Abstract
Acinetobacter spp. have emerged as important nosocomial and multidrug-resistant pathogens in the last decade. A. calcoaceticus, A. baumannii, Acinetobacter genospecies 3, and Acinetobacter genospecies 13TU are genetically closely related and are referred to as the A. calcoaceticus-A. baumannii complex (ACB complex). Distinct Acinetobacter spp. may be associated with differences in antimicrobial susceptibility, so it is important to identify Acinetobacter spp. at the species level. We developed a microsphere-based array that combines an allele-specific primer extension assay and microsphere hybridization for the identification of Acinetobacter spp. This assay can discriminate the 13 different Acinetobacter spp. in less than 8.5 h, and it has high specificity without causing cross-reactivity with 14 other common nosocomial bacterial species. The sensitivity of this assay was 100 A. baumannii cells per ml of blood, and it could discriminate multiple species in various mixture ratios. The developed assay could differentiate clinical Acinetobacter spp. isolates with a 90% identification rate. The antimicrobial susceptibility test showed that A. baumannii isolates were resistant to most antimicrobial agents other than imipenem, while the genospecies 3 and 13TU isolates were more susceptible to most antimicrobial agents, especially ciprofloxacin and ampicillin-sulbactam. These results supported the idea that this assay possibly could be applied to clinical samples and provide accurate species identification, which might be helpful for clinicians when they are treating infections caused by Acinetobacter spp.
During the last decade, Acinetobacter spp. have become the major cause of nosocomial infections. The emergence and spread of multidrug-resistant (MDR) Acinetobacter spp. pose an even greater threat to hospitals. The resistance of MDR Acinetobacter spp. to many commonly used antibiotics, such as aminoglycosides, fluoroquinolones, cephalosporins, β-lactams, and carbapenems, has been increasingly reported worldwide (7, 12, 15, 22, 24). MDR Acinetobacter spp. causing nosocomial infections were first reported in Taiwan in 1998 (12). Furthermore, a significant 3.6-fold increase in nosocomial bloodstream infections and a 2.1-fold increase in all nosocomial infections caused by Acinetobacter spp. were noted from 1991 to 2003 (11). MDR Acinetobacter spp. have become endemic and constitute a therapeutic problem in hospitals in Taiwan (11, 12, 34).
At least 32 named and unnamed Acinetobacter spp. have been described (31). Genospecies 1 (A. calcoaceticus), genospecies 2 (A. baumannii), genospecies 3, and genospecies 13TU are genetically closely related and are referred to as the A. calcoaceticus-A. baumannii complex (ACB complex). Among the members of the ACB complex, A. baumannii, genospecies 3, and genospecies 13TU have been implicated in nosocomial infection outbreaks (2). The proper identification of Acinetobacter spp. at the species level is important for the application of the appropriate therapy to infections, because differences in antimicrobial efficacy against strains belonging to different species have been demonstrated (26, 28).
To identify Acinetobacter spp., many clinical microbiological laboratories routinely use commercial phenotypic methods, but they are unreliable when clinicians are identifying Acinetobacter spp. to the species level (9). Therefore, to substitute for phenotypic methods, several molecular methods have been developed for Acinetobacter species identification, including amplified 16S ribosomal DNA restriction analysis (30); ribotyping (8); randomly amplified polymorphic DNA; the sequencing of various genes, such as the 16S-23S rRNA gene intergenic spacer (ITS) region (3), the recA gene (16), and the rpoB gene (18); and amplified fragment length polymorphism fingerprinting (14). However, these methods usually are labor-intensive, time-consuming, or of low reproducibility. Furthermore, they usually need multiple-tube PCR, thus requiring more DNA.
A novel method with convenient, rapid, and multiplexed properties is desirable to overcome these limitations. The microsphere-based array provides the capacity for conducting up to 100 biological reactions simultaneously in a single reaction vessel, and it combines the specificity and reliability of oligonucleotide hybridization analysis with the speed and sensitivity of a flow cytometer (36). Furthermore, this method has been applied reliably to species identification (23), the genotyping (6) of pathogens, and mutation detection (29). In this study, we developed a microsphere-based array for the identification of Acinetobacter spp. The antimicrobial susceptibilities of the clinical Acinetobacter species isolates also were analyzed.
