
Detection of TTV DNA, TTMDV DNA, and TTMV DNA in 10 umbilical cord blood samples and serum samples from 44 infants of 9 to 364 days of age. For each infant, closed and open circles in the top row represent positivity and negativity for TTV DNA, respectively; closed and open boxes in the middle row represent positivity and negativity for TTMDV DNA, respectively; and closed and open triangles in the bottom row represent positivity and negativity for TTMV DNA, respectively.
Among infants ≤180 days of age, triple infection of TTV, TTMDV, and TTMV was seen in four infants (12.9%), and dual infection of TTV and TTMV or of TTMDV and TTMV was found in six infants (19.3%) (see Fig. S2 in the supplemental material). On the other hand, among infants 181 to 364 days of age, the prevalence of triple infection was 73.9%, comparable with the 78.2% prevalence among subjects 1 to 81 years of age. Although single infections of TTV and TTMV were seen in 22 and 5 subjects, respectively, no case of a single infection of TTMDV was seen in the studied population.
DISCUSSION
The development of methods for specific detection of human anelloviruses has been made more complex by the discovery of isolates classifiable into a third group in the genus Anellovirus, TTMDV. In fact, the present study revealed that TTMDV DNA can be erroneously amplified by previously reported TTMV PCR assays (6, 56). Biagini et al. (3) reported that TTMDV/SAV had a 20% prevalence among French blood donors, which is comparable to the 9% frequency of TTMDV/SAV detection among Italian blood donors (1). In our previous study (28), approximately 40% of healthy individuals had TTMDV DNA in their circulation, detectable by a 3.2-kb long-distance PCR method for TTMDV. It has been reported that the selection of PCR primers and the length of the genomic region for PCR amplification crucially influence the detection of TTV, isolates of which can have extremely divergent genomes (4, 16, 38). Recent surveys using primers specific for individual genotypes or genogroups of TTV, or those that differentiate TTV from TTMV sequences, indicated that approximately 90% of study populations (generally healthy adults) had TTV or TTMV viremia, with coinfection of TTV and TTMV in 44% (6, 21). Therefore, it seemed likely that the actual prevalence of TTMDV DNA is higher than was previously reported. In order to clarify the exact prevalence of TTMDV DNA and to elucidate the prevalence of double or triple infection of TTV, TTMDV, and/or TTMV, the present study was undertaken to develop highly sensitive and specific PCR assays for the differential detection of these three human anelloviruses. Although TTV, TTMDV, and TTMV have a common presumed genomic organization with conserved sequence motifs and transcriptional profiles, they exhibit high dissimilarities with regard to genomic length and sequence. Despite a marked divergence with differences of >50% among TTV genomes (19, 41, 49), differences of up to 33% among TTMDV genomes (28), and differences of up to 40% among TTMV genomes (5, 48), there exists a highly conserved area of 130 nt located just downstream of the TATA box in the TTV, TTMDV, and TTMV genomes. Taking advantage of this particular genomic area that is conserved among known anelloviruses, we developed virus species-specific PCR assays, in which the genomic DNAs of all three anelloviruses are amplified by the first-round PCR with universal primers and TTV DNA, TTMDV DNA, or TTMV DNA is separately amplified by each of the three second-round PCRs (Anello-TTV PCR, Anello-TTMDV PCR, and Anello-TTMV PCR, respectively) with species-specific primers. All 257 molecular clones of the PCR products amplified by universal primers obtained from three subjects (subjects D1 to D3) who were coinfected with TTV, TTMDV, and TTMV were classifiable as TTV, TTMDV, or TTMV by the three differential PCR assays, and the reliability of these assays for classification was confirmed by phylogenetic analysis. When our newly developed PCR assays were applied to serum samples from adults in the general population, high prevalence rates of TTV DNA (100%) and TTMV DNA (82%) were noted, consistent with previously reported rates (4, 7, 25, 34, 42, 49). A high prevalence of TTMDV DNA (75/100 [75%]) was also found, with positivity seen for 35 subjects who had tested negative for TTMDV DNA by 3.2-kb long-distance PCR (28), suggesting the superiority of the present TTMDV PCR method in terms of detection rate, compared with our previous method (3.2-kb long-distance PCR). As for sensitivity, the three PCR assays developed in this study were comparable, being capable of detecting 5.1 to 5.4 copies per test with or without the coexistence of two other anellovirus species clones. In addition, there was no cross-reactivity for TTMDV and TTMV in the Anello-TTV PCR assay, for TTV and TTMV in the Anello-TTMDV PCR assay, and for TTV and TTMDV in the Anello-TTMV PCR assay. Therefore, the results obtained in the present study indicate that the three PCR assays for the differential detection of TTV, TTMDV, and TTMV DNAs can be applied to clinical samples.
