Abstract
The cell surface-associated extracellular adherence protein (Eap) mediates adherence of Staphylococcus aureus to host extracellular matrix components and inhibits inflammation, wound healing, and angiogenesis. A well-characterized collection of S. aureus and non-S. aureus staphylococcal isolates (n = 813) was tested for the presence of the Eap-encoding gene (eap) by PCR to investigate the use of the eap gene as a specific diagnostic tool for identification of S. aureus. Whereas all 597 S. aureus isolates were eap positive, this gene was not detectable in 216 non-S. aureus staphylococcal isolates comprising 47 different species and subspecies of coagulase-negative staphylococci and non-S. aureus coagulase-positive or coagulase-variable staphylococci. Furthermore, non-S. aureus isolates did not express Eap homologs, as verified on the transcriptional and protein levels. Based on these data, the sensitivity and specificity of the newly developed PCR targeting the eap gene were both 100%. Thus, the unique occurrence of Eap in S. aureus offers a promising tool particularly suitable for molecular diagnostics of this pathogen.
Staphylococcus aureus has been usually identified by traditional phenotypic properties based on in-house techniques or commercial assays (30). Assaying biochemical or immunologic reactions, these widely used diagnostic systems present various problems, including unsatisfying sensitivity, specificity, and assay time. The majority of the difficulties associated with these methods result from the variable expression of phenotypic features that are tested in these diagnostic procedures. Furthermore, the lack of objective criteria in the interpretation of these tests, which include slide agglutination, tube coagulation, latex agglutination, and colorimetric assays, may cause additional ambiguity. Molecular methods represent alternatives combining high sensitivity, specificity, speed, and reliability for identification. Several nucleic acid amplification-based assays have been reported for S. aureus that are based on specific gene targets as well as universal sequences, offering different advantages and limitations (2, 6, 12, 13, 27, 29, 31, 38, 42). Thus, additional reliable tools for detection of S. aureus are of major interest.
The anchorless extracellular adherence protein (Eap) of S. aureus, also designated major histocompatibility complex class II analogous protein (Map), selectively recognizes extracellular matrix aggregates but binds promiscuously to monomeric matrix macromolecules (16, 21, 23). This broad-spectrum adhesin shows structural homology to the C-terminal domain of bacterial superantigens but lacks superantigen activity (11, 28). Eap was recently shown to curb acute inflammatory responses, to enhance internalization of the microorganism into eukaryotic cells, to inhibit wound healing, and to function as a potent angiostatic agent (1, 15, 18, 33).
Previously, the presence of the Eap-encoding gene (eap) was determined to occur in only selected human clinical S. aureus isolates (19, 20). While sequencing results suggested that Eap sequences are highly conserved, size differences of various eap genes as a result of different numbers of repeats have been described (Fig. 1) (19). Due to this polymorphism of the eap gene, a novel PCR assay for diagnostic purposes was designed and validated in this study. Subsequently, the feasibility of an eap-based approach was systematically analyzed on the genomic, transcriptional, and protein levels by use of a large collection of human- and veterinary-derived S. aureus and non-S. aureus staphylococcal isolates.
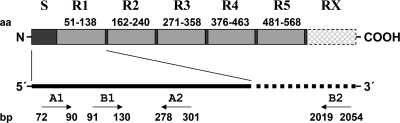
FIG. 1.
Schematic representation (not to scale) of Eap and the encoding gene, showing the amino acid (aa) composition consisting of a 30-aa signal peptide (S) and 110-aa residue domains (R1, R2, … RX) found in multiple copies. The oligonucleotide primer binding sites of EAP-CON1 (A1) and EAP-CON2 (A2) as well as of EAP-P3wt (B1) and EAP-P2wt (B2) are indicated in base pairs (bp); sizes correspond to EAP of the Newman D2C strain.
MATERIALS AND METHODS
Bacterial isolates.
Overall, 597 S. aureus isolates were tested. The collection encompassed a total of 444 German clinical S. aureus isolates comprising human (n = 429) and veterinary (bovine, n = 10; feline, n = 3; canine, n = 1; equine, n = 1) isolates as well as six reference strains of S. aureus (ATCC 13565, ATCC 14458, ATCC 19095, ATCC 23235, ATCC 27664, and ATCC 29213). In addition, S. aureus laboratory strains Newman, Wood 46, and CI7, known to be Eap-producing strains (19), were included, serving also as positive controls for the different Eap isoforms. The human isolates were collected and identified during the course of a German multicenter study comprising 219 blood and 210 nasal isolates (40). The veterinary S. aureus isolates were obtained from the Hannover area (Lower Saxonia, Germany). Furthermore, S. aureus isolates from France (n = 12), Hungary (n = 21), Italy (n = 26), Japan (n = 21), Poland (n = 49), and the United States (n = 15) were included.
