Abstract
Carrion's disease is typically biphasic with acute febrile illness characterized by bacteremia and severe hemolytic anemia (Oroya fever), followed by benign, chronic cutaneous lesions (verruga peruana). The causative agent, Bartonella bacilliformis, is endemic in specific regions of Peru and Ecuador. We describe atypical infection in an expatriate patient who presented with acute splenomegaly and anemia 3 years after visiting Ecuador. Initial serology and PCR of the patient's blood and serum were negative for Bartonella henselae, Bartonella quintana, and B. bacilliformis. Histology of splenic biopsy was suggestive of bacillary angiomatosis, but immunohistochemistry ruled out B. henselae and B. quintana. Bacilli (isolate EC-01) were subsequently cultured from the patient's blood and analyzed using multilocus sequence typing, protein gel electrophoresis with Western blotting, and an immunofluorescence assay (IFA) against a panel of sera from patients with Oroya fever in Peru. The EC-01 nucleotide sequences (gltA and internal transcribed spacer) and protein band banding pattern were most similar to a subset of B. bacilliformis isolates from the region of Caraz, Ancash, in Peru, where B. bacilliformis is endemic. By IFA, the patient's serum reacted strongly to two out of the three Peruvian B. bacilliformis isolates tested, and EC-01 antigen reacted with 13/20 Oroya fever sera. Bacilliary angiomatosis-like lesions were also detected in the spleen of the patient, who was inapparently infected with B. bacilliformis and who presumably acquired infection in a region of Ecuador where B. bacilliformis was not thought to be endemic. This study suggests that the range of B. bacilliformis may be expanding from areas of endemicity in Ecuador and that infection may present as atypical clinical disease.
Human bartonellosis is caused by infection with Bartonella bacilliformis, a motile, aerobic, gram-negative, pleomorphic bacterium (34). B. bacilliformis resides within cells of the human reticuloendothelial systems and initially attaches to and penetrates erythrocytes (16). The organism is transmitted to humans through the bite of the sand fly, Lutzomyia verrucarum (1, 36, 42). Typical bartonellosis is biphasic, characterized by an acute severe febrile illness associated with bacteremia and severe hemolytic anemia (Oroya fever), followed by a cutaneous phase (verruga peruana) which involves chronic skin eruptions (7, 28, 35, 43). The acute phase typically develops soon after infection and, if untreated, may have a 40 to 88% fatality rate in association with secondary infections (salmonellosis, shigellosis, malaria, toxoplasmosis, histoplasmosis, or pneumocystis) (13, 20, 31). Verruga may develop after a period of weeks to months and may persist for months to years.
Historically, Carrion's disease has been endemic in Andean Mountain regions of Peru, Ecuador, and Colombia at elevations of 600 to 3,200 m above sea level (1, 9, 31, 38). Regions of endemicity in Peru have traditionally been localized to river valleys and canyons in the western Andes and inter-Andean valleys in the Central and East Andes. These mountainous areas of endemicity include Ancash, Lima, Cajamarca, Piura, La Libertad, Huancavelica, Huánuco, Ayacucho, Junín, and Ina (reviewed in reference 31). However, during the last 2 decades, emergent disease outbreaks have occurred at lower elevations between the highlands and jungle (Amazonas, Cajamarca, and Huánuco), high forest regions (Chanchamayo and Junín), and in valley regions east of the Andes such as Cuzco (19, 24, 26, 29, 31). In Peru, the highest incidence of bartonellosis has occurred in Ancash, followed by Cajamarca, Amazonas, the Lima highlands, and Cusco (reviewed in 24). Epidemiological studies also suggest that the spectrum of clinical manifestations associated with B. bacilliformis in Peruvian patients is highly variable, ranging from occurrence of either one or both phases to asymptomatic infections characterized by chronic bacteremia (9, 10, 19, 21, 24). In areas of endemicity in Peru, the cutaneous phase is the most common clinical presentation and mainly affects children, while in regions where the disease is both epidemic and endemic, a majority of acute-phase infections are also found in children (reviewed in reference 24).
In Ecuador, typically severe febrile hemolytic diseases have been reported for years from the highland province of Zamora-Chinchipe bordering Peru (11, 12). In contrast, growing numbers of atypical illnesses associated with only chronic verrucous skin lesions have been reported from the coastal lowland provinces of Manabí and Guayas (2). It is speculated that in these areas, the incidence of bartonellosis is highly underreported due to its mild clinical presentation and may be associated with circulation of less virulent isolates of B. bacilliformis (26). Genetic diversity among B. bacilliformis strains has also been suggested as a factor responsible for differences in the clinical progression of human bartonellosis reported from areas of endemicity and newly recognized foci in Peru (21, 26). We report here the isolation of B. bacilliformis from a persistently infected patient who had traveled from the United States to Ecuador 3 years previously and describe preliminary characteristics of this new isolate compared with B. bacilliformis isolates from Peru.
MATERIALS AND METHODS
Case description.
A 34-year-old immunocompetent individual presented to an internist with acute right-sided abdominal pain. Physical examination revealed an enlarged spleen and chronic keloid-like skin lesions on the chest. The patient reported last visiting Ecuador 3 years previously. The patient owned a dog and a recently deceased bird but denied receiving any bites or scratches from pets. Laboratory tests determined the following values: leukocytes, 61,000/μl; neutrophils, 6.5%; lymphocytes, 24.5%; platelets, 194,000/μl; hematocrit, 12.1%; and hemoglobin, 34.9%. Hemolytic anemia was well compensated. Computer-assisted tomography confirmed splenomegaly. The patient was referred to an oncologist who ordered a needle biopsy of the spleen to rule out a diagnosis of lymphoma. Following the splenic biopsy, the patient received oral ciprofloxacin for 5 days. The patient was subsequently referred to an infectious disease specialist. Serological testing and PCR assay of whole blood were performed by a commercial laboratory and showed no evidence of infection with Bartonella quintana or Bartonella henselae. Blood cultures were not done. Because of the patient's country of origin, travel history, and unusual skin lesions, infection with B. bacilliformis was considered. Serum, whole blood, and unstained slides of the formalin-fixed, paraffin-embedded splenic aspirate were submitted to the Centers for Disease Control and Prevention (CDC; Atlanta, GA) for laboratory evaluation. The patient's splenomegaly persisted, and 13 months later, a splenectomy was performed. The excised spleen weighed 461 g, measured 18.0 by 12.0 by 4.0 cm, and contained a solitary, firm, tan mass that measured 6.0 by 5.0 by 4.7 cm. At that time, CDC obtained formalin-fixed, paraffin-embedded tissue sections from the spleen mass for immunohistochemical (IHC) evaluation.
Immunofluorescence assay.
Indirect immunofluorescence assays (IFAs) were performed using antigens of B. bacilliformis, B. henselae, and B. quintana grown in Vero cell culture according to a previously described procedure (8). Antibody titers were determined as the reciprocal of the last dilution of the serum sample showing reactivity with a fluorescein isothiocyanate-conjugated goat anti-human immunoglobulin G (IgG; gamma-chain specific) (Kirkegaard and Perry Laboratories; Gaithersburg, MD) at a dilution of 1/150. The cutoff titer of the IFA was 1/32. Twofold serum dilutions were tested along with appropriate positive and negative control sera. In addition, a panel of antisera from patients in Peru infected with Oroya fever was also tested by IFA against antigens of four B. bacilliformis isolates prepared in Vero cells. Slides were read at a magnification of ×400 using a Zeiss Axiophot epifluorescence microscope (Carl Zeiss Inc., Thornwood, NY) and digitally imaged using Spot, version 4.0.9, software (Diagnostic Instruments Inc., Sterling Heights, MI).
Isolation procedure.
