Abstract
Based on the morphological, physiologic, and molecular (β-tubulin gene) study of 141 isolates of the Pseudallescheria boydii species complex (including several synonyms) and relatives, the new species Scedosporium dehoogii is proposed. Scedosporium apiospermum and P. boydii are considered two different species and the new name Scedosporium boydii is proposed for the anamorph of the latter species. A summary of the key morphological and physiological features for distinguishing the species of Pseudallescheria/Scedosporium is provided.
In recent years molecular phylogenetic analyses based on DNA sequences have promoted a great change in the taxonomy of clinical fungi (4). It is now accepted that numerous common pathogenic species, traditionally considered homogeneous, are indeed polyphyletic (1, 6, 9-12). Pseudallescheria boydii, one of the most common clinical molds, after Aspergillus fumigatus, is another example. Based on a multilocus study, we recently delineated eight phylogenetic species among isolates identified as this species, grouped in five different clades (2). Clades 1 and 2 were described as new species; since only the anamorphic state was observed in isolates of the first clade, it was assigned to Scedosporium (the anamorph genus of Pseudallescheria) as S. aurantiacum, and clade 2 was named Pseudallescheria minutispora because the teleomorph was present. Clade 5 consisted of four subgroups incorporating the type strains of P. boydii, Pseudallescheria angusta, Pseudallescheria ellipsoidea, and Pseudallescheria fusoidea. Clades 3 and 4 remained unnamed. We have phenotypically characterized here these eight phylogenetic species. In order to increase the robustness of the different clades, we have included numerous fresh isolates in the present study identified by sequencing the TUB region of the β-tubulin gene, the most informative molecular marker of the four evaluated previously (2).
A total of 141 isolates was studied, including the available reference or type strains of synonymous species of P. boydii and Scedosporium apiospermum (see the supplemental material). The procedures for DNA extraction, amplification, sequencing, and phylogeny were described previously (2). Morphology was assessed by features observed on potato dextrose agar (PDA) and on oatmeal agar after incubation at 25°C for 2 months.
Fifty-nine physiological tests were performed in duplicate. Inocula, adjusted to 105 conidia/ml by hemacytometer counts, were prepared from 7-day-old PDA plates. Growth, including growth on cycloheximide (0.05 to 0.1%), and assimilation abilities were tested in liquid medium according to the method of Yarrow (15). All of the tests, with the exception of urease, gelatin liquefaction, acid production, and starch formation, were performed in sterile, disposable, multiwell microplates. The medium was dispensed into the wells in 150-μl volumes with a multichannel pipette, and each well was inoculated with 50 μl of the conidial suspension. The microplates were incubated at 25°C in darkness for 14 days. For thermotolerance studies, the isolates were subcultured onto PDA and incubated in darkness for 14 days.
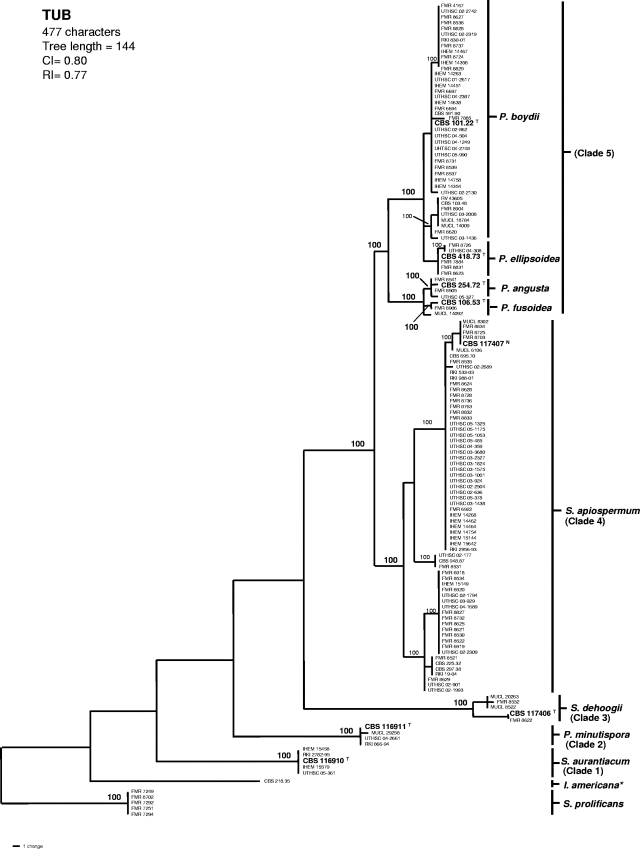
A heuristic search of the partial sequence of the TUB region produced 18 “most-parsimonious trees”; one of them is shown in Fig. 1. The topology of the tree was similar to that obtained in our previous study (2). Most of the new isolates were included in clade 4, which makes it the most common phylogenetic species of the complex. The polymorphic nucleotides for all of the species of the P. boydii complex are shown in Table 1.