MATERIALS AND METHODS
Bacterial strains.
A total of 163 isolates, including 105 Acinetobacter species and 58 non-Acinetobacter species isolates, were used. Of the 105 Acinetobacter species strains, 13 were reference strains, including 4 strains of the ACB complex and 9 strains of other named or unnamed Acinetobacter spp. (Table 1), and 92 strains were clinical isolates identified as A. baumannii using the API 20NE system. Of the 92 Acinetobacter species clinical isolates, each was from a different patient, and no two isolates were from the same outbreak according to clinical history and pulsed-field gel electrophoresis genotyping (data not shown). Fifty-eight non-Acinetobacter species clinical isolates were used to detect the specificity. They belonged to 14 species, including Enterobacter aerogenes (n = 3 strains), Enterobacter cloacae (n = 6), Enterococcus faecalis (n = 6), Enterococcus faecium (n = 6), Escherichia coli (n = 3), Klebsiella pneumoniae (n = 3), Klebsiella oxytoca (n = 5), Proteus mirabilis (n = 4), Pseudomonas aeruginosa (n = 5), Staphylococcus aureus (n = 5), Staphylococcus capitis (n = 2), Staphylococcus epidermidis (n = 3), Staphylococcus haemolyticus (n = 1), and Stenotrophomonas maltophilia (n = 6). Reference strains were obtained from the Bioresource Collection and Research Center (Hsinchu, Taiwan), and all clinical isolates were obtained from the National Taiwan University Hospital (Taipei, Taiwan). All isolates were cultured on tryptic soy agar plates at 37°C for 20 h.
TABLE 1.
Acinetobacter reference strains used in this study
| Species | BCRC strain no.a | ATCC strain no. | ITS size (bp) | Accession no. |
|---|---|---|---|---|
| A. calcoaceticus | 11562 | 14987 | 637 | AY601820 |
| A. baumannii | 10591 | 19606 | 607 | AY601823 |
| Genospecies 3 | 15420 | 17922 | 619 | AY601829 |
| A. haemolyticus | 14852 | 17906 | 614 | AY601831 |
| A. junii | 14854 | 17908 | 706 | AY601832 |
| Genospecies 6 | 15421 | 17979 | 636 | AY601833 |
| A. johnsonii | 14853 | 17909 | 703 | AY601834 |
| A. lwoffii | 14855 | 15309 | 629 | AY601835 |
| Genospecies 10 | 15423 | 17942 | 613 | AY601837 |
| Genospecies 11 | 15424 | 11171 | 593 | AY601838 |
| A. radioresistens | 15425 | 43998 | 632 | AY601839 |
| Genospecies 13TU | 15417 | 17903 | 615 | AY601830 |
| Genospecies 16 | 15883 | 17988 | 595 | AY601844 |
BCRC, Bioresources Collection and Research Center, Hsichu, Taiwan.
Species identification on the microsphere-based array.
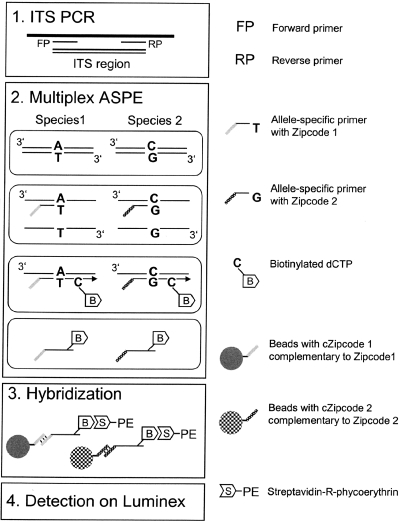
An overview of the microsphere-based array is given in Fig. 1.
FIG. 1.
Overview of species identification by microsphere-based array. (Step 1) ITS PCR. The ITS region of 13 distinct Acinetobacter species was amplified, and the PCR product was treated with shrimp alkaline phosphatase and exonuclease I. (Step 2) Multiplex ASPE. ASPE primers with specific ZipCode sequences overlapped the SNP site in the ITS region, and only the correctly hybridized primer was extended. ASPE primers were extended by Tsp DNA polymerase, and biotinylated dCTP was incorporated into the extended primers. (Step 3) Hybridization. The extended primers with specific ZipCodes were hybridized with specific beads with cZipCodes. Streptavidin-R-phycoerythrin bound to the biotin in the ASPE primers. (Step 4) Detection on the flow cytometer. Specific beads were sorted, and the fluorescent signals of phycoerythrin were measured.
