Abstract
The continuous spread of community-acquired methicillin-resistant Staphylococcus aureus (caMRSA) and the introduction of these highly virulent isolates into hospitals represent increasing threats. The timely recognition of caMRSA strains is crucial for infection control purposes. Thus, we developed a PCR-based assay for the easy and rapid determination of those caMRSA clones that currently are the most prevalent in Germany and Central Europe. This assay was able to correctly identify the majority of the isolates as caMRSA of sequence type 80 (ST80), clonal complex 1 (USA400), and ST8 (USA300). In combination with spa typing-BURP (based upon repeat pattern) analysis and resistance typing, it provides a means for the extensive characterization of suspicious isolates. Thus, this assay represents a reliable tool for monitoring the emergence and spread of different caMRSA clones. The resulting information, in combination with careful interpretation of the epidemiological records, might help to prevent the further spread of those highly virulent caMRSA clones.
Staphylococcus aureus is a facultative pathogenic gram-positive bacterium which is well known as colonizer of the human skin, but it can also cause a variety of diseases, ranging from minor skin and soft tissue infections to life-threatening disease (20). Methicillin-resistant S. aureus (MRSA) and multiresistant S. aureus strains are responsible for a large proportion of nosocomial infections, making treatment difficult (34). Several risk factors for MRSA infection or colonization have been established and include previous hospitalization, residence in a nursing home, antibiotic therapy, and the presence of indwelling medical devices (20).
However, during the last decade an increasing number of reports of MRSA cases among healthy community-dwelling persons without classical risk factors for MRSA acquisition were encountered worldwide (15). The majority of these isolates, referred to as “community-acquired MRSA” (caMRSA) strains (31), are genetically and phenotypically distinct from representative “hospital-acquired” MRSA (haMRSA) strains, as reflected by their narrow resistance patterns, as well as by the distribution of their staphylococcal chromosomal cassette (SCCmec) types (type IV or V) and resistance and toxin determinants (15, 37). Different caMRSA clones were originally shown to be continent specific (37), but recent studies demonstrated their intercontinental spread, with the predominance of particular clones in distinct geographic regions (3, 35, 36). The caMRSA clones currently most predominant in Germany and Central Europe are sequence type 80 (ST80; spa type t044, European clone), ST1 (spa type t127, USA400), and ST8 (spa type t008, USA300) (17, 36, 40, 41).
A matter of particular concern is the threat of introducing highly virulent caMRSA clones from the site of their origin in the community into the hospital, where they meet considerably more compromised patients, potentially leading to markedly increased patient morbidity and mortality (39). Episodes of health care-associated infections due to caMRSA have been already reported in the United States and Europe (14, 18, 32).
Typing is a prerequisite to obtaining knowledge of the epidemiology of S. aureus strains in order to prevent the spread of caMRSA within the community as well as from the community into the hospitals. We have recently shown that spa typing in combination with clustering by BURP (based upon repeat pattern) analysis is a useful tool in S. aureus epidemiology, especially because of its reproducibility and the portability of the typing data (33). However, recent studies (11, 32a) have demonstrated that spa typing-BURP analysis is not always able to discriminate unambiguously between different clones of caMRSA (e.g., between ST80 and ST1 clones) and some common methicillin-susceptible S. aureus (MSSA) clones because similar spa types occur in different clones, but this cannot be explained by large chromosomal replacements (27). Moreover, some spa types occur in different MRSA clones, including caMRSA clones. This was demonstrated for spa type t008, which was found in three different clones (characterized by the possession of different SCCmec types) and which also included caMRSA t008/ST8/USA300 (4).
Therefore, the aim of this study was the development of a rapid and easy PCR assay that can be used to recognize the predominant caMRSA clones based on lineage-specific genetic markers.
MATERIALS AND METHODS
Bacterial strains.