Heparinized and EDTA whole blood and serum from the patient were sent overnight to the CDC on cold packs, maintained at 4°C upon receipt, and immediately cultured for Bartonella. Aliquots of the blood samples were streaked onto 100-mm diameter petri plates with heart infusion agar supplemented with 5% rabbit blood (HIARB) and incubated in plastic bags at 28°C and 5% CO2. The remaining heparinized and EDTA whole blood were divided into equal aliquots and treated as follows. Whole blood (1 ml) was diluted 1:1 with 10 mM phosphate-buffered saline (PBS; pH 7.5), layered onto 3 ml of Hypaque-76 (Sigma, St. Louis, MO), and centrifuged at 800 × g for 10 min at room temperature. Peripheral blood mononuclear cells were harvested and washed once with PBS and inoculated into a 25-cm2 flask containing biphasic medium (BiP) consisting of 5 ml of solidified HIARB overlaid with 5 ml PBS (22). Additionally, 1 ml of whole blood was inoculated into BiP medium; flasks were tightly closed and incubated at 28°C. At 18 days following inoculation, isolate EC-01 was passaged into 75-cm2 flasks containing HIARB medium. The remaining culture supernatant was pelleted, resuspended in SRM freezing medium (0.22 M sucrose, 0.1 M potassium phosphate, 0.005 M sodium l-glutamate, pH 7.0, 0.005 M MgCl2, and 1% Hypaque-76; Nycomed, Inc., Princeton, NJ), and frozen at −70°C. EC-01 stocks used for proteomic, genomic, and serological assays were at low passage, P3 to P6.
Source of Bartonella isolates and reference strains.
Table 1 lists all Bartonella strains and isolates that we cultivated or sequenced at the CDC. Multilocus sequence typing used reference sequences from NCBI for isolates that we did not directly use, and their accession numbers are provided in the phylogenetic trees. All reference isolates were cultivated on HIARB medium at 28°C for B. bacilliformis or at 34°C for other Bartonella spp., and low-passage stocks were preserved in SRM medium and frozen at −70°C. B. bacilliformis isolates were subsequently adapted to growth in BiP in 75-cm2 flasks at 28°C, and low-passage stocks of BiP medium-grown Bartonella strains were also preserved in SRM medium and stored at −70°C.
TABLE 1.
Reference strains of Bartonella used in this study
| Isolate species and strain | Location of isolate
|
Reference and/or source | |
|---|---|---|---|
| Town | Province | ||
| B. bacilliformis | |||
| Car 600-01 | Cusco | Cusco | 8, 19 |
| Choq Col-01 | Caraz | Ancash | 8, 19 |
| Peru 13 | Unknown | Unknown | 8, 19 |
| Peru 358-98 | Caraz | Ancash | 8, 19 |
| Cule III | Culaspampa | Ancash | 8, 19 |
| VAB 9034 | Cascapara | Ancash | Carmen R. Latorrea |
| Cus 005 | Cusco | Cusco | Franca Jonesb |
| Hosp 800-02 | Caraz | Ancash | Franca Jones |
| Hosp 800-31 | Caraz | Ancash | Franca Jones |
| Hosp 800-72 | Caraz | Ancash | Franca Jones |
| Colonia | Caraz | Ancash | Franca Jones |
| Ramirez | Caraz | Ancash | Franca Jones |
| VRB 165 | Unknown | Unknown | Franca Jones |
| Vega | Caraz | Ancash | Franca Jones |
| Vero 97 | Caraz | Ancash | Franca Jones |
| Vero 75 | Unknown | Unknown | Franca Jones |
| KC 583 (ATCC 35685) | 6; ATCC | ||
| B. henselae Houston-1 (ATCC 49882) | 40; ATCC | ||
| B. quintana Fuller (ATCC VR-358) | ATCCc | ||
| B. elizabethae F9251 (ATCC 49927) | 14; ATCC | ||
Departamente de Ancash, Provincia Huarez, Peru.
U.S. Naval Medical Research Center Detachment in Lima, Peru.
Strain is no longer available from ATCC.
Transmission electron microscopy.
Bartonella was grown in BiP medium for 10 days as above and harvested from medium by centrifuging at 2,090 × g for 20 min at 4°C. Glycerol was added to 10% (vol/vol), the tube was gently inverted several times to mix, and bacteria were pelleted as above. The pellet was gently resuspended in 1.5 ml of PBS supplemented with 10% glycerol, transferred to a 1.5-ml screw-top tube, and centrifuged at 420 × g for 5 min at 4°C; the supernatant was discarded, and a washing step was repeated one more time under the same conditions. The pellet was resuspended in 0.5 ml of PBS and gently mixed with an equal volume of 2× fixative solution consisting of 0.2 M sodium cacodylate (pH 7.2) and 4% glutaraldehyde. The fixation was done overnight at room temperature with gentle rocking, after which fixative solution was removed by centrifugation and replaced with 0.2 M cacodylate buffer. Ten-microliter samples were placed on Formvar-coated carbon grids and allowed to incubate for 30 to 60 s at room temperature, and excess liquid was removed using filter paper. The grids were then negatively stained with 2% aqueous uranyl acetate (wt/vol) for 1 min, air dried, and viewed using a JEOL 1200 EX electron microscope operating at 60 kV.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Bartonella bacilli were harvested from supernatants of BiP medium by centrifugation at 23,426 × g for 10 min at 4°C using a Sorvall RC5C preparative centrifuge and SS-34 rotor (Thermo Electron Corporation, Asheville, NC). Bacterial pellets were washed twice with PBS, and the absorbance of intact bacilli at a wavelength of 420 nm was used to estimate the number of milligrams of protein in the bacterial suspension. Bacterial pellets were lysed in CelLytic B-II extraction reagent supplemented with 0.2 mg/ml lysozyme (Bio-Rad, Hercules, CA) to give a final protein concentration of 10 mg/ml. Protein concentration of the bacterial lysates was precisely determined using a bicinchoninic acid protein assay (Pierce Biotechnology, Inc., Rockford, IL). Lysates were mixed 1:1 with 2× Laemmli sample buffer (Bio-Rad Laboratories Inc., Hercules, CA) and heated to 95°C for 5 min; equal protein concentrations were loaded per well and electrophoresed on 20-cm-long, 8 to 16% gradient gels (29:1, acrylamide-bis, at 2.6% crosslinking) overnight at 10 mA/gel. After electrophoresis, protein bands were stained using Coomassie R-250 (Bio-Rad; Hercules, CA). Alternatively for Western blotting, 8 to 16% gradient mini gels were equilibrated in Towbin Transfer Buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, and 20% methanol) and blotted onto immunoblotting polyvinylidene difluoride membrane overnight at 30 V and 4°C using a Bio-Rad Mini Trans-Blot cell (Hercules, CA). The membrane blots were blocked with 5% Blotto (20 mM Tris, pH 7.5, 500 mM NaCl, 5% nonfat dry milk) for 1 h at ambient room temperature (RT). After blots were washed with TBST (20 mM Tris-HCl, 500 mM NaCl [pH 7.5], 0.1% Tween-20) three times for 10 min each time at RT, they were incubated with either polyclonal rabbit anti-B. bacilliformis (1:500) or rabbit anti-B. quintana (1:500) antiserum for 60 min at RT. These two rabbit antisera were previously made at the CDC. Membranes probed with rabbit anti-Bartonella antibodies were washed four times with TBST and then incubated with goat anti-rabbit IgG(H+L)-horseradish peroxidase (1:4,000) (Southern Biotechnology; Birmingham, AL) for 60 min at RT. Following four washes with TBST, blotted proteins were visualized using the colorimetric substrate diaminobenzidine (Bio-Rad, Hercules, CA) as per the manufacturer's directions.
Analysis of protein gel profiles.
Digitalized images of Coomassie blue-stained gels were obtained using the Gel Doc gel documentation system and PDQuest imaging software (Bio-Rad, Hercules, CA). Cluster analysis using BioNumerics image analysis software, version 3.5 (Applied Maths, Saint-Martens-Latem, Belgium) was applied to the protein banding patterns of Bartonella spp., B. bacilliformis isolates, and the new isolate, EC-01, to determine relatedness. Dice binary coefficients that measure the similarity based upon common and different protein bands were used to generate an unweighted pair group method using arithmetic mean dendrogram. A matrix similarity table with the percentage of matching bands between isolates was also generated. Cophenetic correlation analysis was applied to the dendrogram to measure the reliability of the groupings and how well these groupings correlated with the similarity matrix table.