(i) PCR.
The total genomic DNA of the strain was extracted as described previously (35). The bacterium-specific universal primers used were the following: forward primer, 5′-GTCGTAACAAGGTAGCCGTA-3′; reverse primer, 5′-GGGTTYCCCCRTTCRGAAAT-3′ (where Y is C or T and R is A or G). The primers were used to amplify a DNA fragment encompassing part of the 16S rRNA gene region, the ITS, and part of the 23S rRNA gene region as previously described (3).
To remove unincorporated deoxynucleotide triphosphates and primers, we added 1 μl of shrimp alkaline phosphatase (1 U/μl; USB) and 1 μl of exonuclease I (10 U/μl; USB) to each 20 μl of PCR product. The samples were incubated at 37°C for 30 min and then inactivated at 80°C for 15 min. The purified PCR products then were used as templates for the primer extension reactions.
(ii) Multiplex ASPE.
The primers for multiplex allele-specific primer extension (ASPE) and their target species are shown in Table 2. All of the extension primers were designed to possess a melting point of 50 to 57°C, and each primer was appended at the 5′ end with a 25-mer ZipCode oligonucleotide (4, 13). Reverse complements of the 25-mer ZipCode (cZipCode) oligonucleotides were attached to the given bead sets as described above. An Acinetobacter genus-specific extension primer (UniA) was added to each reaction mixture to serve as a measurement of PCR amplification and primer extension success for each sample.
TABLE 2.
ASPE species-specific primers and measurement values
| Primer | Identified species | ZipCodea | Primer sequence | Signal range (MFI)
|
Minimum ratiod | |
|---|---|---|---|---|---|---|
| Negativeb | Positivec | |||||
| P-1 | A. calcoaceticus | 20 | CCTACAAGGAGTAATAAGACATGA | 0-26 | 2,792-3,523 | 107.4 |
| P-2 | A. baumannii | 17 | GATCTTGGTTTATTAACTTCTGTGATTTCAT | 0-445 | 9,831-11,011 | 22.1 |
| P-3 | Genospecies 3 | 27 | ACCCCAAACAGTCGTCAAC | 7-282 | 6,069-6,256 | 21.5 |
| P-4 | A. haemolyticus | 3 | GAATACAGTCTAAGTTGACTGGTTTG | 0-31 | 4,916-5,963 | 158.6 |
| P-5 | A. junii | 14 | GATGAATAATCACAAGCTGCTAG | 0-31 | 14,445-15,698 | 466.0 |
| P-6 | Genospecies 6 | 22 | AACAAGTTGTTCTTCTTGAAGATA | 0-45 | 5,895-7,200 | 131.0 |
| P-7 | A. johnsonii | 24 | TGCTGAATACAGAAAAACAGAG | 0-33 | 11,210-12,082 | 339.7 |
| P-8 | A. lwoffii | 37 | CTCTCCTAGTCTCCACCATC | 57-336 | 3,472-3,979 | 10.3 |
| P-10 | Genospecies 10 | 40 | TCCTAGTCTCCACCACTACTAC | 8-1,686e | 7,457-7,631 | 4.4 |
| P-11 | Genospecies 11 | 41 | GTCAACGGTTCGACTCCGT | 0-48 | 11,084-11,632 | 230.9 |
| P-12 | A. radioresistens | 44 | AGCAGTAATGCAGAAAAACAGATATG | 0-30 | 5,480-9,747 | 182.7 |
| P-13 | Genospecies 13TU | 50 | CACCATGACTTTGACTGGTTA | 0-361 | 12,153-13,457 | 33.7 |
| P-16 | Genospecies 16 | 49 | GTTGCAAACTTAATTGCTACTATTC | 0-91 | 4,962-6,162 | 54.5 |
| UniA | All species | 62 | AACTAAGCTAAGTCCCCAGC | NAf | 9,127-14,703 | NA |
Selected from the Mycobacterium tuberculosis genome (4).