Perinatal transmission of blood-borne viruses, such as HBV, hepatitis C virus, and human immunodeficiency virus type 1, is well documented (30, 31, 40, 46). This mode of transmission plays a pivotal role in maintaining HBV transmission from generation to generation (31, 46). In the present study, however, none of the 10 umbilical cord blood samples had the genomic DNA of TTV, TTMDV, or TTMV, and a rapidly rising prevalence of TTV, TTMDV, and TTMV infections was noted over the subsequent months, reaching a frequency of 100% within the first 2 years of life. Although further studies on larger cohorts with larger population sizes are needed to draw a plausible conclusion, since infection with TTV and/or TTMV in infants has been reported (12, 18, 20, 56), early infection during human infancy may be common among all three anelloviruses, including TTMDV. In support of our view, Kazi et al. (18) reported that 99 (99%) of 100 cord blood samples were negative for TTV DNA and that among infants between 6 to 8 and 12 to 21 months of age, 4 (33%) of 12 and 5 (23%) of 22 infants born to infected and uninfected mothers, respectively, became positive for TTV DNA, respectively, although the difference was not statistically significant. Failure to detect TTV DNA in samples of blood from newborn infants and cord blood has also been reported (54). Therefore, maternal transmission may play only a minor role in early acquisition of TTV and TTMV as well as TTMDV in infants. TTV and TTMV genomes have been detected in feces (5, 32), saliva (5, 15, 56), and breast milk (12, 54), suggesting transmission of anelloviruses, including TTMDV, via horizontal routes. However, studies to investigate the presence of TTMDV in various specimens from infected hosts and to determine the relative importance of each route of transmission are required.
The prevalence of TTMDV infection decreased in the group of individuals 5 to 9 years of age and older age groups, as did the prevalence of TTMV infection in the groups of individuals 10 to 19, 20 to 49, and 50 to 81 years of age. A similar decrease in the prevalence of TTV infection with age has been reported (54, 57), although, in the present study, TTV infection was highly prevalent (97.6 to 100%) in all groups of individuals older than one year of age. The precise reason for this decrease in prevalence remains unknown. However, changes in immunological status with growth from infant to child or adolescent may affect the persistent viremia or viral load of TTMDV, TTMV, or TTV. In support of our speculation, it has been reported that HBV infection acquired during early infancy tends to continue during the rest of the individual's life (9). In addition, a significant association between the CD4 cell count and the TTV/TTMV titer and an inverse relationship between TTV viral load and level of immunosuppression have been reported (44, 50, 53).
Due to the frequent dual or triple infections of TTV, TTMDV, and/or TTMV noted in adults as well as infants in this study and the lack of a cell culture system to support the replication of these viruses, it remains unknown whether each of the three anelloviruses can replicate and maintain the persistent carrier state independently or in concert with one or two of the other anelloviruses. Although single infection of TTMDV was not recognized in the present study, the presence of a single infection of TTV or TTMV in some infected hosts suggests that TTV, TTMDV, and TTMV can independently infect susceptible hosts and replicate in their tissues or organs.
In conclusion, taking advantage of universal and virus species-specific primers derived from a highly conserved area located just downstream of the TATA box of the TTV, TTMDV, and TTMV genomes, we developed a new PCR assay method to separately detect the genomic DNAs of TTV, TTMDV, and TTMV. The PCR method with high sensitivity and reliability revealed high viremia rates for TTV (99.2%), TTMDV (82.4%), and TTMV (89.7%) among subjects 1 to 81 years of age and frequent dual or triple infections of these anelloviruses even in infants. The pathogenetic role of TTMDV infection remains unknown. The changing ratio of three anelloviruses to each other over time, relative viral load, or combination of different genotypes of each anellovirus may be associated with the pathogenicity or the disease-inducing potential of these three human anelloviruses. In this context, further efforts to develop methods to separately or simultaneously quantitate the genomic DNAs of the three anelloviruses and to clarify the extent of genetic variability of TTMDV toward establishing a reliable method of genotyping or genogrouping TTMDV are warranted.