Furthermore, a total of 216 non-S. aureus staphylococcal isolates comprising 164 clinical isolates and 52 type and reference strains of different coagulase-negative as well as coagulase-positive and -variable species and subspecies were studied (Table 1). Except for the clinically derived S. intermedius (3, 5) and S. pettenkoferi (35) isolates, non-S. aureus isolates were (i) recovered from a wide range of clinical specimens derived from, e.g., human skin, respiratory tracts, urogenital tracts, and venous catheters at the University of Münster, Münster, Germany, or were (ii) collected from blood during the course of a German multicenter study and considered etiologically relevant (41). Only one isolate per patient was included in this study.
TABLE 1.
Non-S. aureus staphylococcal (sub-)species studied
| Species and subspecies | No. of clinical isolatesa
|
Type and/or reference strain(s)b | Total no. of isolatesc | |
|---|---|---|---|---|
| Blood | Other | |||
| S. arlettae | 0 | 0 | DSM 20672T | 1 |
| S. auricularis | 0 | 0 | DSM 20609T | 1 |
| S. capitis subsp. capitis | 4 | 1 | DSM 20326T | 6 |
| S. capitis subsp. urealyticus | 0 | 0 | DSM 6717T | 1 |
| S. caprae | 0 | 5 | DSM 20608T | 6 |
| S. carnosus subsp. carnosus | 0 | 0 | DSM 20501T | 1 |
| S. carnosus subsp. utilis | 0 | 0 | DSM 11676T | 1 |
| S. chromogenes | 0 | 0 | DSM 20454T | 1 |
| S. cohnii subsp. cohnii | 4 | 2 | DSM 20260T | 7 |
| S. cohnii subsp. urealyticus | 0 | 0 | DSM 6718T | 1 |
| S. condimenti | 0 | 0 | DSM 11674T | 1 |
| S. delphini | 0 | 0 | DSM 20771T | 1 |
| S. epidermidis | 45 | 5 | DSM 20044T | 51 |
| S. equorum subsp. equorum | 0 | 0 | DSM 20674T, DSM 20675 | 2 |
| S. equorum subsp. linens | 0 | 0 | DSM 15097T | 1 |
| S. felis | 0 | 0 | DSM 7377T | 1 |
| S. fleurettii | 0 | 0 | DSM 13212T | 1 |
| S. gallinarum | 0 | 0 | DSM 20610T | 1 |
| S. haemolyticus | 14 | 1 | DSM 20263T | 16 |
| S. hominis subsp. hominis | 9 | 0 | DSM 20328T | 10 |
| S. hominis subsp. novobiosepticus | 0 | 0 | ATCC 700236T | 1 |
| S. hyicus | 0 | 1 | DSM 20459T | 2 |
| S. intermedius | 1 | 10 | DSM 20373T | 12 |
| S. kloosii | 0 | 0 | DSM 20676T | 1 |
| S. lentus | 0 | 0 | DSM 20352T | 1 |
| S. lugdunensis | 1 | 19 | DSM 4804T | 21 |
| S. lutrae | 0 | 0 | DSM 10244T | 1 |
| S. muscae | 0 | 0 | DSM 7068T | 1 |
| S. nepalensis | 0 | 0 | DSM 15150T, DSM 15151 | 2 |
| S. pasteuri | 0 | 0 | DSM 10656T | 1 |
| S. pettenkoferi | 4 | 1 | CCUG 51270T | 6 |
| S. piscifermentans | 0 | 0 | DSM 7373T | 1 |
| S. pseudintermedius | 0 | 0 | LMG 22219T, LMG 22220, LMG 22221, LMG 22222 | 4 |
| S. saccharolyticus | 0 | 0 | DSM 20359T | 1 |
| S. saprophyticus subsp. bovis | 0 | 0 | CCM 4410T | 1 |
| S. saprophyticus subsp. saprophyticus | 2 | 12 | DSM 20229T | 15 |
| S. schleiferi subsp. coagulans | 0 | 0 | DSM 6628T | 1 |
| S. schleiferi subsp. schleiferi | 0 | 5 | DSM 4807T | 6 |
| S. sciuri subsp. carnaticus | 0 | 0 | ATCC 700058T | 1 |
| S. sciuri subsp. rodentium | 0 | 0 | ATCC 700061T | 1 |
| S. sciuri subsp. sciuri | 2 | 1 | DSM 20345T | 4 |
| S. simulans | 1 | 4 | DSM 20322T | 6 |
| S. succinus subsp. casei | 0 | 0 | DSM 15096T | 1 |
| S. succinus subsp. succinus | 0 | 0 | DSM 14617T | 1 |
| S. vitulinus | 0 | 0 | ATCC 51145T | 1 |
| S. warneri | 5 | 0 | DSM 20316T | 6 |
| S. xylosus | 1 | 4 | DSM 20266T | 6 |
| Total | 93 | 71 | 52 | 216 |
Isolates from blood and from other specimens (skin, wound, respiratory tract, urogenital tract, and venous catheters).