PCR and sequence analysis.
The DNA was extracted from a 200-μl aliquot of EDTA whole blood and 10-μm paraffin-embedded formalin-fixed sections of spleen and from Bartonella grown on blood agar using a QIAamp DNA Mini Kit (Qiagene, Valencia, CA). Clinical specimens were tested using nested PCRs to amplify fragments of htrA (3) and ribC (45) of Bartonella as previously described using PuRe Taq Ready-To-Go PCR beads (Amersham Biosciences, Piscataway, NJ). PCR amplification of the isolate DNA was performed using Qiagen Master Mix reagents in a Gradient Master cycler (Eppendorf, Westbury, NY). The list of oligonucleotide primers used is shown in Table 2; the primers were made by the CDC Core Facility (CDC, Atlanta, GA) and used at a final concentration of 1 μM unless otherwise specified. Thermal cycling conditions were the same as previously reported for the individual PCR assays listed in Table 2. Sequence reactions were prepared using an ABI PRISM 3.0 BigDye Terminator Cycle Sequencing kit as recommended by the manufacturer (Applied BioSystems, Foster City, CA). Sequence reactions were purified with a Qiagen Dye Removal Kit (Qiagen, Valencia, CA) and run on an Applied Biosystems 3100 Nucleic Acid Sequence Analyzer.
TABLE 2.
Oligonucleotide primers used
| Target | Primer name | Primer sequence (5′→ 3′) | Reference or source |
|---|---|---|---|
| 16S-23S rRNA intergenic region | QHVE1 | TTCAGATGATGATCCCAA | 23 |
| QHVE4 | AACATGTCTGAATATATC | ||
| 16S rRNA gene | Rick16SF1 | GTATGCTTAACACATGCAAGTCGAAC | 39 |
| Rick16SR4 | TCCGCGATTACTAGCGATTCC | ||
| ftsZ | FtsZ F (Bfp1) | ATTAATCTGCAYCGGCCAGA | 44 |
| FtsZ R (BatfsZR) | GCTGGTATTTCCAAYTGATCT | ||
| rpoB | rpoB F (1400F) | CGCATTGGCTTACTTCGTATG | 41 |
| rpoB R (2200R) | GTAGACTGATTAGAACGCTG | ||
| gltA | BhCS.781p | GGGGACCAGCTCATGGTGG | 37 |
| BhCS.1137n | AATGCAAAAAGAACAGTAAACA | ||
| ribC | RibC-1F | CGGATATCGGTTGTGTTGAA | 45 |
| RibC-1R | CATCAATRTGACCAGAAACCA | ||
| RibC-2F | GCATCAATTGCGTGTTCA | ||
| RibC-2R | CCCATTTCATCACCCAAT | ||
| htrA | CAT-1 | GATTCAATTGGTTTGAA(A/G)GAGGCT | 3 |
| CAT-2 | TCACATCACCAG G(A/G)CGTATTC | ||
| CAT-FN | AAGCTGGTATCAAGGCAG | J. Sumner, personal communication | |
| CAT-RN | CCCATCATCAGAAGGAGC |
The CAP (contig assembly program) sequence assembler (www.infobiogen.fr) and ClustalW multiple sequence alignment (Bioinformatics Center Institute for Chemical Research, Kyoto University, Kyoto, Japan; http://clustalw.genome.jp//) programs were used to analyze the sequences. A phylogenetic analysis and DNA sequence similarities were calculated using MEGA software, version 2.0 (http://www.megasoftware.net) (27). Sequencing of the corresponding fragments of reference strains of B. bacilliformis, B. quintana, and B. henselae was performed to ensure the quality and accuracy of the GenBank sequence data (accession numbers are given below). GenBank accession numbers of reference strains of Bartonella used for the phylogenetic analysis are provided on the corresponding figures.
Immunohistochemistry and histopathology.
Unstained, fixed sections of the splenic aspirate were sent to the CDC and were evaluated by an immunoalkaline phosphatase staining technique for B. henselae and B. quintana, as described previously (15). The primary antibodies included a monoclonal anti-B. henselae antibody (diluted 1/100) and a polyclonal rabbit anti-B. quintana antibody (1/100). After the patient's splenectomy, fixed sections of the solitary fibrotic tumor in the spleen were obtained by the CDC and stained with hematoxylin and eosin and Steiner's silver stain.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained during this study for isolate EC-01 were deposited in the NCBI GenBank under the following accession numbers: DQ179109 for the gltA fragment, DQ179110 for the rpoB gene fragment, DQ179107 for the 16S-23S rRNA intergenic region (internal transcribed spacer [ITS]), DQ179108 for the 16S rRNA gene fragment, and DQ179112 and DQ179111 for the ftsZ fragments. Related sequences for other B. bacilliformis isolates were deposited under the following accession numbers: isolate VRB 165 gltA, DQ200877; isolate Hsp800-31 gltA, DQ200878; isolate Vega gltA, DQ200879; isolate Vero97 gltA, DQ200880; isolate Vero75 gltA, DQ200881; isolate VRB 165 ITS, DQ200882; isolate Hsp800-1 ITS, DQ200883; isolate Vega ITS, DQ200884; isolate Vero97 ITS, DQ200886; and isolate Vero75 ITS, DQ200887.
RESULTS
Diagnostic testing of clinical specimens from patient.
Initial PCR and serological testing of the patient's blood by a commercial laboratory was negative for DNA and IgG/IgM antibodies to B. henselae and B. quintana, respectively. The computer-assisted tomography scan done at the hospital confirmed that the patient had an enlarged spleen. Following needle biopsy by an oncologist, histopathologic examination of fixed slides of splenic aspirate was interpreted as bacillary angiomatosis with tiny aggregates of bacilli identified by using Warthin-Starry stain. After subsequent referral to an infectious disease specialist, which was 10 weeks following onset of the patient's splenomegaly, serum, whole blood, and unstained slides of the formalin-fixed, paraffin-embedded splenic aspirate were submitted to the CDC for evaluation. Nested PCRs using htrA primers for B. henselae and B. quintana and conserved ribC primers for all Bartonella species failed to detect DNA gene sequences for Bartonella in patient whole blood and fixed tissue sections of splenic aspirate. By indirect IFA, patient serum was negative (titer of <32) for IgG antibodies to B. bacilliformis, B. henselae, and B. quintana. IHC evaluation of fixed slides of splenic aspirate was also negative for B. quintana and B. henselae. IHC for B. bacilliformis was not done because the CDC did not have a suitable primary antibody to specifically detect this bacterial organism in the IHC assay. At the CDC, histology of the fixed splenic aspirate tissue sections indicated lymphoid depletion, aggregates of small vascular channels, and scattered histiocytes. No viral inclusions, fungal elements, or parasitic organisms were seen. Silver staining for bacilli on fixed tissue sections of splenic aspirate were not repeated at the CDC due to limited numbers of sections.
Isolation of Bartonella and electron microscopy.
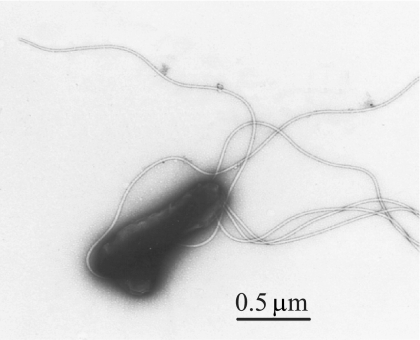
No bacterial growth was detected on petri plates of HIARB medium that were directly inoculated with the patient's blood. However, growth of nonadherent bacteria was observed in the supernatant phase of BiP medium inoculated with either peripheral blood mononuclear cells or whole blood on day 18 following inoculation. Bacteria were observed to aggregate and form clumps during growth in the liquid phase of BiP medium. They also did not stain well with classic Gram staining and Wright-Giemsa staining procedures (data not shown). Transmission electron microscopy detected bacillary-shaped organisms of 1 to 1.5 μm in length with polar flagella (Fig. 1), indicating cellular morphology consistent with identification of the new isolate as B. bacilliformis or another species of flagellated Bartonella.