Range of MFI for all strains showing negative MFI after subtracting the MFI of the PCR negative-control value for the given primer.
Range of MFI for all strains showing positive MFI after subtracting the MFI of the PCR negative-control value for the given primer.
The lowest recorded positive MFI divided by the highest recorded negative MFI.
The genospecies 5 strain cross-reacted with P-10 at about 23% of the signal of the genospecies 10 strain.
NA, not applicable.
(a) Coupling cZipCode oligonucleotides to microspheres.
A total of 2.5 × 106 carboxylated beads (Luminex, TX) per assay were pelleted, resuspended in 50 μl 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer, pH 4.5 (Sigma), and mixed with 1 mM cZipCode oligonucleotide, which contained a 5′ amino 12-carbon linker as previously described (4). A 3-μl aliquot of fresh 1-ethyl-3-3(3-3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) solution (10 mg/ml) (Pierce Biotechnology), with amine-modified cZipCodes attached to the carboxylated beads, was added to the bead-cZipCode mixture and incubated at room temperature in the dark for 30 min. Another 3-μl aliquot of fresh EDC solution was added and incubated at room temperature in the dark for 30 min. After incubation with EDC, 0.5 ml of 0.02% Tween 20 was added; the beads then were vortexed and centrifuged at 8,000 rpm for 2 min. The supernatant was aspirated, and 0.5 ml of 0.1% sodium dodecyl sulfate was added. The beads were vortexed and then centrifuged again at 8,000 rpm for 2 min. The supernatant was aspirated, and the beads were resuspended in 50 μl Tris-EDTA. The prepared beads were stored in the dark at 4°C until use.
(b) Primer extension.
The primer extension reactions were carried out in 20-μl volumes of ASPE buffer (2 mM Tris-HCl, pH 8.4, 5 mM KCl) containing 2.0 mM MgCl2; 0.75 U Tsp DNA polymerase (Invitrogen, Carlsbad, CA); 5 μM (each) dATP, dTTP, and dGTP; 5 μM biotin-dCTP (Invitrogen); and 25 nM of each extension primer. The primer extension reaction steps were the following: preheating at 96°C for 2 min, and then 35 cycles consisting of 96°C for 30 s, 55°C for 1 min, and extension at 72°C for 2 min. The samples then were held at 4°C until hybridization took place.
(iii) Hybridization.
The bead sets were diluted using 1.5× tetramethylammonium chloride (TMAC) solution (Sigma, St. Louis, MO) that contained 4.5 M TMAC, 0.15% Sarkosyl, 75 mM Tris-HCl (pH 8.0), and 6 mM EDTA (pH 8.0), such that a total of 33 μl of 1.5× TMAC solution contained 5,000 beads. The bead solution was added to 17 μl of the extension products and mixed by being pipetted up and down five times, and then it was incubated in the dark at an initial denaturation temperature of 95°C for 5 min, followed by 30 min of incubation at 40°C. After being incubated, the mixture was centrifuged at 8,000 rpm and the supernatant carefully discarded. Seventy microliters of 1× TMAC solution containing 10 ng/μl streptavidin-R-phycoerythrin (Molecular Probes, Eugene, OR) then was added to each sample and incubated for 10 min in the dark at 40°C.
(iv) Detection on flow cytometer and calculation.
Samples were measured on the basis of fluorescence intensity in a Bio-Plex 200 suspension array system (Bio-Rad Laboratories, Inc., Hercules, CA). The median fluorescence intensities (MFI) were calculated from 100 replicate measurements with a digital signal processor and Bio-Plex Manager 4.1.1 software. The minimal ratio is the lowest recorded positive MFI divided by the highest recorded negative MFI. Values twice the minimal ratio were used as a threshold for defining positive events.
Sensitivity and multiple-species detection.
The sensitivity of the microsphere-based array was tested by spiking pooled blood from three healthy individuals with serial dilutions of A. baumannii cells ranging from 1 to 104 CFU per ml and serial dilutions of genomic DNA ranging from 10 to 10−4 ng. The total DNA of each sample was extracted using a QIAamp DNA blood kit.