Type or reference strains of the respective (sub-)species as deposited in the American Type Culture Collection (ATCC); Czech Collection of Microorganisms (CCM); Culture Collection, University of Göteborg, Sweden (CCUG); German Collection of Microorganisms and Cell Cultures (DSM); or Belgian Coordinated Collections of Microorganisms (LMG).
Values represent total numbers of clinical isolates plus type and/or reference strain isolates.
Clinical isolates were identified on the species level by use of an ATB32 Staph biochemical test kit (BioMerieux, Marcy l'Etoile, France). When the identification of coagulase-negative staphylococci (CoNS) was ambiguous or categorized as unacceptable, universal target gene sequencing was performed as previously described (2, 7, 29). For differentiation of coagulase-positive species, in-house coagulase tube tests using rabbit plasma and rapid slide agglutination tests (Pastorex Staph-Plus, Sanofi Diagnostics Pasteur, Paris, France) were additionally performed. PCR assays for detection of S. aureus- and S. intermedius-specific nuc genes were applied for all included strains to confirm the differentiation of both species from other coagulase-positive or -variable and clumping factor-positive or -variable staphylococcal species (5, 6).
In addition, 30 type and reference strains of nonstaphylococcal gram-positive species mainly occurring in clusters were tested, comprising members of the genera Dermacoccus (D. abyssi DSM 17573T; D. barathri DSM 17574T; D. nishinomiyaensis DSM 20448T; and D. profundi DSM 17575T), Kocuria (K. aegyptia DSM 17006T; K. carniphila DSM 16004T; K. himachalensis DSM 44905T; K. kristinae DSM 20032T; K. marina JCM 13363T; K. palustris DSM 11925T; K. polaris DSM 14382T; K. rosea DSM 20447T; and K. varians DSM 20033T), Kytococcus (K. schroeteri DSM 13884T and K. sedentarius DSM 20547T), Macrococcus (M. bovicus DSM 15607T; M. brunensis CCM 4811T; M. carouselicus DSM 15608T; M. caseolyticus DSM 20597T; M. equipercicus DSM 15609T; M. hajekii CCM 4809T; and M. lamae CCM 4815T), and Micrococcus (M. antarcticus JCM 11467T; M. flavus JCM 14000T; M. luteus DSM 14234 and DSM 14235; and M. lylae DSM 20315T).
Oligonucleotide primer design.
Use of the published eap-derived primer pair P2 and P3 was hampered by the fact that amplicons of different sizes were yielded due to various repeat numbers of the eap gene, resulting in size variability of the gene product (19). Therefore, new primers, referred as EAP-CON1 and EAP-CON2, were designed (Fig. 1), aligning the sequences of eap-N, eap-7, and eap-W (EMBL accession numbers AJ290973, AJ243790, and AJ245439) originating from the strain Newman D2C (ATCC 25904), strain 7 (a clinical isolate), and strain Wood 46, respectively. These primers were compared with the primers EAP-P2wt and EAP-P3wt, which are characterized by the same target nucleotide sequences as described for the published primer pair P2 and P3 but without the added enzyme restriction site sequences (Table 2).
TABLE 2.
eap-targeting oligonucleotide primers used in this study
| Name | Oligonucleotide sequence (5′→3′) | Amplicon size (bp) | Source or reference |
|---|---|---|---|
| EAP-CON1 | TAC TAA CGA AGC ATC TGC C | 230 | This study |
| EAP-CON2 | TTA AAT CGA TAT CAC TAA TAC CTC | ||
| EAP-P3wt | GCA GCT AAG CCA TTA GAT AAA TCA TCA AGT TGC TTA CAC C | Variablea | Derived from primer P3 (19)b |
| EAP-P2wt | TTA AAA TTT AAT TTC AAT GTC TAC TTT TTT AAT GTC |
Strain-specific amplicon size depending on the number of tandem repeats in the eap gene.
The primer sequences were modified by omitting the originally introduced restriction sites.
For verification purposes, the five clinical S. aureus isolates tested previously and shown to be eap negative by using the primer pair P2 and P3 (19) were reanalyzed by applying the EAP-CON1 and EAP-CON2 primer pair.
PCR procedures.
DNA was isolated as previously described (4). The diagnostic PCR assays were performed for both target genes in a thermal cycler with a hot bonnet (iCycler; Bio-Rad, Munich, Germany). The PCR mixture consisted of 1 μg DNA, 10 mM Tris-HCl [pH 8.3], 10 mM KCl, 3 mM MgCl2, 1 μM primer, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 5 U of Taq DNA polymerase (Qbiogene, Heidelberg, Germany), and double-distilled water added to achieve a final volume of 50 μl. A total of 30 PCR cycles were run under the following conditions: DNA denaturation at 95°C for 1 min (5 min for the first cycle), primer annealing at 50°C for 1 min, and DNA extension at 72°C for 2 min. After the final cycle, the reaction was terminated by holding at 72°C for 10 min. A 10-μl volume of the PCR product was analyzed using a 1% ethidium bromide-stained agarose gel.
Southern blot analysis.