The S. aureus isolates investigated in this study (n = 125) were sent to the German Reference Centre for Staphylococci for further characterization and typing. Isolates originated from microbiological laboratories from throughout Germany, as well as from other Central European countries. The isolates included in this study were BURP analysis-defined relatives of spa types t044 (ST80) and t127 (ST1), as well as isolates of type t008/t024 (potentially ST8/USA300) and isolates from other clonal lineages (clonal complex 5 [CC5], CC22, CC30, CC45, CC121, ST59, ST152, and ST154), which were supposed to be of community origin on the basis of epidemiological information. Strains were isolated from patients with skin and soft tissue infections (n = 55), conjunctivitis (n = 2), and otitis (n = 1); 9 isolates were collected from patients with invasive infections (bacteremia, pneumonia, urinary tract infections). Twenty-one strains were isolated from nasal swabs. For 39 isolates no information concerning the medical history of the patient was available (most of them were selected because of their relatedness to spa types t044 and t127).
All isolates were cultured on sheep blood agar and were confirmed to be S. aureus by colony morphology and a positive plasma coagulase reaction. They were subjected to susceptibility testing by the broth microdilution method, as described by DIN (5). Previously characterized reference isolates for the most prevalent caMRSA clones in Germany and Central Europe were the following: 05-01290, ST1/t127 (USA400), seh lukPV; 06-01172, ST8/t008 (USA300), arcA lukPV; and 06-00300, ST80/t044 (European caMRSA), etd lukPV.
DNA extraction.
Genomic DNA was isolated from 2 ml overnight culture with a DNeasy tissue kit (Qiagen, Hilden, Germany) by using lysostaphin (100 mg/liter; Sigma, Taufkirchen, Germany) to achieve bacterial lysis.
Selection of lineage-specific loci.
The determinants most likely to be specific for particular caMRSA clones were selected from the literature as well as from published genomes (23, 36). The following determinants were chosen for amplification by a multiplex approach: the enterotoxin H gene (seh) as a marker for caMRSA of clonal lineage ST1/USA400 (12, 30), the arginine deiminase gene (arcA) as part of the ACME (arginine catabolic mobile element) cluster for ST8/t008/USA300 (6, 9), and the gene for exfoliative toxin D (etd) for European caMRSA clones of ST80 (42, 43). In addition, the Panton-Valentine leukocidin gene (lukPV) was selected as a marker as it is often epidemiologically associated with the caMRSA clones prevalent in Central Europe and the United States (1).
caMRSA-MP.
The primers used for the multiplex PCR (MP) for caMRSA detection (caMRSA-MP) were designed to facilitate the concomitant amplification of all putative PCR products in a single reaction. Therefore, all oligonucleotides had similar melting temperatures of approximately 60°C and yielded PCR products of 200 to 600 bp (Table 1). All primers were selected from public databases by using the freely available software Primer 3 (29) and were synthesized by Metabion (Munich, Germany). Single PCR amplifications as well as MP amplifications were performed with Ready-to-Go-PCR beads (GE Healthcare, Munich, Germany) in a 25-μl reaction mixture containing approximately 10 ng of template DNA and 2.5 pmol of each primer. Initial denaturation at 94°C for 3 min was followed by 30 cycles of amplification with 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s (except for the final cycle, which had an extension step of 4 min). The PCR products were analyzed on a 2% agarose gel. Initially, the PCR products were confirmed by sequencing. Sequencing reactions were carried out with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA), as specified by the manufacturer. Comparison of the sequences to the published sequence data was performed with the DNAStar software package (DNAStar Inc., Madison, WI).
TABLE 1.
Primers used in this study
| Primera | Sequence (5′-3′) | Product size (bp) | Position (GenBank accession no.) | Reference |
|---|---|---|---|---|
| BSetd f | CCC GTT GAT TAG TCA TGC AG | 607 | 5468-5487 (AB057421) | This study |
| BSetd r | TCC AGA ATT TCC CGA CTC AG | 6074-6055 (AB057421) | ||
| WWarcA f | TTG CTC AAA CTT TGA GAG ATG AA | 215 | 74182-74160 (CP000255.1) | This study |
| WWarcA r3 | TTA CGT ACG CCA GCC ATG AT | 73966-73985 (CP000255.1) | ||
| seh f | CAA CTG CTG ATT TAG CTC AG | 358 | 60475-60494 (BX571857.1) | 22 |
| seh r | GTC GAA TGA GTA ATC TCT AGG | 60833-60813 (BX571857.1) | ||
| lukPV f | ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A | 432 | 1640-1670 (X72700) | 26 |
| lukPV r | GCA TCA AGT GTA TTG GAT AGC AAA AGC | 2072-2046 (X72700) |
f, forward; r, reverse.