FIG. 1.
Transmission electron microphotograph of isolate EC-01. Scale bar, 0.5 μm.
Multiple locus sequence characterization of a new isolate.
To determine genus and species identity of the patient bacterial isolate EC-01, several gene fragments were amplified and sequenced. Accordingly, nucleotide sequences were determined for a 1,157-bp fragment of the 16S rRNA gene, 707-bp and 607-bp fragments corresponding to 5′ and 3′ ends of ftsZ, a 502-bp fragment of rpoB, a 338-bp fragment of gltA, and 503 bp of the 16S rRNA-23S rRNA ITS. Nucleotide sequences determined for EC-01 had 99 to 100% of sequence identity with respective gene fragments of the majority of B. bacilliformis isolates. The gltA sequence of EC-01 had 98% sequence identity with strain Caraz SC (AY114119) and 96% sequence identity with strains LA6.3, Vega, and Vero97, corresponding, respectively, to 3 and 10 to 11 nucleotide differences. The gltA sequence identity of EC-01 to other Bartonella spp. was >86%. In addition to gltA and ITS fragments that we generated by PCR from the DNA of B. bacilliformis isolates from Peru and Ecuadorian isolate EC-01, our phylogenetic comparison included gltA and ITS fragments of other B. bacilliformis isolates available in the GenBank. Thus, not all isolates had both gltA and ITS fragments available for phylogenetic analysis. The ribC primers used to evaluate the patient blood gave an amplicon with the EC-01 isolate while the htrA primers did not.
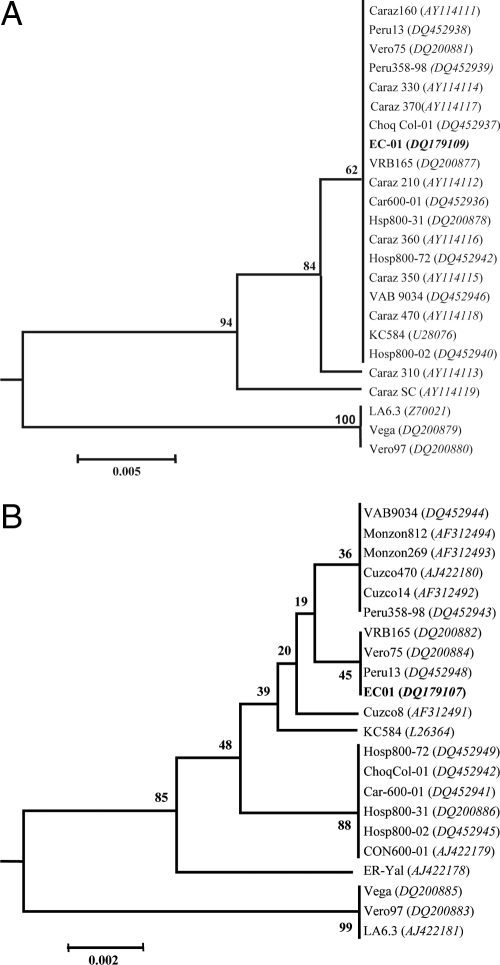
Based on phylogenetic analysis of gltA nucleotide sequences, isolate EC-01 clustered with the majority of B. bacilliformis isolates. Most of these isolates had been obtained from patients around Caraz, Ancash, in Peru (Fig. 2A). This cluster also included isolate KC584 which is a standard reference, highly passaged, laboratory strain of B. bacilliformis. A second cluster included two human isolates, Vero97 and Vega, from the Caraz area and strain LA6.3, isolated from a patient in the Haillacayan Valley in the department of Ancash (5). Divergence for gltA nucleotide sequences was 0.5% (Fig. 2A). Also, although ITS sequences were not available for all isolates of B. bacilliformis that were analyzed for gltA, the phylogenetic tree that was constructed based on alignment of the ITS region was similar (Fig. 2B). A majority of the isolates from the Cusco area formed a very tight cluster. This cluster also included the new isolate EC-01 from Ecuador and strain KC584. The second cluster consisted of strain LA6.3 and the same two human isolates from the Caraz, Ancash, area. Isolate ER-Yal isolated from a patient in Amazonas was found to be the most divergent among B. bacilliformis included in this analysis (Fig. 2B). ITS nucleotide sequences were 0.2% divergent.
FIG. 2.
The genetic relationships between the new isolate EC-01 and other isolates of B. bacilliformis. Neighbor-joining phylogenetic trees based on gltA (A) and ITS (B) sequence similarities were drawn using MEGA2 software. The distance matrix was calculated using Jukes-Cantor parameters and 231 sites for gltA and 361 sites for the ITS. The scale bar represents 0.5% and 0.2% divergence, respectively, for panels A and B. The numbers at nodes are the proportions of 1,000 bootstrap resamplings that support the topology shown. The NCBI accession numbers of the Bartonella sequences used in this analysis are shown.
Serological characterization of the EC-01 isolate.
Human antisera were collected from patients with Oroya fever from the departments of Cusco and Ancash in Peru during 2001 to 2002. Of these patients, only Ramirez and Colonia had both antisera and B. bacilliformis isolates available so that titers to homologous antigen could be determined. Cule III antigen is the reference antigen used at the CDC to detect antibody to B. bacilliformis by IFA. Antibody titers of antisera from the patient EC-01 and from the panel of Peruvian patients were detected using whole-cell antigens from patient isolates EC-01, Ramirez, Colonia, and Cule III (Table 3). While EC-01 patient antiserum reacted with homologous antigen at only a titer of 1/64, it cross-reacted with heterologous Ramirez and Colonia antigens at titers of 1/1,028 and 1/256, respectively. In addition, the EC-01 antiserum did not react to Cule III antigen whereas 20 other infected patient antisera from Peru had titers ranging from 1/64 to >1/2,056. Seven out of these 20 antisera were negative with EC-01 antigen (endpoint titers of <1/32), and the remaining 13 antisera had relatively low antibody titers ranging from 1/32 to 1/128. Like EC-01 antigen, Colonia antigen also exhibited low to moderate binding to other Peruvian patient antisera. In contrast, Ramirez and Cule III antigen bound these patient antisera at significantly higher titers. These data suggested that while isolate EC-01 was related to other patient isolates of B. bacilliformis from Peru by serology, there was antigenic variation among these isolates.
TABLE 3.
IFA titers of patient and control antisera against EC-01 antigen and antigens from B. bacilliformis isolated in Perua
| Antiserumb | Antibody titer to indicated antigen
|
|||
|---|---|---|---|---|
| EC-01 | Ramirez | Colonia | Cule III | |
| Cus 807 | <32 | >2,056 | <32 | 512 |
| Cus 1002 | <32 | 512 | <32 | 512 |
| Cus 1003 | <32 | >2,056 | <32 | >2,056 |
| Cus 1006 | <32 | 256 | 64 | 256 |
| Cus 1007 | 64 | >2,056 | 32 | 512 |
| Cus 1008 | 128 | 256 | 128 | 1,028 |
| Cus 1011 | <32 | <32 | 32 | >2,056 |
| Cus 1202 | 32 | 64 | 64 | 1,028 |
| Cus 1203 | 32 | >2,056 | 32 | >2,056 |
| Cus 3405 | 32 | >2,056 | 64 | 256 |
| Cus 3601 | 64 | 1,028 | 64 | >2,056 |
| Cus 3602 | 64 | 512 | 128 | 512 |
| Cus 3802 | 64 | >2,056 | 128 | 64 |
| Cus 3803 | <32 | 32 | 64 | 256 |
| Cus 4003 | <32 | 256 | <32 | >2,056 |
| Elizabeth | 32 | 256 | 64 | 128 |
| Garcia | 64 | 128 | 64 | >2,056 |
| Vega | 128 | 1,028 | 64 | >2,056 |
| Ramirez | 64 | 64 | 32 | 1,028 |
| Colonia | 32 | 512 | 32 | 512 |
| EC-01 | 64 | 1,028 | 256 | <32 |
| B. henselae (+) | <32 | 1,028 | <32 | <32 |
| Pooled human sera (−) | <32 | <32 | <32 | <32 |
Antisera was collected in Peru from 2001 to 2002 from patients with Oroya fever and were provided by Franca Jones, U.S. Naval Medical Research Center Detachment in Lima, Peru.