DNA was restricted with EcoRI, separated on 1% agarose gel, blotted onto a nylon membrane, and probed with digoxigenin (DIG)-labeled PCR products of eap.
RNA isolation and Northern blot analysis.
For total RNA isolation, bacteria were grown in 5 ml brain heart infusion broth for 18 h at 37°C with shaking at 160 rpm. Bacteria were pelleted by centrifugation at 4,000 rpm for 10 min at 4°C followed by RNA isolation using an RNeasy kit (Qiagen) according to the manufacturer's recommendations. Isolated total RNA was separated on a 1% agarose gel containing 15% formaldehyde, blotted onto a polyvinylidene difluoride membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and fixed with UV light. The membrane was probed with an eap-PCR amplicon generated by primers EAP-P2wt and EAP-P3wt and prepared using DIG-labeled deoxynucleoside triphosphates. Subsequently, blots were exposed to anti-DIG antibodies (Roche) and developed with a color reaction according to the manufacturer's recommendations.
SDS-PAGE and Western blot analysis.
Eap protein was detected as described earlier (19, 20). Briefly, to prepare cell surface proteins, staphylococci were grown in 5 ml brain heart infusion broth at 37°C for 18 h and then centrifuged at 10,000 × g for 2 min. The pellet was resuspended in extraction buffer (125 mM Tris-HCl [pH 7.0]-2% sodium dodecyl sulfate [SDS; Sigma-Aldrich]), heated at 95°C for 3 min, and then centrifuged at 10,000 × g for 3 min. The supernatant was passed through a Nap-10 column (Amersham Pharmacia Biotech Europe, Freiburg, Germany) containing Sephadex G-25 to remove SDS, and the eluate was stored at −20°C. A 50-μl volume of 5× sample buffer (60 mM Tris-HCl [pH 6.8], 25% glycerol, 2% SDS, 14.4 mM 2-mercaptoethanol, and 0.1% bromophenol blue [Merck]) was added to 20 μl of cell surface extract, and the mixture was heated at 95°C for 3 min and then separated in an SDS-polyacrylamide gel electrophoresis (SDS-PAGE) minigel. For Western blot analysis, proteins separated by SDS-PAGE were electrophoretically transferred (Trans-blot SD; Bio-Rad, Munich, Germany) onto a nitrocellulose membrane (Schleicher & Schüll, Dassel, Germany), and then the membrane was blocked with 3% bovine serum albumin fraction V powder (Sigma, Taufkirchen, Germany). For probing blocked blots, anti-Eap was used, and detection was subsequently performed using alkaline-phosphatase-conjugated anti-rabbit antibody (Dako, Hamburg, Germany) developed in sheep and an alkaline phosphatase color reaction kit (Bio-Rad, Munich, Germany).
Polyclonal antibodies.
Recombinant Eap (rEap) was expressed and purified as described earlier (19, 20). Polyclonal antibodies against the recombinant protein were raised separately in two rabbits by standard procedures (17). After preimmune serum samples were collected, each rabbit was immunized subcutaneously with 50 μg of rEap derived from strain Newman in complete Freund's adjuvant (Sigma). Second and third injections of antigen in incomplete adjuvant were given subcutaneously 2 and 4 weeks later, respectively. Blood was obtained 2 weeks after the last antigen injection. Naturally occurring antistaphylococcal antibodies were complexed by mixing serum with 100 volumes of cell surface protein extract (prepared by heating whole cells in 2% SDS at 95°C for 3 min and by pelleting bacteria by centrifugation) from S. aureus strain AH 12 (isogenic eap disruption mutant of strain Newman), which does not express Eap.
RESULTS
Prevalence of eap in S. aureus.
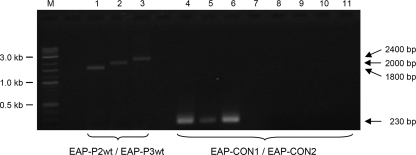
By applying the newly designed primers EAP-CON1 and EAP-CON2 in the PCR assay, a single amplification product of the eap gene showing the expected size of 230 bp was detected in all 588 clinical isolates and nine reference and laboratory strains of S. aureus tested (Fig. 2). Thus, the sensitivity of the newly developed PCR approach was 100%. Due to the primer selection targeting the first repeat domain only, different sizes of the PCR products known to be targeted by primer pairs P2 and P3 and primer pairs EAP-P2wt and EAP-P3wt, respectively, were not observed.
FIG. 2.
Agarose gel (1%) electrophoresis patterns showing PCR products amplified with eap primers by use of genomic DNA of S. aureus ATCC 29213 (lanes 1 and 4), S. aureus Newman (lanes 2 and 5), and S. aureus Wood 46 (lanes 3 and 6) as well as genomic DNA of S. epidermidis DSM 20044 (lane 7), S. haemolyticus DSM 20263 (lane 8), S. hominis subsp. hominis DSM 20238 (lane 9), S. intermedius DSM 20373 (lane 10), and S. lugdunensis DSM 4804 (lane 11). Lane M, DNA molecular mass marker (1-kb/100-bp DNA ladder); lanes 1 to 3, eap gene amplified with primers EAP-P2wt and EAP-P3wt; lanes 4 to 11, eap gene amplified with newly designed primers EAP-CON1 and EAP-CON2 (see Table 2).