To test the multiple-species detection capability, 10 pg of genomic DNA from genospecies 3 and 13TU was used, while the amount of A. baumannii was increased gradually from 100- to 10,000-fold to simulate the inconsistent ratios of mixed infections.
Antimicrobial susceptibility testing.
MICs of the antimicrobial agents ciprofloxacin, imipenem, meropenem, ampicillin-sulbactam, aztreonam, gentamicin, cefazidime, and cefepime for the clinical isolates were determined by an agar dilution method (32) according to guidelines of the Clinical and Laboratory Standards Institute (5).
RESULTS
Confirmation of ITS PCR products.
ITS products were electrophoresed on 2% agarose gels to confirm the success of PCR amplification. ITS fragments of 593 to 706 bp were successfully amplified from 13 reference strains belonging to different species as well as 92 clinical Acinetobacter species isolates (data not shown).
ASPE assay and specificity.
Table 2 shows the negative and positive ranges for all species-specific ASPE primers and minimum ratios. The positive MFI of 13 species-specific primers ranged from 2,792 to 15,698, with minimum ratios from 4.4 to 466. The Acinetobacter genus-specific primer (UniA) was positive for all 13 reference strains, and the MFI ranged from 9,127 to 14,703. The species-specific primer P-10 generated a low minimum ratio of 4.4, because it partially cross-reacted with genospecies 5. In addition, we also tested the specificity of species-specific ASPE primers for the other 14 non-Acinetobacter bacterial species that frequently cause nosocomial infections (see Fig. S1 in the supplemental material). No cross-reactivity with the 58 non-Acinetobacter isolates was found. This suggested that all species-specific ASPE primers have a high specificity for the identification of Acinetobacter spp.
Sensitivity and multiple-species detection.
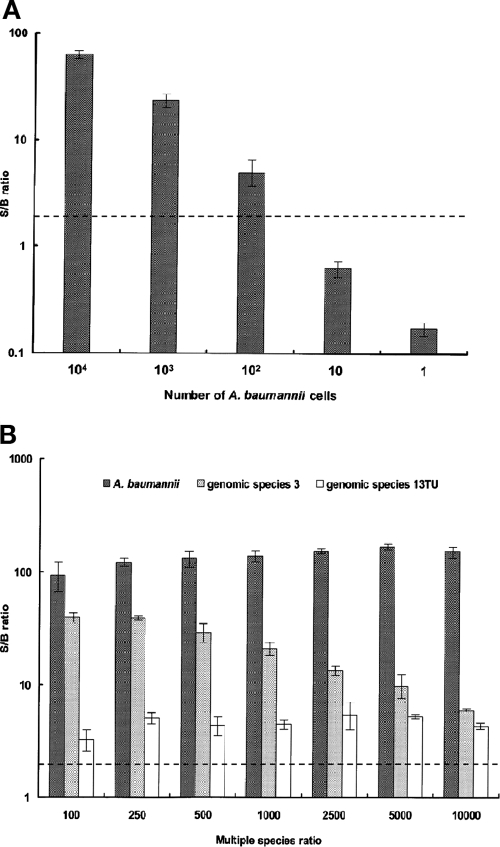
To determine the sensitivity of the ASPE assay, a distinct number of A. baumannii cells (from 1 to 104 CFU per assay) were spiked into pooled human blood and detected by the ASPE assay (Fig. 2A). By relying on the 2× minimal ratio, the sensitivity of the ASPE assay was as few as 100 CFU per ml of blood, and the smallest amount of genomic DNA required for detection for all four species of the ACB complex was 1 pg (data not shown). To evaluate whether the ASPE assay could accurately detect individual species in a mixture of multiple species, genomic DNA from three major Acinetobacter clinical strains, A. baumannii, genospecies 3, and genospecies 13TU, were mixed in various ratios, and PCR amplification and an ASPE assay were conducted (Fig. 2B). Despite the fluorescence signals of genospecies 3 and 13TU being partially influenced by the presence of DNA from other species, the results of the ASPE assay clearly discriminated the three Acinetobacter species from the multiple DNA sources at inconsistent ratios, all exceeding the 2× minimal ratio.
FIG. 2.