+, positive IFA control; −, negative IFA control.
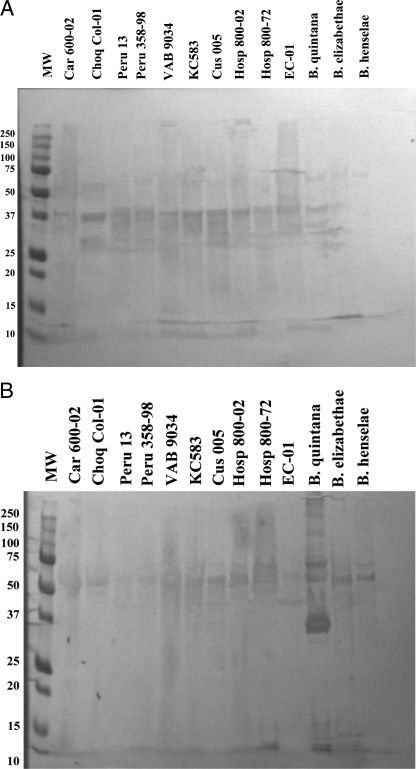
CDC-produced rabbit polyclonal antibodies against intact B. bacilliformis and B. quintana, respectively, were used to assess the antigenic relatedness between specific proteins of isolate EC-01 and other Peruvian isolates of B. bacilliformis by Western blotting. EC-01 proteins bound hyperimmune anti-B. bacilliformis antiserum in a pattern similar to that of other B. bacilliformis isolates from Peru (Fig. 3A), whereas there were significant differences in the binding of this antiserum to the antigens of other Bartonella species. Figure 3B also demonstrated the significant cross-reactivity of polyclonal rabbit antiserum made against B. quintana to antigens of Peruvian B. bacilliformis isolates, isolate EC-01, and other Bartonella species. Here, more variation in antibody binding to antigens of EC-01 and other B. bacilliformis isolates was observed.
FIG. 3.
Immunoreactivity of proteins from EC-01 and Peruvian B. bacilliformis isolates to polyclonal anti-Bartonella antibodies. Western blots of protein lysates from bacterial isolates were probed with either rabbit polyclonal antibody to B. bacilliformis (A) or rabbit polyclonal antibody to B. quintana (B) and visualized using horseradish peroxidase-conjugated secondary antibody and diaminobenzidine substrate. Anti-B. bacilliformis antiserum reacted commonly with proteins having molecular weights between 27 to 37 kDa whereas cross-reactive anti-B. quintana antiserum bound predominantly to a 50-kDa protein common to all isolates. EC-01 shared antigenic proteins with other B. bacilliformis isolates and with B. quintana. MW, molecular weight (kDa).
Protein fingerprinting by gel electrophoresis.
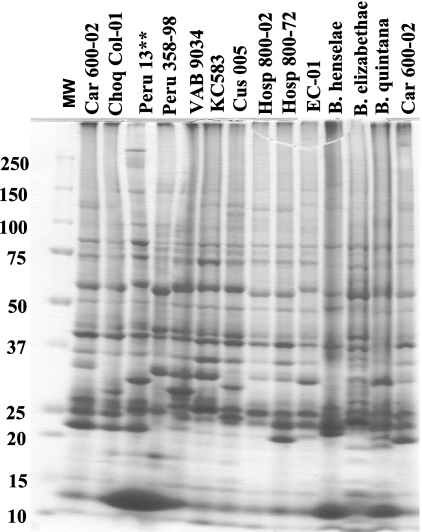
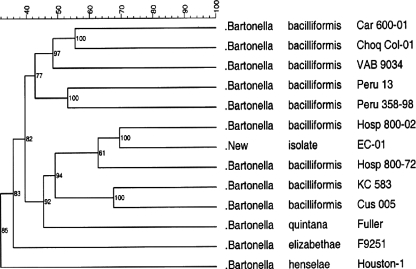
SDS-PAGE protein profiles were determined for nine isolates of B. bacilliformis including ATCC type strain KC583. Three other Bartonella species pathogenic for humans, B. henselae, B. elizabethae, and B. quintana, were also included in this analysis and compared with the protein profile of isolate EC-01 from Ecuador (Fig. 4). Distinctive similarities and differences in the protein fingerprints between EC-01 and the other Bartonella species were observed. For instance, EC-01 and the other Bartonella isolates contained similar molecular weight proteins of approximately 87.5 kDa, 75 kDa, 52 kDa, 43.5 kDa, 40 kDa, 25 kDa, and 12 kDa. In contrast, protein band heterogeneity was observed at 58 kDa, 30 to 35 kDa, and 21 to 24 kDa. In order to determine the relatedness of the protein banding patterns between EC-01 and the other Bartonella species and B. bacilliformis isolates, BioNumerics cluster analysis with a band-matching algorithm was used to generate a proteodendrogram (Fig. 5). Compared to B. bacilliformis isolates from Peru, EC-01 was most closely clustered with isolate Hosp 800-02, Hosp 800-72, KC583, Cus 005, and B. quintana in one clade. The other B. bacilliformis isolates were clustered in a second clade (Fig. 4). In the proteodendrogram, both B. henselae and B. elizabethae were clustered more distantly from isolate EC-01 and other B. bacilliformis isolates. In a similarity matrix table, the percentage of band matches between EC-01 and other B. bacilliformis isolates was 69% for Hosp 800-02, 57% for Hosp 800-72, 49% for KC583, and 52% for Cus 005 (Table 4). Surprisingly, band matching between EC-01 and B. quintana were 54%. In contrast, band matching between EC-01 and B. henselae and B. elizabethae was only 33% and 38%, respectively. High cophenetic values shown in the dendrogram indicated good reliability between the groupings within the dendrogram.
FIG. 4.
Protein banding patterns of Bartonella isolates. Whole-cell lysates were resolved on an 8 to 16% gradient SDS-PAGE gel and stained with Coomassie blue. Lane 1, molecular weight (MW) marker; lane 2, Car 600-01; lane 3, Choq Col-01; lane 4, Peru 13; lane 5, Peru 358-98; lane 6, VAB 9034; lane 7, KC583; lane 8, Cus 005; lane 9 Hosp 800-02; lane 10, Hosp 800-72; lane 11, EC-01; lane 12, B. henselae; lane 13, B. elizabethae; lane 14, B. quintana; lane 15, Car 600-01. Numbers on the left are molecular weights (kDa).
FIG. 5.
Cluster analysis comparing protein fingerprints of EC-01 with Peruvian B. bacilliformis strains and other Bartonella species associated with human disease. Similarity coefficients were generated by the different-bands algorithm and used to construct a dendrogram based on the unweighted pair group method using arithmetic averages for B. bacilliformis isolates and isolate EC-01. Cophenetic values (small numbers in the dendrogram branches) expressed how well the dendrogram correlated with band matching coefficients. Caraz is an area of endemicity for bartonellosis whereas Cusco is not considered to be an area endemicity.
TABLE 4.