Moreover, the five clinical S. aureus isolates tested to be eap negative in previous study by using the primer pair P2 and P3 (19) were now found to be eap positive by applying the newly designed primer pair (data not shown).
Prevalence of eap in non-S. aureus bacteria.
To analyze the specificity of the newly designed eap-specific primers, the prevalence of the eap gene among 216 non-S. aureus staphylococcal isolates comprising 47 different CoNS and coagulase-positive or -variable staphylococcal (sub-)species was investigated at the DNA level. The PCR presented here showed 100% specificity; i.e., the eap gene was not detected in 216 clinical isolates, including a large number of CoNS isolated from blood and considered etiologically relevant, or in 52 staphylococcal type and reference strains obtained from various strain collections (Fig. 2).
In addition, all other gram-positive cocci included in this study (micrococcal and macrococcal species) were tested to be eap negative.
Southern blot analysis.
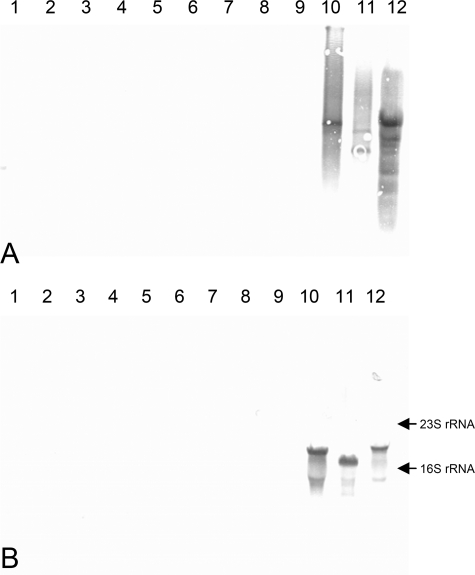
To exclude the possibility that DNA sequence heterogeneities of the primer binding sites might cause false-negative results with non-S. aureus isolates, Southern blot analyses were performed. Genomic DNAs of 20 representative non-S. aureus staphylococcal strains, including S. epidermidis (n = 10), S. saprophyticus subsp. saprophyticus (n = 3), S. haemolyticus (n = 3), S. carnosus subsp. carnosus (n = 1), S. hominis subsp. hominis (n = 1), S. capitis (n = 1), and S. xylosus (n = 1) were restricted with EcoRI, separated on 1% agarose gel, blotted onto a nylon membrane, and probed with a DIG-labeled PCR product of eap from strain Newman by use of primers EAP-P3wt and EAP-P2wt. All non-S. aureus isolates tested showed negative results with DIG-labeled eap PCR products, but positive results were seen with genomic DNA of S. aureus control strains Newman, Mu-7, and Wood 46 (Fig. 3A).
FIG. 3.
Southern blot analysis of genomic DNA (A) and Northern blot of total RNA (B). Lanes 1 to 4, S. epidermidis; lane 5, S. carnosus subsp. carnosus; lane 6, S. haemolyticus; lane 7, S. hominis subsp. hominis; lanes 8 to 9, S. saprophyticus subsp. saprophyticus; lanes 10 to 12, S. aureus (lane 10, strain Newman; lane 11, strain CI7; lane 12, strain Wood 46). Blots were prepared as described in Materials and Methods.
Transcription analysis of eap gene.
The same 20 non-S. aureus strains used for Southern blot analysis were selected for Northern blot analysis of eap gene transcripts. Whereas eap mRNA signals for strains Newman, CI7, and wood 46 were positive, mRNA signals of all non-S. aureus strains were absent (Fig. 3B).
SDS-PAGE and Western blot analysis.
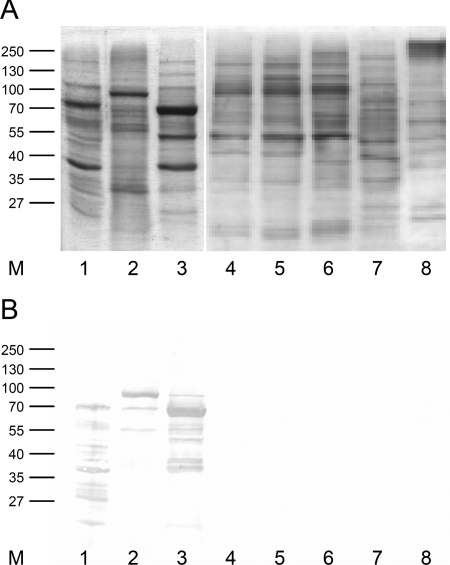
A set of 20 representative strains was analyzed for the expression of Eap protein. In Coomassie blue-stained SDS-PAGE gels, non-S. aureus strains tested produced a range of proteins in quantities similar to those seen with S. aureus strains Newman, CI7, and Wood 46. However, the band corresponding to Eap was not detected in any of the 20 tested non-S. aureus strains (Fig. 4A).