Molecular typing.
Spa typing and BURP analysis, as well as multilocus sequence typing (MLST) and eBURST analysis, were conducted as described elsewhere (32a).
RESULTS
Molecular characterization of isolates and validation of MP results.
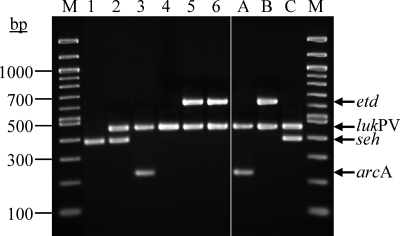
Individual primer pairs as well as the combination of optimized primer sets were tested with previously characterized isolates representing caMRSA clones ST1/USA400, ST8/USA300, and ST80 before they were used for the molecular characterization of the study isolates (Fig. 1). A total of 125 isolates were examined by caMRSA-MP. Additionally, all isolates were characterized by spa typing. A subset of isolates was also typed by MLST analysis. The resulting types were grouped by BURP and eBURST analyses. The results of the strain characterization are summarized in Table 2. As demonstrated in other studies (11, 32a), grouping by BURP analysis was not always sufficient to group the isolates unambiguously into definite groups associated with caMRSA, e.g., into CC1 and ST80. Thus, BURP group A contains a mixture of isolates belonging to CC1, CC7, CC15, CC80, and CC97 (Table 2, BURP group A c6). Adjustment of the default parameters of the BURP algorithm to a more stringent group definition, as proposed in a recent study by Mellmann et al. (21), only partly solved this problem (Table 2, BURP group A c4).
FIG. 1.
caMRSA-MP characterization of clinical S. aureus isolates. A 2% agarose gel stained with ethidium bromide is shown. Lanes: M, marker, 100-bp ladder; 1, 07-00812, t127/ST1/seh; 2, 07-00821, t127/ST1/seh lukPV; 3, 05-01197-2, t008/ST8/arcA lukPV; 4, 06-00468, t311/ST5/lukPV; 5, 04-02349, t131/ST80/etd lukPV; 6, 05-02914, t044/ST80/etd lukPV; A, control isolate 06-01172, t008/ST8/arcA lukPV; B, control isolate 06-00300, t044/ST80/etd lukPV; C, control isolate 05-01290, t127/ST1/seh lukPV.
TABLE 2.
Characteristics of isolates in this study
| BURP group (c6)a | BURP group (c4)b | Spa type | MLST (no. of isolates) | MLST CCc | MP profile (no. of isolates) | Methicillin resistance phenotype (no. of isolates) |
|---|---|---|---|---|---|---|
| A (t044) | A1 (t044) | t042 | ST080 (1d) | 80 | etd lukPV | MRSA |
| t044 | ST080 (12; 8d) | 80 | etd lukPV | MRSA | ||
| t131 | ST080 (4; 2d) | 80 | etd lukPV | MRSA | ||
| t376 | ST080 (2d) | 80 | etd lukPV | MRSA | ||
| t434 | ST080 (1d) | 80 | etd lukPV | MRSA | ||
| t455 | ST080 (1d) | 80 | etd lukPV | MRSA | ||
| t1198 | ST080 (1d) | 80 | etd lukPV | MSSA | ||
| t267 | ST097 (2) | 97 | —e | MSSA | ||
| t359 | ST097 (2d) | 97 | — | MRSA | ||
| t521 | ST097 (1d) | 97 | — | MSSA | ||