Matrix similarity values comparing protein band relatedness between isolate EC-01, B. bacilliformis isolates from Peru, and other Bartonella speciesa
| Isolate identifier | Isolate | % Matching protein bands
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | ||
| A | Car 600-01 | 100.0 | ||||||||||||
| B | Choq Col-01 | 55.4 | 100.0 | |||||||||||
| C | VAB 9034 | 47.5 | 49.3 | 100.0 | ||||||||||
| D | Peru 13 | 39.8 | 47.8 | 45.3 | 100.0 | |||||||||
| E | Peru 358-98 | 35.4 | 39.3 | 48.8 | 53.1 | 100.0 | ||||||||
| F | Hosp 800-02 | 41.0 | 40.8 | 44.9 | 33.6 | 43.9 | 100.0 | |||||||
| G | EC-01 | 31.0 | 37.3 | 49.4 | 37.1 | 43.6 | 69.4 | 100.0 | ||||||
| H | Hosp 800-72 | 43.2 | 37.9 | 48.2 | 34.7 | 30.9 | 67.7 | 57.5 | 100.0 | |||||
| I | KC583 | 39.9 | 41.4 | 53.7 | 39.3 | 41.2 | 51.0 | 49.0 | 43.3 | 100.0 | ||||
| J | Cus 005 | 40.6 | 45.6 | 40.9 | 41.2 | 41.6 | 50.6 | 51.6 | 48.7 | 67.5 | 100.0 | |||
| K | Bq Fuller | 37.0 | 24.7 | 37.0 | 30.9 | 32.1 | 44.9 | 53.7 | 45.3 | 40.8 | 42.0 | 100.0 | ||
| L | Be F9251 | 29.4 | 34.4 | 42.7 | 28.7 | 32.5 | 39.9 | 36.0 | 43.3 | 34.1 | 38.4 | 32.8 | 100.0 | |
| M | Bh Houston-1 | 33.8 | 27.0 | 28.8 | 25.5 | 32.5 | 33.9 | 35.0 | 39.0 | 29.6 | 27.7 | 34.1 | 33.0 | 100.0 |
The matrix table was constructed using cluster analysis and the different bands algorithm of BioNumerics software. Bq, B. quintana; Be, B. elizabethae; Bh, B. henselae. All other isolates listed are B. bacilliformis. A band matching value of 100% indicates comparison of an isolate to itself. Values are the percentages of matched protein bands. Values in boldface represent the B. bacilliformis isolates in Peru that most closely matched the protein fingerprint of Ecuadorian isolate EC-01.
Histology.
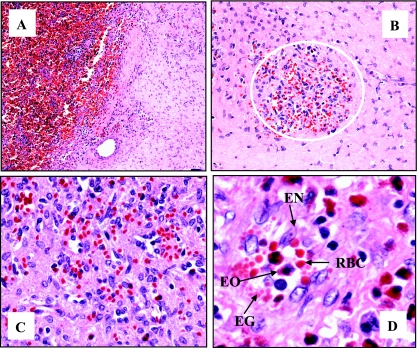
Pathological evaluation of tissue sections of the splenic aspirate from the enlarged spleen that were sent to the CDC showed aggregates of small vascular channels and scattered histiocytes. No immunohistochemical evidence of infection with B. henselae or B. quintana was obtained by using IHC assays for these agents. After splenectomy, histology of tissue sections of the excised splenic mass submitted to the CDC showed a circumscribed fibrotic lesion containing small scattered aggregates of plump atypical endothelial cells arranged around slit-like vascular channels. The interstitium contained dense fibrous tissue and multiple mineralized foci. Eosinophils, lymphocytes, and occasionally small eosinophilic globules were abundantly distributed around endothelial proliferations (Fig. 6). No obvious inclusions were identified in any of the endothelial cells, and no individual or clumped bacteria were identified in the tissue by use of hematoxylin and eosin and Steiner's stain.
FIG. 6.
Histology of patient splenic mass discovered during splenectomy. Thirteen months following the onset of splenomegaly, the patient underwent splenectomy. Gross examination of the excised spleen revealed a solitary fibrotic mass which was diagnosed as a benign vascular lesion of the spleen. Formalin-fixed, paraffin-embedded tissue sections of the mass were stained with hematoxylin and eosin. (A) Normal red pulp of the spleen on the left (dark red) with demarcated area of the fibrotic lesion on the right (pink). (B) Within the fibrotic lesion is a representative nodule at 10-fold magnification consisting of small vasculature with focal cellular growth (area within white circle). (C) Fourfold magnification of the nodule. (D) Eightfold magnification of the nodule showing small aggregates of proliferating endothelial cells (EN) forming slit-like vascular spaces (white areas) with eosinophils (EO), eosinophilic globules (EG), and red blood cells (RBC).
DISCUSSION
Using morphological, genetic, proteomic, and antigenic criteria, isolate EC-01 was characterized as B. bacilliformis. Electron microscopy confirmed the presence of polar flagella on the bacteria, thus confirming morphology consistent with that of B. bacilliformis. The gene sequences of isolate EC-01 were nearly identical to a subset of B. bacilliformis isolates from the region of Caraz, Ancash, in Peru, where B. bacilliformis is endemic. The protein fingerprint data likewise indicated that EC-01 was most closely related to B. bacilliformis isolates (Hosp 800-02 and Hosp 800-72) from Caraz, Ancash, thus correlating with genomic data.
IFA and Western immunoblotting data indicated that the B. bacilliformis isolates shared a spectrum of antigens, but the IFA and protein fingerprint data also confirmed that the isolates were antigenically heterogeneous. Thus, confirmation of cases of infection of B. bacilliformis by serology may be suspect if a subset of antigen types is not included in diagnostic IFA testing. Disease presentation in this patient was unusual because clinical signs of Bartonella infection occurred after the patient's prolonged residence in the United States and 3 years after the patient last visited Ecuador. After lymphoma was ruled out as a cause of the patient's splenomegaly, bartonellosis was suspected, given the patient's country of origin, travel to Ecuador, and skin lesions. However, a presumptive diagnosis of bartonellosis could not initially be confirmed for the patient during the acute phase of illness (splenomegaly) based on serology and PCR testing for B. henselae and B. quintana by commercial laboratories. Similarly, we did not detect B. bacilliformis, B. henselae, or B. quintana in the patient blood or serum by IFA or PCR or in splenic aspirate by IHC assay. Only when we were able to culture bacilli from the patient's blood and subsequently to identify this isolate (EC-01) as B. bacilliformis was the case fully confirmed. The inability to detect Bartonella infection in the patient's blood initially by PCR and IFA led us to investigate the reasons for this. The serum sample we analyzed might be considered to be convalescent-phase since it was obtained 10 weeks following onset of splenomegaly. Our IFA is 82% sensitive in detecting B. bacilliformis antibodies in acute-phase blood samples of laboratory-confirmed bartonellosis patients and 93% positive for convalescent-phase serum (8). Two possible factors could explain the negative IFA results: either (i) the patient's antibodies did not react to the antigen used in our B. bacilliformis IFA or (ii) infection with B. bacilliformis did not stimulate a detectable antibody response. When screened against prototypic B. bacilliformis Cule III antigen used in our IFA and against antigens from two other patient isolates (Ramirez and Colonia), the patient antiserum failed to react with Cule III antigen by IFA whereas it did react weakly against EC-01 patient antigen and strongly against antigens of Ramirez and Colonia isolates, thus supporting the first explanation. The weak reactivity of antigen from isolate EC-01 to patient serum (titer, 1/64) has similar correlates to other Bartonella infections in which low or negative antibody reactivity and variation in antibody titers with different strains were well documented. For example, there have been isolate-positive but seronegative cases of infection with B. quintana (17) and B. henselae (18, 32). Parallel with our findings with B. bacilliformis antigens, B. henselae infection was detected only with Marseille strain antigens but not with standard Houston-1 strain antigens by IFA (32). In addition, IFA-positive infections of human cat scratch disease have been shown to exhibit significant variation in the specific antigens recognized by immune sera (33). As shown in Table 3, there was a wide range of patient antiserum reactivity to both homologous and heterologous bacterial antigens, and this may be related to differences in virulence between bacterial isolates and symptomatology in each patient. We were unable to further investigate the differences in antiserum titers to homologous and heterologous bacterial antigens for the rest of the antisera listed in Table 3 because bacterial isolates were not available from most of the patients.