FIG. 4.
SDS-PAGE (A) and Western immunoblot (B) analysis of SDS surface protein extracts. Lane M, marker; lanes 1 to 3, S. aureus strains CI7 (lane 1), Wood 46 (lane 2), and Newman (lane 3); lanes 4 to 8, strains of non-S. aureus staphylococcal species S. epidermidis (lanes 4 and 5), S. carnosus subsp. carnosus (lane 6), S. haemolyticus (lane 7), and S. hominis subsp. hominis (lane 8). Blots were prepared as described in Materials and Methods.
In Western blot analyses, anti-Eap antiserum used at a dilution of 1:20,000 recognized a 60-kDa Eap protein in cell surface extracts from strains Newman and CI7 and a 70-kDa Eap protein in cell surface extracts of strain Wood 46. In contrast, non-S. aureus staphylococcal isolates did not produce any proteins detectable by anti-Eap antiserum (Fig. 4B).
DISCUSSION
For S. aureus, secreted proteins such as Eap have pleiotropic effects, mediating adherence to host extracellular matrix components (e.g., fibronectin, fibrinogen, vitronectin, collagens, and elastin) and thereby potentially contributing to bacterial colonization of host tissues (10) and, in part, to invasion of nonprofessional phagocytes (19-21). In addition, Eap has anti-inflammatory and immunomodulatory functions in the host, indicating that Eap constitutes a potent virulence factor in the course of staphylococcal infections (1, 8, 15, 18, 25). Thus, Eap acts as a secretable expanded repertoire adhesive molecule (9).
In this study, the Eap-encoding gene (eap) was studied as a diagnostic target for specific identification of S. aureus by PCR amplification. Whereas sequencing results suggest that the eap sequences are in general highly conserved, size differences are known for the encoding gene, as a result of different numbers of repeats, as well as for the translated product, as additionally determined by assays using an insertional point mutation or ribosomal slippage at the site of a poly(A) region located at the 3′ end of the respective repeat region (19). Thus, the described PCR primer pairs generated to study the size variants (19) do not fulfill requirements (i) to detect eap homologs in other staphylococcal species and (ii) to use this gene as a target for specific and reliable identification of S. aureus. Consequently, a novel primer pair more suitable for diagnostic purposes was generated based on all known eap nucleotide sequences.
Based on previous observations that this gene is highly conserved among S. aureus isolates and that it was shown to be absent in a small collection of S. epidermidis isolates (19, 20), large collections of staphylococcal species and subspecies comprising different CoNS as well as coagulase-positive or -variable and/or clumping factor-positive or -variable staphylococci from various clinical and geographical sources were analyzed for the presence of the eap gene and gene product homologs on the DNA, transcriptional, and protein levels. To our knowledge, this is currently one of the most comprehensive collections of staphylococcal isolates tested for validation of a molecular diagnostic target. It is noteworthy that the clinical isolates tested previously to be eap negative (19) have now in fact been shown to harbor the eap gene, demonstrating the superiority of the newly designed primer pair to former applied primers. It also stresses the so far unexceptional possession of this gene by S. aureus.
For detection of putative differences between CoNS strains recovered from the physiological skin floras and those strains involved in clinical significant infections, isolates were included from a German multicenter study in which only CoNS strains considered etiologically relevant were collected. However, when the newly designed PCR primers were used, all of the non-S. aureus staphylococci were shown to be eap negative. In order to exclude negative PCR results due to potential DNA sequence heterogeneities of the primer binding sites, Southern blot analyses were performed with DIG-labeled eap PCR amplicons. Of particular interest, Southern blot analyses as well as Northern and Western blot analyses gave no positive results for non-S. aureus isolates. Hence, the existence of Eap homologs in other staphylococcal species is highly unlikely. Given the pleiotropic biologic effects of Eap and the prevalence value of 100% for the eap gene, one may speculate that this secretable expanded repertoire adhesive molecule may be of crucial importance to S. aureus. However, currently it is unclear why only 70% of the isolates tested expressed Eap under in vitro conditions (19) and whether this correlates with specific requirements in a given ecological niche.