| t657f | ST772 (1d) | 1 | lukPV | MSSA | ||
| A2 | t209 | ST109 (1d) | — | MSSA | ||
| A3 | t189 | ST188 (2d) | — | MSSA | ||
| A4 | t175 | ST001 (2d) | 1 | seh lukPV | MRSA | |
| A5 (t127) | t114 | ST001 (1d) | 1 | seh | MSSA | |
| t127 | ST001 (11, 5d) | 1 | seh (10), seh lukPV (1) | MRSA (8), MSSA (3) | ||
| t127 | ST081 (1d) | 1 | seh | MSSA | ||
| t127 | ST748 (1d) | 1 | seh | MSSA | ||
| t127 | ST852 (1d) | 1 | seh lukPV | MSSA | ||
| t128 | ST001 (1d) | 1 | seh lukPV | MRSA | ||
| t321 | ST001 (1d) | 1 | seh | MSSA | ||
| t591 | ST001 (1d) | 1 | seh | MRSA | ||
| t1383 | ST001 (1d) | 1 | seh | MRSA | ||
| t1388 | ST081 (1d) | 1 | seh | MSSA | ||
| A6 (t273) | t273 | ST001 (1d) | 1 | seh | MSSA | |
| t343 | ST001 (1d) | 1 | seh | MSSA | ||
| t1491 | ST001 (1d) | 1 | seh | MSSA | ||
| t1775 | ST001 (1d) | 1 | seh | MSSA | ||
| t1931 | ST001 (1d) | 1 | seh | MSSA | ||
| A7 (t084/t346) | t084 | ST015 (1d) | 15 | — | MSSA | |
| t085 | ST015 (1d) | 15 | — | MSSA | ||
| t094 | ST015 (1d) | 15 | — | MSSA | ||
| t346 | ST015 (1d) | 15 | — | MSSA | ||
| t499 | ST015 (1d) | 15 | — | MSSA | ||
| t091 | ST007 (1d) | 7 | — | MSSA | ||
| B (t305) | B1 (t024) | t008 | ST008 (31, 6d) | 8 | — (5), arcA lukPV (16), arcA (7), lukPV (3) | MRSA (28), MSSA (3) |
| t024 | ST008 (5) | 8 | — | MRSA (3), MSSA (2) | ||
| t305 | ST617 (7, 2d) | 45 | — | MRSA | ||
| B2 | t1105 | ST617 (1d) | 45 | — | MRSA | |
| C (t002) | C | t002 | ST005 (2d) | 5 | lukPV (1) | MRSA |
| t311 | ST005 (1d) | 5 | lukPV | MRSA | ||
| D | D | t435 | ST121 (4) | 121 | lukPV | MSSA |
| t645 | ST121 (1d) | 121 | — | MSSA | ||
| E | E | t012 | ST030 (1d) | 30 | etd | MRSA |
| t318 | ST030 (1) | 30 | lukPV | MRSA | ||
| F | F | t310 | ST022 (3) | 22 | lukPV | MRSA |
| G | G | t355 | ST152 (2d) | lukPV | MRSA | |
| H | H | t437 | ST059 (1d) | 59 | lukPV | MRSA |
| A (t044)I | I | t1028 | ST154 (1d) | — | MRSA |
Results of BURP grouping with calculated cost between members of a group less than or equal to 6. Types in parentheses indicate the putative ancestor of the group as defined by BURP analysis.
Results of BURP grouping with calculated cost between members of a group less than or equal to 4. Types in parentheses indicate the putative ancestor of the group as defined by BURP analysis.
CCs as defined by eBURST analysis with a stringent group definition with six of seven loci.
Number of isolates with confirmed MLST types; all other MLST data were inferred on the basis of the spa-MLST mappings done previously at our institute.
—, negative for the relevant determinants.
Boldface indicates a false classification of the isolates on the basis of the MP profile.