Supplementary Material
Acknowledgments
This study was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 19 December 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Andreoli, E., F. Maggi, M. Pistello, S. Meschi, M. Vatteroni, L. C. Nelli, and M. Bendinelli. 2006. Small anellovirus in hepatitis C patients and healthy controls. Emerg. Infect. Dis. 121175-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biagini, P., R. N. Charrel, P. de Micco, and X. de Lamballerie. 2003. Association of TT virus primary infection with rhinitis in a newborn. Clin. Infect. Dis. 36128-129. [DOI] [PubMed] [Google Scholar]
- 3.Biagini, P., P. de Micco, and X. de Lamballerie. 2006. Identification of a third member of the anellovirus genus (“small anellovirus”) in French blood donors. Arch. Virol. 151405-408. [DOI] [PubMed] [Google Scholar]
- 4.Biagini, P., P. Gallian, H. Attoui, J. F. Cantaloube, M. Touinssi, P. de Micco, and X. de Lamballerie. 2001. Comparison of systems performance for TT virus detection using PCR primer sets located in non-coding and coding regions of the viral genome. J. Clin. Virol. 2291-99. [DOI] [PubMed] [Google Scholar]
- 5.Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J. Cantaloube, P. de Micco, and X. de Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82379-383. [DOI] [PubMed] [Google Scholar]
- 6.Biagini, P., P. Gallian, J. F. Cantaloube, H. Attoui, P. de Micco, and X. de Lamballerie. 2006. Distribution and genetic analysis of TTV and TTMV major phylogenetic groups in French blood donors. J. Med. Virol. 78298-304. [DOI] [PubMed] [Google Scholar]
- 7.Biagini, P., P. Gallian, M. Touinssi, J. F. Cantaloube, J. P. Zapitelli, X. de Lamballerie, and P. de Micco. 2000. High prevalence of TT virus infection in French blood donors revealed by the use of three PCR systems. Transfusion 40590-595. [DOI] [PubMed] [Google Scholar]
- 8.Biagini, P., D. Todd, M. Bendinelli, S. Hino, A. Mankertz, S. Mishiro, C. Niel, H. Okamoto, S. Radial, B. W. Ritchie, and G. C. Teo. 2005. Anellovirus, p. 335-341. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, San Diego, CA.
- 9.Edmunds, W. J., G. F. Medley, D. J. Nokes, A. J. Hall, and H. C. Whittle. 1993. The influence of age on the development of the hepatitis B carrier state. Proc. Biol. Sci. 253197-201. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 11.Forns, X., P. Hegerich, A. Darnell, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J. Med. Virol. 59313-317. [PubMed] [Google Scholar]
- 12.Gerner, P., R. Oettinger, W. Gerner, J. Falbrede, and S. Wirth. 2000. Mother-to-infant transmission of TT virus: prevalence, extent and mechanism of vertical transmission. Pediatr. Infect. Dis. J. 191074-1077. [DOI] [PubMed] [Google Scholar]
- 13.Hino, S., and H. Miyata. 2006. Torque teno virus (TTV): current status. Rev. Med. Virol. 1745-57. [DOI] [PubMed] [Google Scholar]
- 14.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 1011-12. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa, T., Y. Hamano, and H. Okamoto. 1999. Frequent detection of TT virus in throat swabs of pediatric patients. Infection 27298. [PubMed] [Google Scholar]
- 16.Itoh, K., M. Takahashi, M. Ukita, T. Nishizawa, and H. Okamoto. 1999. Influence of primers on the detection of TT virus DNA by polymerase chain reaction. J. Infect. Dis. 1801750-1751. [DOI] [PubMed] [Google Scholar]
- 17.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 798230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazi, A., H. Miyata, K. Kurokawa, M. A. Khan, T. Kamahora, S. Katamine, and S. Hino. 2000. High frequency of postnatal transmission of TT virus in infancy. Arch. Virol. 145535-540. [DOI] [PubMed] [Google Scholar]
- 19.Khudyakov, Y. E., M. E. Cong, B. Nichols, D. Reed, X. G. Dou, S. O. Viazov, J. Chang, M. W. Fried, I. Williams, W. Bower, S. Lambert, M. Purdy, M. Roggendorf, and H. A. Fields. 2000. Sequence heterogeneity of TT virus and closely related viruses. J. Virol. 742990-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, H. H., J. H. Kao, P. I. Lee, and D. S. Chen. 2002. Early acquisition of TT virus in infants: possible minor role of maternal transmission. J. Med. Virol. 66285-290. [DOI] [PubMed] [Google Scholar]