In the light of the ongoing increase of the prevalence of methicillin-resistant S. aureus strains in hospitals and other health care facilities worldwide (14) and the advent, dissemination, and emergence of the highly virulent Panton-Valentine leukocidin-positive S. aureus clones (39), rapid and accurate identification of S. aureus infections and/or colonization is therefore a prerequisite for adequate and timely therapy, disease control, and effective epidemiological surveillance. Presently, phenotypic detection and identification of S. aureus are standard procedures in routine clinical microbiology. However, identification based on morphological characteristics, metabolic pathways, growth factor requirements, or antigenic composition depends fundamentally on expression of genetic information influenced by various factors and often involves nonobjective criteria for interpretation of results. Consequently, manifold molecular strategies have been developed to overcome the limitations of the traditional identification approaches. In addition to universal gene targets based on the ribosomal gene cluster or on the RNA polymerase B (rpoB) (2, 29, 31, 34), several specific targets, such as the staphylococcal coagulase gene (coa), a thermostable nuclease gene (nuc), a superoxide dismutase gene (sodM), the HSP60 gene, specific genomic DNA fragments of unknown function, and the fem factor-encoding genes (6, 13, 24, 26, 32, 37), were used for molecular detection and identification of S. aureus. However, some of these studies concentrated on only a limited number of staphylococcal species. Additionally, target gene-negative S. aureus strains as well as target gene homologs in CoNS were observed, hampering both the sensitivity and the specificity of these molecular approaches (22, 24, 36). Here, we demonstrate that targeting the eap gene allows appending the diagnostic toolbox. This gene appears to be particularly suitable for molecular diagnostics of S. aureus. Based on these data, the sensitivity and specificity values for the eap gene were both remarkably high (i.e., 100%).
In conclusion, Eap was shown to be a virulence factor specifically restricted to S. aureus and occurring in all isolates of this species tested. In contrast, CoNS and other coagulase-positive or -variable staphylococcal species were shown to miss the encoding gene as well as homologs of this factor as shown at the DNA, transcriptional, and protein levels. Thus, the presence of Eap seems to be a core trait of S. aureus, offering a promising target for sensitive and specific detection of S. aureus.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Collaborative Research Center 492, project B9, and in part by a research grant from the German Federal Ministry for Education and Science (BMBF) in the context of the Pathogenomic Plus Network (PTJ-BIO/0313801B), by DFG Priority Programme 1130 (He 1850/7-2), and by a grant of the Medical Faculty of the University of Saarland (HOMFOR).
We are grateful to the participants of the various German multicenter studies. We thank also Z. Berek (Budapest, Hungary), G. Blaiotta (Portici-Naples, Italy), J. Etienne (Lyon, France), A. Heide (Hannover, Germany), K. Hiramatsu (Tokyo, Japan), G. Lina (Lyon, France), R.A. Proctor (Madison, WI), and B. Sadowska (Lódź, Poland) for providing S. aureus strains.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Athanasopoulos, A. N., M. Economopoulou, V. V. Orlova, A. Sobke, D. Schneider, H. Weber, H. G. Augustin, S. A. Eming, U. Schubert, T. Linn, P. P. Nawroth, M. Hussain, H. P. Hammes, M. Herrmann, K. T. Preissner, and T. Chavakis. 2006. The extracellular adherence protein (Eap) of Staphylococcus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood 1072720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S rDNA-based identification of Staphylococcus species. J. Clin. Microbiol. 424988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, K., B. Keller, C. von Eiff, M. Brück, G. Lubritz, J. Etienne, and G. Peters. 2001. Enterotoxigenic potential of Staphylococcus intermedius. Appl. Environ. Microbiol. 675551-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 362548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K., C. von Eiff, B. Keller, M. Brück, J. Etienne, and G. Peters. 2005. Thermonuclease gene as a target for specific identification of Staphylococcus intermedius isolates: use of a PCR-DNA enzyme immunoassay. Diagn. Microbiol. Infect. Dis. 51237-244. [DOI] [PubMed] [Google Scholar]
- 6.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 301654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkopf, C., D. Hammel, H. H. Scheld, G. Peters, and K. Becker. 2005. Impact of a molecular approach to improve the microbiological diagnosis of infective heart valve endocarditis. Circulation 1111415-1421. [DOI] [PubMed] [Google Scholar]
- 8.Chavakis, T., M. Hussain, S. M. Kanse, G. Peters, R. G. Bretzel, J. I. Flock, M. Herrmann, and K. T. Preissner. 2002. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 8687-693. [DOI] [PubMed] [Google Scholar]
- 9.Chavakis, T., K. Wiechmann, K. T. Preissner, and M. Herrmann. 2005. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb. Haemost. 94278-285. [DOI] [PubMed] [Google Scholar]
- 10.Flock, J. I. 1999. Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol. Med. Today 5532-537. [DOI] [PubMed] [Google Scholar]
- 11.Geisbrecht, B. V., B. Y. Hamaoka, B. Perman, A. Zemla, and D. J. Leahy. 2005. The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens. J. Biol. Chem. 28017243-17250. [DOI] [PubMed] [Google Scholar]