Our caMRSA-MP approach facilitated the unambiguous assignment of isolates to caMRSA/caMSSA clones CC1, ST8, and ST80 in the majority of cases. All ST80 isolates examined in this study (n = 22; seven different spa types) were positive for etd and carried lukPV; all but one of the isolates were MRSA. In contrast, isolates of CC1 were more heterogeneous (n = 28; 14 different spa types, 4 different MLST types, 15 MSSA and 13 MRSA isolates) and only 5 isolates carried lukPV; However, all isolates except one (ST772, single-locus variant ST1) were positive for seh. Two more distantly related isolates with spa type t189 (ST188, double-locus variant ST1) were also negative for seh. All other isolates clustered into BURP group A but did not belong to CC1 or ST80 (ST97, ST15, ST7, ST109, ST188) and so were negative for the determinants etd and seh, thus indicating their affiliation to alternative clonal lineages not associated with caMRSA.
The caMRSA-MP profiles of isolates of type t008/ST8 (n = 31; 3 MSSA and 28 MRSA isolates) were found to be the most heterogeneous. While the majority of isolates tested (n = 16) contained arcA as well as lukPV, indicating their affiliation with the USA300 caMRSA clone, we also found isolates that carried only arcA (n = 7) or lukPV (n = 3). Five isolates of spa type t008, as well as all spa type t024 isolates, lacked any determinant for which analyses were carried out. Interestingly, six of seven type t008 isolates that carried arcA only were isolated within the same hospital.
Except for one isolate (t012, ST30, etd positive), all isolates not belonging to the caMRSA clones of interest were negative for the respective determinants (etd, seh, arcA). In contrast, lukPV was detected in isolates of all clonal lineages examined except ST45 and ST154 (Table 2). Thus, on the basis of caMRSA-MP alone, one isolate of ST30 would have been classified as a lukPV-negative ST80 isolate; on the other hand, one isolate of CC1 (ST772) would have been classified as a lukPV-positive caMRSA isolate not belonging to the predominant caMRSA lineages, leading to two falsely classified isolates among a total of 127 (1.6%) isolates tested.
DISCUSSION
Monitoring the epidemiology of caMRSA clones is crucial to prevent their spread within the community as well as their introduction into hospitals. In particular, the emergence of “new” clones and the acquisition of additional resistance determinants by already circulating clones pose an ongoing challenge for infection control authorities (10, 36).
Recent studies (11, 32a) demonstrated that spa typing and BURP analysis, which are, in general, accepted helpful tools in studies of the short-term as well as the long-term epidemiology of S. aureus, are not able to discriminate unambiguously between particular clones of caMRSA and common MSSA clones. In addition, the emergence and spread of “new” caMRSA clones in Central Europe (especially the spread of caMRSA t008/ST8/USA300) cannot be monitored efficiently, because this caMRSA lineage exhibits ambiguous spa type t008, which also occurs in other clones, especially in ST8-haMRSA-SCCmec type IV (epidemic MRSA clones 2 and 6) and ST8-haMRSA-SCCmec type II (“Irish-1”), as well as MSSA strains.
Therefore, the aim of this study was the development and validation of a rapid and easy PCR tool for the detection of the caMRSA clones currently most prevalent in Germany and Central Europe. The genetic determinants most likely specific for the three clones were selected on the basis of previous studies, with a focus on the distribution of particular virulence determinants within the S. aureus population (seh, etd, and arcA). In addition, we selected lukPV as a determinant, as it is often epidemiologically associated with caMRSA. LukPV occurs in both MRSA and MSSA strains (24), but its role in the virulence of S. aureus is currently controversial (1, 8, 38). Although lukPV is not generally associated with caMRSA isolates (25, 28), it seems to be widespread among European and American caMRSA populations (36). In the present study we detected lukPV in all ST80 isolates, while the determinant was variably present in community-acquired S. aureus isolates of ST1 and ST8. We also found lukPV in caMRSA isolates of ST5, ST30, ST22, ST152, and ST59 but not in isolates of CC45 (ST617) and ST154, which is in agreement with the findings of other studies published previously (36). Thus, we consider lukPV to be a useful marker for the detection of caMRSA in this multiplex approach, in particular, because of its putative role in invasive infections like necrotizing pneumonia (1, 16); however, it cannot replace the careful interpretation of epidemiological records for the classification of isolates as community or hospital acquired.
caMRSA isolates of ST8 (USA300) were previously shown to be positive for arcA and lukPV in the majority of cases (9); however, we found a high degree of variability of multiplex profiles within spa type t008, once again highlighting the diversity of clones exhibiting this spa type. Seven isolates carried arcA but not lukPV. Interestingly, six of them were collected within a single hospital, indicating the local spread of a new caMRSA clone descending from USA300 by the loss of the lukPV determinant.