- 21.Maggi, F., E. Andreoli, L. Lanini, C. Fornai, M. Vatteroni, M. Pistello, S. Presciuttini, and M. Bendinelli. 2005. Relationships between total plasma load of torquetenovirus (TTV) and TTV genogroups carried. J. Clin. Microbiol. 434807-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata, H., H. Tsunoda, A. Kazi, A. Yamada, M. A. Khan, J. Murakami, T. Kamahora, K. Shiraki, and S. Hino. 1999. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J. Virol. 733582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muljono, D. H., T. Nishizawa, F. Tsuda, M. Takahashi, and H. Okamoto. 2001. Molecular epidemiology of TT virus (TTV) and characterization of two novel TTV genotypes in Indonesia. Arch. Virol. 1461249-1266. [DOI] [PubMed] [Google Scholar]
- 24.Mushahwar, I. K., J. C. Erker, A. S. Muerhoff, T. P. Leary, J. N. Simons, L. G. Birkenmeyer, M. L. Chalmers, T. J. Pilot-Matias, and S. M. Dexai. 1999. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc. Natl. Acad. Sci. USA 963177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niel, C., and E. Lampe. 2001. High detection rates of TTV-like mini virus sequences in sera from Brazilian blood donors. J. Med. Virol. 65199-205. [PubMed] [Google Scholar]
- 26.Niel, C., F. L. Saback, and E. Lampe. 2000. Coinfection with multiple TT virus strains belonging to different genotypes is a common event in healthy Brazilian adults. J. Clin. Microbiol. 381926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninomiya, M., T. Nishizawa, M. Takahashi, F. R. Lorenzo, T. Shimosegawa, and H. Okamoto. 2007. Identification and genomic characterization of a novel human torque teno virus of 3.2 kb. J. Gen. Virol. 881939-1944. [DOI] [PubMed] [Google Scholar]
- 28.Ninomiya, M., M. Takahashi, T. Shimosegawa, and H. Okamoto. 2007. Analysis of the entire genomes of fifteen torque teno midi virus variants classifiable into a third group of genus Anellovirus. Arch. Virol. 1521961-1975. [DOI] [PubMed] [Google Scholar]
- 29.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 24192-97. [DOI] [PubMed] [Google Scholar]
- 30.Ohto, H., S. Terazawa, N. Sasaki, N. Sasaki, K. Hino, C. Ishiwata, M. Kako, N. Ujiie, C. Endo, A. Matsui, H. Okamoto, S. Mishiro, et al. 1994. Transmission of hepatitis C virus from mothers to infants. N. Engl. J. Med. 330744-750. [DOI] [PubMed] [Google Scholar]
- 31.Okada, K., I. Kamiyama, M. Inomata, M. Imai, and Y. Miyakawa. 1976. e antigen and anti-e in the serum of asymptomatic carrier mothers as indicators of positive and negative transmission of hepatitis B virus to their infants. N. Engl. J. Med. 294746-749. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, H., Y. Akahane, M. Ukita, M. Fukuda, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1998. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J. Med. Virol. 56128-132. [PubMed] [Google Scholar]
- 33.Okamoto, H., T. Nishizawa, N. Kato, M. Ukita, H. Ikeda, H. Iizuka, Y. Miyakawa, and M. Mayumi. 1998. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol. Res. 101-16. [Google Scholar]
- 34.Okamoto, H., T. Nishizawa, M. Takahashi, A. Tawara, Y. Peng, J. Kishimoto, and Y. Wang. 2001. Genomic and evolutionary characterization of TT virus (TTV) in tupaias and comparison with species-specific TTVs in humans and non-human primates. J. Gen. Virol. 822041-2050. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, H., T. Nishizawa, A. Tawara, Y. Peng, M. Takahashi, J. Kishimoto, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Species-specific TT viruses in humans and nonhuman primates and their phylogenetic relatedness. Virology 277368-378. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto, H., T. Nishizawa, M. Ukita, M. Takahashi, M. Fukuda, H. Iizuka, Y. Miyakawa, and M. Mayumi. 1999. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology 259437-448. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto, H., M. Takahashi, and T. Nishizawa. 2004. Torque teno virus (TTV): molecular virology and clinical implications, p. 241-254. In I. K. Mushahwar (ed.), Viral hepatitis: molecular biology, diagnosis, epidemiology and control. Elsevier, Amsterdam, The Netherlands.