- 12.Goh, S. H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 301642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 353116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 15.Haggar, A., M. Hussain, H. Lonnies, M. Herrmann, A. Norrby-Teglund, and J. I. Flock. 2003. Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect. Immun. 712310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, U., M. Hussain, D. Villone, M. Herrmann, H. Robenek, G. Peters, B. Sinha, and P. Bruckner. 2006. The anchorless adhesin Eap (extracellular adherence protein) from Staphylococcus aureus selectively recognizes extracellular matrix aggregates but binds promiscuously to monomeric matrix macromolecules. Matrix Biol. 25252-260. [DOI] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Harraghy, N., M. Hussain, A. Haggar, T. Chavakis, B. Sinha, M. Herrmann, and J. I. Flock. 2003. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology 1492701-2707. [DOI] [PubMed] [Google Scholar]
- 19.Hussain, M., K. Becker, C. von Eiff, G. Peters, and M. Herrmann. 2001. Analogs of Eap protein are conserved and prevalent in clinical Staphylococcus aureus isolates. Clin. Diagn. Lab. Immunol. 81271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 1836778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrmann. 2002. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 702933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonas, D., H. Grundmann, D. Hartung, F. D. Daschner, and K. J. Towner. 1999. Evaluation of the mecA femB duplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18643-647. [DOI] [PubMed] [Google Scholar]
- 23.Jönsson, K., D. McDevitt, M. H. McGavin, J. M. Patti, and M. Höök. 1995. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol. Chem. 27021457-21460. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, N., H. Wu, K. Kojima, K. Taniguchi, S. Urasawa, N. Uehara, Y. Omizu, Y. Kishi, A. Yagihashi, and I. Kurokawa. 1994. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol. Infect. 113259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, L. Y., Y. J. Miyamoto, B. W. McIntyre, M. Höök, K. W. McCrea, D. McDevitt, and E. L. Brown. 2002. The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell-mediated responses. J. Clin. Investig. 1101461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1998. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 36618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason, W. J., J. S. Blevins, K. Beenken, N. Wibowo, N. Ojha, and M. S. Smeltzer. 2001. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 393332-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massey, R. C., T. J. Scriba, E. L. Brown, R. E. Phillips, and A. K. Sewell. 2007. Use of peptide-major histocompatibility complex tetramer technology to study interactions between Staphylococcus aureus proteins and human cells. Infect. Immun. 755711-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellmann, A., K. Becker, C. von Eiff, U. Keckevoet, P. Schumann, and D. Harmsen. 2006. Sequencing and staphylococci identification. Emerg. Infect. Dis. 12333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology, vol. 1 and 2, p. 1-2310. ASM Press, Washington, DC. [Google Scholar]
- 31.Saruta, K., S. Hoshina, and K. Machida. 1995. Genetic identification of Staphylococcus aureus by polymerase chain reaction using single-base-pair mismatch in 16S ribosomal RNA gene. Microbiol. Immunol. 39839-844. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz, F. J., C. R. MacKenzie, B. Hofmann, J. Verhoef, M. Finken-Eigen, H. P. Heinz, and K. Köhrer. 1997. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by a multiplex PCR. J. Med. Microbiol. 46773-778. [DOI] [PubMed] [Google Scholar]
- 33.Sobke, A. C., D. Selimovic, V. Orlova, M. Hassan, T. Chavakis, A. N. Athanasopoulos, U. Schubert, M. Hussain, G. Thiel, K. T. Preissner, and M. Herrmann. 2006. The extracellular adherence protein from Staphylococcus aureus abrogates angiogenic responses of endothelial cells by blocking Ras activation. FASEB J. 202621-2623. [DOI] [PubMed] [Google Scholar]
- 34.Straub, J. A., C. Hertel, and W. P. Hammes. 1999. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Food Prot. 621150-1156. [DOI] [PubMed] [Google Scholar]
- 35.Trülzsch, K., B. Grabein, P. Schumann, A. Mellmann, U. Antonenka, J. Heesemann, and K. Becker. 2007. Staphylococcus pettenkoferi sp. nov., a novel coagulase-negative staphylococcal species isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 571543-1548. [DOI] [PubMed] [Google Scholar]
- 36.Ünal, S., J. Hoskins, J. E. Flokowitsch, C. Y. Wu, D. A. Preston, and P. L. Skatrud. 1992. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J. Clin. Microbiol. 301685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valderas, M. W., J. W. Gatson, N. Wreyford, and M. E. Hart. 2002. The superoxide dismutase gene sodM is unique to Staphylococcus aureus: absence of sodM in coagulase-negative staphylococci. J. Bacteriol. 1842465-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valderas, M. W., and M. E. Hart. 2001. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J. Bacteriol. 1833399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters (Study Group). 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 34411-16. [DOI] [PubMed] [Google Scholar]
- 41.von Eiff, C., R. R. Reinert, M. Kresken, J. Brauers, D. Hafner, and G. Peters for the Multicenter Study on Antibiotic Resistance in Staphylococci and Other Gram-Positive Cocci Study (MARS) Group. 2000. Nationwide German multicenter study on prevalence of antibiotic resistance in staphylococcal bloodstream isolates and comparative in vitro activities of quinupristin-dalfopristin. J. Clin. Microbiol. 382819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, I. G., J. E. Cooper, and A. Gilmour. 1991. Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Appl. Environ. Microbiol. 571793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]