Proteomic analysis of the EC-01 protein band fingerprint using BioNumerics software confirmed that isolate EC-01 from Ecuador was a unique B. bacilliformis strain that was most highly related to Peruvian isolate Hosp 800-02 from Caraz, Ancash, where infection with B. bacilliformis is endemic. However, the patient had never traveled to Peru or to areas of Ecuador where infection with B. bacilliformis is endemic. Patient EC-01's exposure to B. bacilliformis presumably occurred within the area bounded by Quito, Esmeraldas, and Coca in Ecuador, where infections resulting from B. bacilliformis have not previously been reported in the scientific literature. However, the lowland province of Manabi, where Carrion's disease has been endemic since pre-Colombian times (1), is within 50 to 100 miles of Quito and Esmeraldas. From 1987 to 1995 in south-central Manabi, 21% of people surveyed were seropositive, and 11/224 (4.7%) presented with monophasic verrucous cutaneous disease (2). Thus, Manabi has been a focal point for asymptomatic infection and mild clinical disease, and it is quite possible that the distribution of B. bacilliformis has spread northward to the coastal lowlands of Amazonas where the patient traveled. Conceivably, the patient's infection was either asymptomatic or characterized by a mild case of verruga acquired during travel to the lowland province of Esmeraldas, Ecuador, 3 years before, which subsequently resurfaced as acute splenomegaly. The association of verruga with splenomegaly was confirmed by a case study in Peru in which 5/77 verruga patients presented with splenomegaly as a clinical sign (30). Although the skin lesions of our patient could not be confirmed as verruga because skin biopsy was declined, their presence was suggestive of chronic, subclinical infection with B. bacilliformis. At this time, we cannot explain the epidemiology of isolate EC-01 in Ecuador. Several hundred miles geographically separate isolate EC-01 from related isolates in Caraz, Peru. For several decades, hotspots of B. bacilliformis infection in Peru have occurred in the provinces of Ancash, Cajamarca, Amazonas, Lima, and Cusco with new areas of endemicity of infection occurring in adjacent provinces of Cajamarca, Huánuco, and Junín (reviewed in reference 24). Taken together, bartonellosis in Peru ranges from Cusco in the south all the way to Amazonas in the north, which borders Ecuador. In all likelihood, the incidence of B. bacilliformis infection in Ecuador is more widespread than is presently recognized and underdiagnosed if infection is subclinical or atypical.
The cutaneous pathology of verruga peruana resulting from B. bacilliformis infection has been well documented (4, 7). However, there are no contemporary descriptions of the splenic pathology of verruga peruana. Indeed, it has been stated that verruga peruana does not involve internal organs (35). Nonetheless, early investigators of this condition described involvement of various internal tissues and organs, including the mucous membranes of the gastrointestinal and genitourinary tracts, and the central nervous system, lungs, liver, pancreas, kidneys, and spleen. In these locations, verrugae are typically miliary and associated with interstitial connective tissue (38). Typically, bacillary angiomatosis of internal organs is associated with disseminated infection of B. henselae or B. quintana in immunocompromised individuals (25). In contrast, the histopathology of the splenic nodule in the patient described in this report shares characteristics of cutaneous verruga and bacillary angiomatosis, but no bacteria were definitively identified by either Steiner staining or broad-range 16S rRNA gene PCR in a section of this lesion. This is not surprising, particularly since limited sections of the splenic lesion were available for bacterial analyses and because the patient was initially treated with a short course of antibiotics during the acute phase of splenomegaly, which was 12 months prior to splenectomy. We suggest that the fibrotic tissue and mineralized foci observed within the interstitium in the splenic lesion may be evidence of previous bacterial colonization in the spleen that was self-limiting or resolved by the previous antibiotic treatment. In addition, even a diligent search for bacteria in active cutaneous lesions using special stains often fails to demonstrate B. bacilliformis (4). The present case clearly is not typical of classic acute or chronic infection with B. bacilliformis. We could not unequivocally determine that the patient's splenic lesions were associated with B. bacilliformis infection. However, expanding spectrums of atypical infection with B. henselae and B. quintana have been documented, so we fully expect that other presentations of B. bacilliformis infection will also be found.
The epidemiology of B. bacilliformis infection in Ecuador is unknown at this time. In addition, tourism companies in Peru report that large numbers of tourists are visiting areas such as Cusco, Macchu Pichu, Urubamba, and Ollantaytambo (personal communication with Andean Odyssey and Condor Travel). These tourist destinations are in geographic regions where infection with B. bacilliformis is endemic and emergent. Thus, large numbers of susceptible individuals may potentially be exposed to, and infected with, B bacilliformis while visiting these areas. For native populations living in Peru and Ecuador, Carrion's disease constitutes a continuing public health threat. Whether infected individuals remain asymptomatic or develop acute-phase Oroya fever or eruptive skin disease may likely depend upon host-pathogen interactions. Important factors in these interactions include virulence of B. bacilliformis, host immunity, susceptibility and resistance to reinfection, species-specific differences in arthropod vectors, and the ability of these vectors to transmit disease. In this paper, the patient's epidemiological data and atypical symptomatology support the hypotheses that B. bacilliformis infection may be more widespread in Ecuador than is presently recognized and that mild disease may present with atypical or vague clinical signs leading to underdiagnosis. The data also argue for transmission of bacilli by an alternative vector, such as the related sand fly, Lutzomyia columbiana, that is found in areas where the patient traveled in Ecuador (42). There is a need to further investigate the potential for disease transmission by other species of Lutzomyia, particularly in areas where the disease is emergent, and to better understand the spectrum of disease associated with infection by B. bacilliformis. Because IFA and PCR tests initially did not detect B. bacilliformis infection in this patient, there is also a need to reevaluate diagnostic strategies to recognize atypical disease.
Acknowledgments
Human antisera from patients with Oroya fever in Peru were kindly provided by Franca Jones of the U.S. Naval Medical Research Center Detachment in Lima, Peru. We also thank Elizabeth Bosserman (CDC) for assistance with PCR and sequencing of Bartonella, the CDC Core Facility for primer preparation, Maria Ah (CDC) for cultivation and harvesting of B. bacilliformis isolates, and Perry Comegys (University of Maryland) for assistance with electron microscopy and photography.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Alexander, B. 1995. A review of bartonellosis in Ecuador and Colombia. Am. J. Trop. Med. Hyg. 52354-359. [DOI] [PubMed] [Google Scholar]
- 2.Amano, Y., J. Rumbea, J. Knobloch, J. Olson, and M. Kron. 1997. Bartonellosis in Ecuador: serosurvey and current status of cutaneous verrucous disease. Am. J. Trop. Med. Hyg. 57174-179. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, B., K. Sims, R. Regnery, L. Robinson, M. J. Schmidt, S. Goral, C. Hager, and K. Edwards. 1994. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J. Clin. Microbiol. 32942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias-Stella, J., P. H. Lieberman, R. A. Erlandson, and J. Aria-Stella, Jr. 1986. Histology, immunohistochemistry, and ultrastructure of the verruga in Carrion's disease. Am. J. Surg. Pathol. 10595-610. [DOI] [PubMed] [Google Scholar]