The characterization of community-acquired S. aureus isolates of CC1 also revealed a high degree of heterogeneity with a high number of MSSA isolates and only a few lukPV-positive isolates, which is in agreement with the findings of previous studies (2, 24). However, the large number of lukPV-negative MSSA isolates might represent a putative community reservoir, in which isolates are waiting for the acquisition of SCCmec, lukPV, and other virulence or resistance determinants to become caMRSA clones in the future.
The determinants included in our assay are located on mobile genetic elements and thus are subject to putative horizontal transfer (19). Although Holtfreter et al. (13) demonstrated a strong association of mobile genetic elements with a clonal background, they found remarkable variations in gene profiles, indicating horizontal gene transfer within clonal lineages as well as between isolates of different lineages, finally leading to the occurrence of particular markers within unrelated lineages. This was demonstrated for etd, which was found in MSSA CC25 lineages in that study as well as for seh, which was found in MSSA isolates of ST1 and a second genetic background (ST34, spa type t089). Other studies demonstrated the rare detection of arcA in genetic backgrounds different from t008/ST8/USA300 (7, 9). In this study, we unexpectedly found one isolate of ST30 carrying etd. However, since these “different” genetic backgrounds are characterized by clearly distinct spa types (13) they can be distinguished unambiguously in most instances. This is also the case for caMRSA isolates of clonal lineages different from ST80, CC1, and ST8. Thus, we advise the use of a combination of spa typing and caMRSA-MP to detect suspicious isolates rapidly.
In conclusion, we present the development of an easy MP assay for the rapid detection of the most common caMRSA clones in Central Europe. Our assay facilitated the unambiguous assignment of caMRSA/caMSSA isolates to the currently most prevalent clones in the majority of cases. In combination with spa typing-BURP analysis and resistance testing, this assay facilitates the rapid detection of isolates suspected of being community-acquired S. aureus and MRSA isolates. Monitoring of the emergence and spread of caMRSA clones might assist with the prevention of the further introduction of virulent caMRSA strains into hospitals.
Acknowledgments
We are grateful to all the laboratories that delivered isolates to the National Reference Centre for Staphylococci. We highly appreciate strain donations from Hendrik Westh, Hviodore Hospital, Copenhagen, Denmark; Alexander W. Friedrich, University of Muenster, Muenster, Germany; Stephan Harbarth, Hopital Cantonal Universitaire, Geneva, Switzerland; Werner Ruppitsch, AGES, Vienna, Austria, Karina Krziwanek, Krankenhaus der Elisabethinen, Linz, Austria; and Michal A. Borg, St. Luke's Hospital, G'Mangia, Malta. We highly appreciate the excellent technical assistance of our sequencing unit at the Robert Koch Institute in Berlin, Germany.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Boyle-Vavra, S., and R. S. Daum. 2006. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab. Investig. 873-9. [DOI] [PubMed] [Google Scholar]
- 2.Coombs, G. W., J. C. Pearson, F. G. O'Brien, R. J. Murray, W. B. Grubb, and K. J. Christiansen. 2006. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg. Infect. Dis. 12241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denis, O., A. Deplano, H. De Beenhouwer, M. Hallin, G. Huysmans, M. G. Garrino, Y. Glupczynski, X. Malaviolle, A. Vergison, and M. J. Struelens. 2005. Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leucocidin genes in Belgium. J. Antimicrob. Chemother. 561103-1106. [DOI] [PubMed] [Google Scholar]
- 4.Deurenberg, R. H., C. Vink, S. Kalenic, A. W. Friedrich, C. A. Bruggeman, and E. E. Stobberingh. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13222-235. [DOI] [PubMed] [Google Scholar]
- 5.Deutsches Institut für Normung. 2004. DIN 58940. Medical microbiology—susceptibility testing of pathogens to antimicrobial agents. Part 8. Microdilution. General method specific requirements, p. 342-353. In Deutsches Institut für Normung eV (ed.), DIN-Taschenbuch 222: medizinische Mikrobiologie und Immunologie—diagnostische Verfahren. Beuth-Verlag, Berlin, Germany.