FIG. 1.
One of the 18 most-parsimonious trees obtained from heuristic searches based on TUB sequences. Bootstrap support values of 100% are indicated at the nodes. Type strains are indicated with boldface type and with a superscript “T”. Neotype is indicated by boldface type and with a superscript “N”. Strains of S. prolificans were used as outgroups. CI, consistency index; RI, retention index; I, Indiella; P., Pseudallescheria; S., Scedosporium. *, The reference strain of Indiella americana.
TABLE 1.
Polymorphic sites in the TUB locus (exons 5 and 6) identified by using DnaSP v.4.10.3
| Species | Nucleotide at polymorphic site at indicated positiona
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47 | 74 | 47 | 74 | 113 | 116 | 119 | 125 | 128 | 131 | 146 | 155 | 182 | 209 | 230 | 250 | 254 | 257 | 258 | 261 | 262 | 263 | 268 | 273 | 275 | 282 | 283 | 284 | 285 | 286 | 290 | 291 | 296 | 297 | 298 | 300 | 302 | 303 | 305 | 306 | 307 | 320 | 321 | 322 | 329 | 332 | 333 | 335 | 336 | 337 | 358 | 359 | 360 | 361 | 363 | 364 | 368 | 369 | 370 | 379 | 380 | 382 | 383 | 384 | 385 | 389 | 392 | 400 | 415 | 439 | 496 | 499 | 508 | 511 | 532 | 535 |
| P. boydii | C | T | G | T | T | T | T | T | T | G | T | C | C | A | T | A | T | C | T | A | — | — | — | — | — | — | — | C | A | A | C | T | A | G | T | C | C | T | G | C | G | A | C | T | C | C | C | — | G | T | G | A | G | A | C | T | C | A | G | A | G | T | T | C | T | C | T | G | C | T | T | T | C | T | |
| S. apiospermum | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | C/· | · | — | — | — | — | — | — | — | · | · | G | · | · | · | · | · | · | · | · | · | A/G | · | · | · | · | · | T | — | — | · | C | · | G | · | · | · | · | · | · | A | G | · | G | G | T | · | · | · | · | · | · | A | · | · | · | |
| S. aurantiacum | G | · | · | C | · | · | · | · | · | · | · | · | T | G | · | T | C | — | — | — | G | C | A | C | C | C | C | · | C | C/— | · | C | C | A | A | · | A | C | A | · | · | G | · | · | G | T | G | T | — | — | A | G | · | G | — | C | · | — | · | G | · | C | · | T | C | T | · | · | T | C | C | C | · | C | |
| S. dehoogii | · | C | T | · | G | · | C | · | · | · | C | · | · | · | · | T | · | T/· | · | T/— | — | — | — | — | T | T | T | · | T | · | · | · | · | A | A | · | · | G | A | T | A | G | G | C | G | T | — | — | A | · | · | · | A | C | — | · | T | G | T | G/C | T | · | · | — | · | T | · | · | · | · | C | · | · | · | |
| P. angusta | · | · | · | · | · | C | · | · | C | · | · | · | · | · | · | · | · | T | · | · | — | — | — | — | — | — | — | A | · | G | T | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | T | — | · | · | · | · | · | · | T | · | · | · | · | G | · | G | · | T | · | · | · | · | · | · | · | · | · | · | |
| P. ellipsoidea | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | — | — | — | — | — | — | — | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | T | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| P. fusoidea | G | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | T | · | · | — | — | — | — | — | — | — | A | · | G | T | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | T | — | · | · | · | · | · | · | T | · | · | · | · | G | · | G | · | T | · | · | · | · | · | · | · | · | · | · | |
| P. minutispora | G | · | · | · | · | · | · | C | · | T | · | · | · | · | G | T | · | — | — | — | G | G | T | T | T | T | T | · | — | — | — | — | — | — | — | — | — | — | — | — | — | — | G | — | — | — | — | — | — | — | · | · | A | G | — | · | · | C | · | G | T | G | · | · | · | T | C | A | · | C | C | C | T | · | |
Sites with alignment gaps were included if there was a polymorphism. The sequence of type strain of P. boydii CBS 101.22 (GenBank accession no. AJ890121) was used as the master sequence. Nucleotides identical to the corresponding nucleotides in the P. boydii type strain sequence are shown as dots. A dash (—) denotes a gap. The data were adapted from a study by Rozas et al. (12a).