- 38.Okamoto, H., M. Takahashi, T. Nishizawa, M. Ukita, M. Fukuda, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1999. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology 259428-436. [DOI] [PubMed] [Google Scholar]
- 39.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 40.Peckham, C., and D. Gibb. 1995. Mother-to-child transmission of the human immunodeficiency virus. N. Engl. J. Med. 333298-302. [DOI] [PubMed] [Google Scholar]
- 41.Peng, Y. H., T. Nishizawa, M. Takahashi, T. Ishikawa, A. Yoshikawa, and H. Okamoto. 2002. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch. Virol. 14721-41. [DOI] [PubMed] [Google Scholar]
- 42.Saback, F. L., S. A. Gomes, and C. Niel. 2002. High frequency of mixed TT virus infections in healthy adults and children detected by a simplified heteroduplex mobility assay. J. Virol. Methods 101117-125. [DOI] [PubMed] [Google Scholar]
- 43.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 44.Shibayama, T., G. Masuda, A. Ajisawa, M. Takahashi, T. Nishizawa, F. Tsuda, and H. Okamoto. 2001. Inverse relationship between the titre of TT virus DNA and the CD4 cell count in patients infected with HIV. AIDS 15563-570. [DOI] [PubMed] [Google Scholar]
- 45.Simmonds, P. 2002. TT virus and related human circoviruses, p. 613-621. In D. D. Richman, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology, 2nd ed. ASM Press, Washington, DC.
- 46.Stevens, C. E., R. P. Beasley, J. Tsui, and W. C. Lee. 1975. Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 292771-774. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, K., H. Hoshino, Y. Ohta, N. Yoshida, and S. Mishiro. 1998. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol. Res. 12233-239. [Google Scholar]
- 48.Takahashi, K., Y. Iwasa, M. Hijikata, and S. Mishiro. 2000. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 145979-993. [DOI] [PubMed] [Google Scholar]
- 49.Thom, K., C. Morrison, J. C. Lewis, and P. Simmonds. 2003. Distribution of TT virus (TTV), TTV-like minivirus, and related viruses in humans and nonhuman primates. Virology 306324-333. [DOI] [PubMed] [Google Scholar]
- 50.Thom, K., and J. Petrik. 2007. Progression towards AIDS leads to increased Torque teno virus and Torque teno minivirus titers in tissues of HIV infected individuals. J. Med. Virol. 791-7. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokita, H., S. Murai, H. Kamitsukasa, M. Yagura, H. Harada, M. Takahashi, and H. Okamoto. 2002. High TT virus load as an independent factor associated with the occurrence of hepatocellular carcinoma among patients with hepatitis C virus-related chronic liver disease. J. Med. Virol. 67501-509. [DOI] [PubMed] [Google Scholar]
- 53.Touinssi, M., P. Gallian, P. Biagini, H. Attoui, B. Vialettes, Y. Berland, C. Tamalet, C. Dhiver, I. Ravaux, P. De Micco, and X. De Lamballerie. 2001. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J. Clin. Virol. 21135-141. [DOI] [PubMed] [Google Scholar]
- 54.Toyoda, H., M. Naruse, S. Yokozaki, K. Morita, I. Nakano, A. Itakura, M. Okamura, Y. Fukuda, and T. Hayakawa. 1999. Prevalence of infection with TT virus (TTV), a novel DNA virus, in healthy Japanese subjects, newborn infants, cord blood and breast milk. J. Infect. 38198-199. [DOI] [PubMed] [Google Scholar]
- 55.Ukita, M., H. Okamoto, T. Nishizawa, A. Tawara, M. Takahashi, H. Iizuka, Y. Miyakawa, and M. Mayumi. 2000. The entire nucleotide sequences of two distinct TT virus (TTV) isolates (TJN01 and TJN02) remotely related to the original TTV isolates. Arch. Virol. 1451543-1559. [DOI] [PubMed] [Google Scholar]
- 56.Vasconcelos, H. C., M. Cataldo, and C. Niel. 2002. Mixed infections of adults and children with multiple TTV-like mini virus isolates. J. Med. Virol. 68291-298. [DOI] [PubMed] [Google Scholar]
- 57.Yamada-Osaki, M., R. Sumazaki, E. Noguchi, M. Shibasaki, and A. Matsui. 1998. Transfusion transmitted virus. Lancet 3521309-1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.