- 6.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367731-739. [DOI] [PubMed] [Google Scholar]
- 7.Ellington, M. J., L. Yearwood, M. Ganner, C. East, and A. M. Kearns. 2007. Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J. Antimicrob. Chemother. doi: 10.1093/jac/dkm422. [DOI] [PubMed]
- 8.Ellington, M. J., R. Hope, M. Ganner, M. Ganner, C. East, G. Brick, and A. M. Kearns. 2007. Is Panton-Valentine leucocidin associated with the pathogenesis of Staphylococcus aureus bacteraemia in the UK? J. Antimicrob. Chemother. 60402-405. [DOI] [PubMed] [Google Scholar]
- 9.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 451981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graber, C. J., M. K. Wong, H. A. Carleton, F. Perdreau-Remington, B. L. Haller, and H. F. Chambers. 2007. Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerg. Infect. Dis. 13491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallin, M., A. Deplano, O. Denis, R. De Mendonca, R. de Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holden, M. T. G., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. J. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtfreter, S., D. Grumann, M. Schmudde, H. T. T. Nguyen, P. Eichler, B. Strommenger, K. Kopron, J. Kolata, S. Giedrys-Kalemba, I. Steinmetz, W. Witte, and B. M. Broker. 2007. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 452669-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klevens, R. M., M. A. Morrison, S. K. Fridkin, A. Reingold, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, G. Fosheim, L. K. McDougal, and F. C. Tenover. 2006. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg. Infect. Dis. 121991-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluytmans-Vandenbergh, M. F., and J. A. Kluytmans. 2006. Community-acquired methicillin-resistant Staphylococcus aureus: current perspectives. Clin. Microbiol. Infect. 12(Suppl. 1)9-15. [DOI] [PubMed] [Google Scholar]
- 16.Labandeira-Rey, M., F. Couzon, S. Boisset, E. L. Brown, M. Bes, Y. Benito, E. M. Barbu, V. Vazquez, M. Hook, J. Etienne, F. Vandenesch, and M. G. Bowden. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 3151130-1133. [DOI] [PubMed] [Google Scholar]
- 17.Larsen, A., M. Stegger, R. Goering, M. Sorum, and R. Skov. 2007. Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000-2005). Euro Surveill. http://www.eurosurveillance.org/em/v12n02/1202-222.asp.
- 18.Linde, H., F. Wagenlehner, B. Strommenger, I. Drubel, J. Tanzer, U. Reischl, U. Raab, C. Holler, K. G. Naber, W. Witte, F. Hanses, B. Salzberger, and N. Lehn. 2005. Healthcare-associated outbreaks and community-acquired infections due to MRSA carrying the Panton-Valentine leucocidin gene in southeastern Germany. Eur. J. Clin. Microbiol. Infect. Dis. 24419-422. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 21.Mellmann, A., T. Weniger, C. Berssenbruegge, M. Sammeth, J. Stoye, A. W. Friedrich, H. Grundmann, and D. Harmsen. 2007. Determination of the clonal relatedness of the natural population of Staphylococcus aureus using a calibrated BURP (based upon repeat patterns) algorithm based on spa typing data, abstr. C-322. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 22.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 373411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monecke, S., P. Slickers, H. Hotzel, G. Richter-Huhn, M. Pohle, S. Weber, W. Witte, and R. Ehricht. 2006. Microarray-based characterisation of a Panton-Valentine leukocidin-positive community-acquired strain of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12718-728. [DOI] [PubMed] [Google Scholar]