A relatively large number of carbon and nitrogen sources were assimilated by all of the species studied (see the supplemental material). The assimilation patterns for P. boydii, P. angusta, P. ellipsoidea, and P. fusoidea were similar, with no significant differences. Assimilation of sucrose, maltose, d-ribose, l-arabinitol, and ribitol and growth at 40 and 45°C were the most useful characteristics for discriminating the species included in the study (Table 2).
TABLE 2.
Morphological and physiological key characters for differentiating among species of the Pseudallescheria/Scedosporium complex and S. prolificansa
| Species | Conidiogenous cells | Sessile conidia | Colony reverse in orange shadesb | Yellow diffusible pigmentb | Assimilation of:
|
Growth at:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ribitol | l-Arabinitol | Sucrose | Maltose | d-Ribose | 40°C | 45°C | |||||
| P. boydii and relativesc (clade 5) | Cylindrical | Globose to subglobose, thick-walled | − | V | + | + | + | + | + | + | − |
| P. minutispora (clade 2) | Cylindrical | Ellipsoidal to obovoid, thin-walled | − | − | + | + | + | − | − | + | − |
| S. apiospermum (clade 4) | Cylindrical | Globose to subglobose, thick-walled | − | V | + | + | + | + | − | + | − |
| S. aurantiacum (clade 1) | Cylindrical or slightly flask-shaped | Mostly obovoid, thick-walled | + | + | + | + | − | + | + | + | + |
| S. dehoogii (clade 3) | Cylindrical or slightly flask-shaped | Mostly obovoid, thin-walled | − | − | + | + | + | + | − | − | − |
| S. prolificans | Flask-shaped | Globose to subglobose, thick-walled | − | − | − | − | − | + | − | + | V |
Abbreviations and symbols: −, all strains of the species displayed a negative response; +, all strains of the species displayed a positive response; V, variable (i.e., some strains were positive and others were negative).
That is, on PDA at 25°C.
That is, P. angusta, P. ellipsoidea, and P. fusoidea. These species can be identified by other molecular and morphological characteristics.
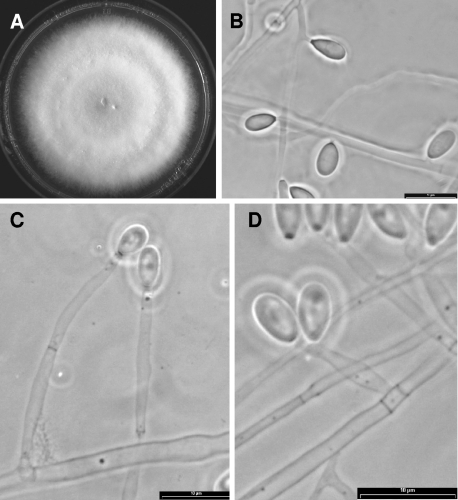
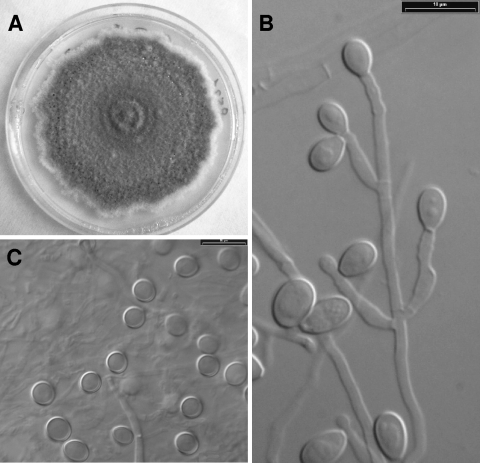
The morphology of the conidiophores and the sessile conidia (conidia borne individually along the sides of the vegetative hyphae), and the appearance of the colonies on PDA were the most useful phenotypic features for separating the different clades (Table 2). None of the isolates of clades 3 and 4 developed the teleomorph (sexual state) after 2 months, while all of them developed the Scedosporium anamorph. Synnematous conidiophores (Graphium anamorph) were usually absent in isolates from clade 3, whereas they were present in more than 90% of the isolates in clade 4. Clade 3 was characterized by solitary and usually unbranched conidiophores, subhyaline to pale gray and thin-walled, sessile conidia, and pale gray colonies (Fig. 2), whereas members of clade 4 showed branched conidiophores, brownish and thick-walled sessile conidia, and brownish colonies (Fig. 3).
FIG. 2.
Sedosporium dehoogii (CBS 117406). (A) Colony growing on PDA after 14 days at 25°C. (B) Sessile conidia. (C and D) Conidiogenous cells and conidia.