- 5.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46891-897. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, D. J., S. P. O'Connor, D. G. Hollis, R. E. Weaver, and A. G. Steigerwalt. 1991. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J. Clin. Microbiol. 291299-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres-Rios, H., J. Rodriquez-Tafur, F. Barvo-Puccio, C. Maguina-Vargas, C. S. Diaz, D. C. Ramos, and R. Patarca. 1995. Verruga peruana: an infectious endemic angiomatosis. Crit. Rev. Oncol. 647-56. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlin, J., L. Laughlin, S. Gordon, S. Romero, N. Solorzano, and R. L. Regnery. 2000. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: test development and application to a population in an area of bartonellosis endemicity. J. Clin. Microbiol. 384269-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlin, J., L. W. Laughlin, S. Romero, N. Solorzano, S. Gordon, R. G. Andre, P. Pachas, H. Friedman, C. Ponce, and D. Watts. 2002. Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J. Infect. Dis. 186983-990. [DOI] [PubMed] [Google Scholar]
- 10.Chomel, B. B., R. W. Kasten, J. E. Skyes, H. J. Boulouis, and E. B. Breitschwerdt. 2003. Clinical impact of persistent Bartonella bacteremia in humans and animals. Ann. N. Y. Acad. Sci. 990267-278. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, P., R. Guderian, W. Paredes, R. Daniels, D. Perera, M. Espinel, M. Valdez, and G. Griffin. 1996. Bartonellosis in Zamora Chinchipe province in Ecuador. Trans. R. Soc. Trop. Med. Hyg. 90241-243. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, P., R. Guderian, P. Orellana, C. Sandoval, H. Olalla, M. Valdez, M. Calvopina, A. Guevara, and G. Griffin. 1997. An outbreak of bartonellosis in Zamora Chinchipe province in Ecuador. Trans. R. Soc. Trop. Med. Hyg. 91544-546. [DOI] [PubMed] [Google Scholar]
- 13.Cuadra, M. 1956. Salmonellosis complication in human bartonellosis. Tex. Rep. Biol. Med. 1497-113. [PubMed] [Google Scholar]
- 14.Daly, J. S., M. G. Worthington, D. J. Breener, C. W. Moss, D. G. Hollis, R. S. Weyant, A. G. Steigerwalt, R. E. Weaver, and M. I. Daneshvar. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daybell, D., C. D. Paddock, S. R. Zaki, J. A. Comer, R. D. Woodruff, K. J. Hansen, and J. E. Peacock. 2004. Disseminated infection with Bartonella henselae as a cause of spontaneous splenic rupture. Clin. Infect. Dis. 39e21-e24. [DOI] [PubMed] [Google Scholar]
- 16.Dehio, C. 2004. Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58365-390. [DOI] [PubMed] [Google Scholar]
- 17.Drancourt, M., V. Moal, P. Brunet, B. Dussol, Y. Berland, and D. Raoult. 1996a. Bartonella (Rochalimaea) quintana infection in a seronegative hemodialyzed patient. J. Clin. Microbiol. 341158-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996b. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347441-443. [DOI] [PubMed] [Google Scholar]
- 19.Ellis, B. A., L. D. Rotz, J. A. Leake, F. Samalvides, J. Bernable, G. Ventura, C. Padilla, P. Villascea, L. Beati, R. Regnery, J. E. Childs, J. G. Olson, and C. P. Carrillo. 1999. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am. J. Trop. Med. Hyg. 61344-349. [DOI] [PubMed] [Google Scholar]
- 20.Gray, G. C., A. A. Johnson, S. A. Thornton, W. A. Smith, J. Knobloch, P. W. Kelley, L. O. Escudero, M. A. Huayda, and F. S. Wignall. 1990. An epidemic of Oroya fever in the Peruvian Andes. Am. J. Trop. Med. Hyg. 42215-221. [DOI] [PubMed] [Google Scholar]
- 21.Hambuch, T. M., S. A. Handley, B. Ellis, J. Chamberlin, S. Romero, and R. Regnery. 2004. Population genetic analysis of Bartonella bacilliformis isolates from areas of Peru where Carrion's disease is endemic and epidemic. J. Clin. Microbiol. 423675-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrix, L. R. 2000. Contact-dependent hemolytic activity distinct from deforming activity of Bartonella bacilliformis. FEMS Microbiol. Lett. 182119-124. [DOI] [PubMed] [Google Scholar]
- 23.Houpikian, P., and D. Raoult. 2001. 16S/23S rRNA intergenic spacer regions for phylogenetic analysis, identification, and subtyping of Bartonella species. Clin. Microbiol. 392768-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huarcaya, E., C. Maguina, R. Torres, J. Rupay, and L. Fuentes. 2004. Bartonellosis (Carrion's disease) in the pediatric population of Peru: an overview and update. Brazilian J. Infect. Dis. 8331-339. [DOI] [PubMed] [Google Scholar]
- 25.Koehler, J. E., M. A. Sanchez, C. S. Garrido, M. J. Whitfield, F. M. Chen, T. G. Berger, M. C. Rodriguez-Barradas, P. E. LeBoit, and J. W. Tappero. 1997. Molecular epidemiology of Bartonella infections in patients with bacillary angiomatosis-peliosis. N. Engl. J. Med. 3371876-1883. [DOI] [PubMed] [Google Scholar]
- 26.Kosek, M. R., R. H. Laverello, R. H. Gilman, J. Delgado, C. Maguina, M. Verastegui, A. G. Lescano, V. Mallqui, J. C. Kosek, S. Recavarren, and L. Cabrera. 2000. Natural history of infection with Bartonella bacilliformis in a nonendemic population. J. Infect. Dis. 182865-872. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, K., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Loutit, J. S. 1997. Bartonella infections. Curr. Clin. Top. Infect. Dis. 17269-290. [PubMed] [Google Scholar]
- 29.Maco, V., C. Maguina, A. Tirado, C. Maco, and J. E. Vidal. 2004. Carrion's disease (Bartonella bacilliformis) confirmed by histopathology in the high forest of Peru. Rev. Inst. Med. Trop. S. Paulo. 46171-174. [DOI] [PubMed] [Google Scholar]
- 30.Maguina, C., P. J. Garcia, E. Gotuzzo, L. Cordero, and D. H. Spach. 2001. Bartonellosis (Carrion's disease) in the modern era. Clin. Infect. Dis. 33772-779. [DOI] [PubMed] [Google Scholar]
- 31.Maguina, C., and E. Gotuzzo. 2000. Bartonellosis. New and Old. Dis. Clin. N. Am. 141-22. [DOI] [PubMed] [Google Scholar]
- 32.Mainardi, J.-L., C. Figlioni, F. W. Goldstein, P. Blanche, M. Baret-Rigoulet, N. Galezowski, P-.E. Fournier, and D. Raoult. 1998. Cat scratch disease due to Bartonella henselae serotype Marseille (Swiss cat) in a seronegative patient. J. Clin. Microbiol. 362800. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCool, T. L., J. G. Hoey, F. Montileone, H. B. Goldenberg, E. Mordechai, and M. E. Adelson. 2008. Discovery and analysis of Bartonella henselae antigens for use in clinical serologic assays. Diagn. Microbiol infect. 6017-23. [DOI] [PubMed] [Google Scholar]
- 34.Minnick, M. F., and B. E. Anderson. 22 June 2004, posting date. The genus Bartonella in M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.6. Springer-Verlag, NY. http://link.springer-ny.com/link/service/books/10125.
- 35.Montgomery, E. A., and F. U. Garcia. 1997. Bartonellosis-infection with Bartonella bacilliformis, p. 431-439. In D. H. Connor, F. W. Chandler, D. A. Schwartz, H. J. Manz, E. E. Lack (ed.), Pathology of infectious diseases vol. 1. Appleton and Lange, Samford, CT. [Google Scholar]
- 36.Noguchi, H., R. C. Shannon, E. B. Tilden, and J. Tyler. 1929. Etiology of Oroya fever. XIV. The insect vectors of Carrion's disease. J. Exp. Med. 49993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 331797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odriozola, E. 1898. La maladie de Carrión ou la verruga péruvienne. Carré et Naud, Paris, France.
- 39.Reeves, W. K. 2005. Molecular genetic evidence for a novel bacterial endosymbiont of Icosta americana (Diptera: Hippoboscidae). Entomol. News 116263-265. [Google Scholar]
- 40.Regnery, R. L., B. E. Anderson, J. E. Clarridge III, M. C. Rodriquez-Barradas, D. C. Jones, and J. H. Carr. 1992. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J. Clin. Microbiol. 30265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutledge, L. C., and R. K. Gupta. 2002. Moth flies and sand flies (Psychodidae), p. 153-154. In G. Mullins and L. Durden (ed.), Medical and veterinary entomology. Academic Press, New York, NY.
- 43.Schultz, M. G. 1968. A history of bartonellosis (Carrion's disease). Am. J. Trop. Med. Hyg. 17503-515. [DOI] [PubMed] [Google Scholar]
- 44.Zeaiter, Z., Z. Liang, and D. Raoult. 2002. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J. Clin. Microbiol. 403641-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeaiter, Z., P.-E. Fournier, G. Greub, and D. Raoult. 2003. Diagnosis of Bartonella endocarditis by a real-time nested PCR assay using serum. J. Clin. Microbiol. 41919-922. [DOI] [PMC free article] [PubMed] [Google Scholar]