- 24.Mongkolrattanothai, K., S. Boyle, M. D. Kahana, and R. S. Daum. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 371050-1058. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien, F. G., T. T. Lim, F. N. Chong, G. W. Coombs, M. C. Enright, D. A. Robinson, A. Monk, B. Said-Salim, B. N. Kreiswirth, and W. B. Grubb. 2004. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. J. Clin. Microbiol. 423185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prevost, G., B. Cribier, P. Couppie, P. Petiau, G. Supersac, V. Finck-Barbancon, H. Monteil, and Y. Piemont. 1995. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect. Immun. 634121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 1861060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossney, A. S., A. C. Shore, P. M. Morgan, M. M. Fitzgibbon, B. O'Connell, and D. C. Coleman. 2007. The emergence and importation of diverse genotypes of MRSA harboring the Panton-Valentine leukocidin gene pvl reveals that pvl is a poor marker for community-acquired MRSA in Ireland. J. Clin. Microbiol. 452554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 30.Said-Salim, B., B. Mathema, and B. N. Kreiswirth. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect. Control Hosp. Epidemiol. 24451-455. [DOI] [PubMed] [Google Scholar]
- 31.Salgado, C. D., B. M. Farr, and D. P. Calfee. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin. Infect. Dis. 36131-139. [DOI] [PubMed] [Google Scholar]
- 32.Stam-Bolink, E. M., D. Mithoe, W. H. Baas, J. P. Arends, and A. V. Moller. 2007. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur. J. Clin. Microbiol. Infect. Dis. 26723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Strommenger, B., C. Braulke, D. Heuck, C. Schmidt, B. Pasemann, U. Nübel, and W. Witte. 2008. Spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 46574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 442533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiemersma, E. W., S. L. Bronzwaer, O. Lyytikainen, J. E. Degener, P. Schrijnemakers, N. Bruinsma, J. Monen, W. Witte, and H. Grundman. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg. Infect. Dis. 101627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tietz, A., R. Frei, and A. F. Widmer. 2005. Transatlantic spread of the USA300 clone of MRSA. N. Engl. J. Med. 353532-533. [DOI] [PubMed] [Google Scholar]
- 36.Tristan, A., M. Bes, H. Meugnier, G. Lina, B. Bozdogan, P. Courvalin, M. E. Reverdy, M. C. Enright, F. Vandenesch, and J. Etienne. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voyich, J. M., M. Otto, B. Mathema, K. R. Braughton, A. R. Whitney, D. Welty, R. D. Long, D. W. Dorward, D. J. Gardner, G. Lina, B. N. Kreiswirth, and F. R. DeLeo. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 1941761-1770. [DOI] [PubMed] [Google Scholar]
- 39.Wenzel, R. P., G. Bearman, and M. B. Edmond. 2007. Community-acquired methicillin-resistant Staphylococcus aureus (MRSA): new issues for infection control. Int. J. Antimicrob. Agents 30210-212. [DOI] [PubMed] [Google Scholar]
- 40.Witte, W., C. Braulke, C. Cuny, B. Strommenger, G. Werner, D. Heuck, U. Jappe, C. Wendt, H. J. Linde, and D. Harmsen. 2005. Emergence of methicillin-resistant Staphylococcus aureus with Panton-Valentine leukocidin genes in central Europe. Eur. J. Clin. Microbiol. Infect. Dis. 241-5. [DOI] [PubMed] [Google Scholar]
- 41.Witte, W., C. Cuny, B. Strommenger, C. Braulke, and D. Heuck. 2004. Emergence of a new community acquired MRSA strain in Germany. Euro. Surveill. 91-2. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi, T., K. Nishifuji, M. Sasaki, Y. Fudaba, M. Aepfelbacher, T. Takata, M. Ohara, H. Komatsuzawa, M. Amagai, and M. Sugai. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 705835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki, O., A. Tristan, T. Yamaguchi, M. Sugai, G. Lina, M. Bes, F. Vandenesch, and J. Etienne. 2006. Distribution of the exfoliative toxin D gene in clinical Staphylococcus aureus isolates in France. Clin. Microbiol. Infect. 12585-588. [DOI] [PubMed] [Google Scholar]