FIG. 3.
Scedosporium apiospermum (CBS 117407). (A) Colony growing on PDA after 14 days at 25°C. (B) Conidiogenous cells and conidia. (C) Sessile conidia.
Given the genotypic and phenotypic uniqueness of clade 3, we therefore propose the new species Scedosporium dehoogii.
Scedosporium dehoogii Gilgado, Cano, Gené et Guarro, sp. nov. = Clade 3 sensu Gilgado et al. (2).
Coloniae dilute griseae coloratae. Conidiophora solitaria plerumque non-ramosa. Conidia sessilis subhyalina vel dilute grisea, tenuitunicata, plerumque obovata, 5 vel 8 per 5 vel 6 μm. Teleomorphosis ignota. Assimilantur ribitolum, l-arabinitolum, sucrosum et maltosum. Non assimilantur d-ribosum. Augmentum fit in temperatura 37°C.
The colonies on PDA attained a diameter of 45 to 60 mm at 25°C after 14 days. They were cottony and white to pale gray with a colorless reverse. Solitary conidiophores were usually reduced to conidiogenous cells, which were subhyaline, smooth-walled, usually cylindrical, 6 to 50 μm long by 1 to 1.5 μm wide, and produced pale brown, obovoid or ellipsoidal conidia measuring 6 to 11 μm long by 4 to 5 μm wide. Synnematous conidiophores were erect, 80 to 450 μm long, and terminated in a slimy head of conidia. Those conidia were cylindrical or claviform, 6 to 11 μm long by 3 to 4 μm wide, with a wide truncate base. Sessile conidia were subhyaline to pale gray, thin-walled, mostly obovate, 5 to 8 μm long by 5 to 6 μm wide. A teleomorph was not observed for any isolate after 2 months. Maximum growth temperature was at 37°C (5 to 10 mm in diameter after 14 days). The fungus was able to assimilate ribitol, l-arabinitol, sucrose and maltose, but not d-ribose.
Etymology.
Derived from the name of the mycologist G. Sybren de Hoog.
A dry culture of the strain isolated from garden soil (Barcelona, Spain) has been deposited in the International Mycological Institute-CABI Bioscience (Egham, England) as IMI 394089 (holotype). A living culture of the isolate has been deposited in the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands) as CBS 117406.
The members of clade 4 are morphologically indistinguishable from the anamorph of Pseudallescheria boydii (clade 5), but they can be separated by the response to d-ribose test and by the absence of a teleomorph (Table 2). Clade 4 also included the type strains of Acremonium suis, Sporocybe borzinii, and Polycytella hominis and the only reference strain of Scedosporium apiospermum. Therefore, the clade 4 must be identified as S. apiospermum, since this name has priority from a nomenclatural point of view over the other species included in the clade. A previous name is Monosporium apiospermum, which was proposed by Saccardo (13) in 1911 based on a mycetoma isolate, but later Monosporium was considered as a nomen illegitimum (5). One of the most important aspects of this study has been to precisely demonstrate that S. apiospermum (clade 4) and P. boydii (clade 5) are really two different species. Up to now, the former had been considered as the anamorph of the latter (7, 8).
Since the type of S. apiospermum is apparently lost, choosing a neotype for this species is required taxonomically. It may be possible to select the reference strain CBS 225.32 since, according to the CBS curators (unpublished data), this strain is presumed to be a subculture of the original strain from which Saccardo based the description of the species. However, over the years this strain has degenerated, and it sporulates very poorly in all of the media tested. We therefore preferred to choose as a neotype an isolate of the same clade which showed a better sporulation in order to illustrate the most important morphological features of S. apiospermum (Fig. 3). A dry culture of the strain isolated from human keratitis (São Paulo, Brazil) has been deposited in the CABI Bioscience (Edgham, England) database as IMI 394090 (neotype). A living culture of the isolated has been deposited in the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands) database as CBS 117407.
A new name is then required for the anamorph of P. boydii. Because it was originally described as Cephalosporium boydii by Shear in 1922 (14) and later defined as a Scedosporium species, it can be classified as Scedosporium boydii (Shear) Gilgado, Gené, Cano et Guarro comb. nov. (Basionym: Cephalosporium boydii Shear, Mycologia 14: 242, 192).
The study of the type strains of Acladium castellanii, Sporocybe chartoikoon, and Sporotrichum councilmanii (see the supplemental material), also considered synonyms of S. apiospermum/P. boydii, revealed that they are morphologically incompatible with the anamorphs of the P. boydii complex. The internal transcribed spacer regions of these strains were sequenced and, when compared to the sequences of the GenBank database, none matched the species in the P. boydii complex. The only synonymous species that was morphologically compatible with anamorphs of Pseudallescheria was Indiella americana. However, the TUB sequence placed this species in a branch phylogenetically distant to the species of the P. boydii complex (Fig. 1). Because the taxonomy of I. americana is ill defined, an extensive study with more strains would be required for the proper delineation of this species.
Traditionally, the classification of the fungi of clinical interest has been based on morphology. However, molecular methods, based mainly on sequencing rRNA genes, have recently evolved as useful tools for this purpose. Combining both approaches is not yet a common practice but seems to be the best approach for obtaining a more natural classification. In recent years, different authors have used multigene analyses to demonstrate the existence of numerous cryptic species in important clinical fungi (6, 9, 11). In our study, we used a polyphasic approach that combines morphological, physiologic, and molecular data sets to characterize the species of the genus Pseudallescheria and its relatives.
Although it has been shown that the TUB region is a good marker for delimiting the different species of the complex, many clinical laboratories may not have the capability for molecular characterization of isolates. The fact that these species can also be identified by using simple and inexpensive phenotypic methods has made their delineation in routine laboratories possible. Now that phylogenetic species of the P. boydii complex can be recognized and their different responses to the antifungals drugs have been demonstrated (3), it will be of interest to see whether they also elicit different clinical manifestations.
Supplementary Material
Acknowledgments
We are grateful to G. Sybren de Hoog (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands) for useful comments on the taxonomy of the fungi included in the study.
This study was supported by Spanish Ministerio de Ciencia y Tecnología grants CGL 2004-00425/BOS and CGL2005-7394.
Footnotes
Published ahead of print on 12 December 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
Communication of the ECMM Working Group on Pseudallescheria and Scedosporium.
REFERENCES
- 1.Bard, C. B., J. L. Carter, S. P. Keely, L. Huang, N. J. Pieniazek, I. N. Moura, J. M. Roberts, A. W. Hightower, M. S. Bens, A. R. Freeman, S. Lee, J. R. Stringer, J. S. Duchin, C. del Rio, D. Rimland, R. P. Baughman, D. A. Levy, V. J. Dietz, P. Simon, and T. R. Navin. 2000. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg. Infect. Dis. 6265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilgado, F., J. Cano, J. Gené, and J. Guarro. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J. Clin. Microbiol. 434930-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilgado, F., C. Serena, J. Cano, J. Gené, and J. Guarro. 2006. Antifungal susceptibilities of the species of the Pseudallescheria boydii complex. Antimicrob. Agents Chemother. 504211-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarro, J., J. Gené, and A. M. Stchigel. 1999. Developments in fungal taxonomy. Clin. Microbiol. Rev. 12454-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes, S. J. 1958. Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Can. J. Bot. 36727-836. [Google Scholar]
- 6.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 945478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malloch, D. 1970. New concepts in the Microascaceae illustrated by two new species. Mycologia 62727-739. [Google Scholar]
- 8.McGinnis, M. R., A. A. Padhye, and L. Ajello. 1982. Pseudallescheria Negroni et Fischer, 1943 and its later synonym Petriellidium Malloch, 1970. Mycotaxon 1494-102. [Google Scholar]
- 9.Mostert, L., J. Z. Groenewald, R. C. Summerbell, V. Robert, D. A. Sutton, A. A. Padhye, and P. W. Crous. 2005. Species of Phaeoacremonium associated with infections in humans and environmental reservoirs in infected woody plants. J. Clin. Microbiol. 431752-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell, K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92919-938. [Google Scholar]
- 11.O'Donnell, K., D. A. Sutton, M. G. Rinaldi, K. C. Magnon, P. A. Cox, S. G. Revankar, S. Sanche, D. M. Geiser, J. H. Juba, J. A. van Burik, A. Padhye, E. J. Anaissie, A. Francesconi, T. J. Walsh, and J. S. Robinson. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 425109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 591886-1899. [PubMed] [Google Scholar]
- 12a.Rozas, J., J. C. Sanchez del Barrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 192496-2497. [DOI] [PubMed] [Google Scholar]
- 13.Saccardo, P. A. 1911. Notae mycologicae (mycological notes). Annals Mycol. 9249-257. [Google Scholar]
- 14.Shear, C. L. 1922. Life history of an underscribed ascomycete isolated from a granular mycetoma of man. Mycologia 14239-243. [Google Scholar]
- 15.Yarrow, D. 1998. Methods for the isolation, maintenance, and identification of yeasts, p. 95-97. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts: a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.