Sensitivity and detection of distinct DNA ratios of multiple species by the ASPE assay. (A) A distinct number of A. baumannii cells (from 104 to 1 cell per assay) were spiked into pooled human blood and detected by the ASPE assay. The dotted line indicates the 2× minimal ratio. (B) Genomic DNAs of A. baumannii, genospecies 3, and genospecies 13TU were mixed in distinct ratios. The amounts of DNA of genospecies 3 and genospecies 13TU were invariable (10 pg), while the amount of DNA of A. baumannii was increased progressively from 100- to 10,000-fold. PCR amplification and the ASPE assay then were conducted to detect the distinct DNA ratios of multiple species. Each column represents the mean (error bars indicate standard deviations). The S/B ratio is the MFI after subtracting the PCR negative-control value and dividing by the PCR negative-control value.
Species identification of clinical isolates.
To further explore the feasibility of the ASPE assay for the identification of clinical Acinetobacter species isolates, 92 clinical isolates previously assigned to A. baumannii by the API 20NE system were analyzed by ITS sequencing and the ASPE assay (Table 3). The species identities of clinical isolates were assigned according to the maximum similarities between the ITS sequences and the ITS database of the NCBI. A total of 56 isolates were identified as A. baumannii by the ASPE assay as well as by ITS sequencing. Among the 36 non-A. baumannii isolates identified by the ASPE assay, 10 were found to exhibit results discrepant from those of ITS sequencing.
TABLE 3.
Clinical isolates of Acinetobacter spp. identified by the ASPE assay, ITS sequencing, and antimicrobial susceptibility tests
| Identified species | No. of clinical isolates identified by:
|
No. (%) of strains resistant toa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ASPE assay | ITS sequencing | CIP | IPM | MEM | SAM | ATM | GEN | CAZ | FEP | |
| A. baumannii | 56 | 56 | 53 (95) | 18 (32) | 33 (59) | 30 (54) | 53 (95) | 52 (93) | 54 (96) | 38 (69) |
| Genospecies 3 | 13b | 15 | 0 | 6 (40) | 5 (33) | 1 (7) | 7 (50) | 5 (33) | 6 (40) | 4 (27) |
| Genospecies 13TU | 19c | 17 | 0 | 2 (12) | 4 (24) | 1 (6) | 6 (35) | 5 (29) | 3 (18) | 1 (6) |
| Genospecies 10 | 1 | 1 | 0 | 1 (100) | 1 (100) | 0 | 1 (100) | 0 | 1 (100) | 1 (100) |
| Acinetobacter sp. strain ADP1 | NAd | 1 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 |
| Other speciese | 3 | 2 | 0 | 1 (50) | 0 | 0 | 0 | 1 (50) | 1 (50) | 0 |
The drug abbreviations and their MICs are the following: CIP, ciprofloxacin (≥4 mg/liter); IPM, imipenem (≥16 mg/liter); MEM, meropenem (≥16 mg/liter); SAM, ampicillin-sulbactam (≥16 mg/liter); ATM, aztreonam (≥32 mg/liter); GEN, gentamicin (≥32 mg/liter); CAZ, ceftazidime (≥32 mg/liter); and FEP, cefepime (≥32 mg/liter).
Five isolates yielded results that were discrepant from those determined by sequencing, with four isolates that were assigned to genospecies 3 by sequencing being assigned to genospecies 13TU by ASPE and one isolate with genospecies 3 and genospecies 13TU signals concurrently.
Four discrepant isolates, with three isolates that were assigned to genospecies 13TU by sequencing being assigned to genospecies 3 by ASPE and one with concurrent genospecies 3 and genospecies 13TU signals.
NA, not applicable.
Isolates identified as not belonging to the 13 reference Acinetobacter species.
Among the 15 isolates identified as genospecies 3 by ITS sequencing, 10 isolates were assigned to genospecies 3, 4 isolates were assigned to genospecies 13TU, and 1 isolate was identified as belonging to genospecies 3 and 13TU concurrently. Among the 17 isolates identified as genospecies 13TU by ITS sequencing, 13 isolates were assigned to genospecies 13TU, 3 isolates were assigned to genospecies 3, and 1 isolate was identified to be simultaneously genospecies 3 and genospecies 13TU by the ASPE assay. Three isolates were nonidentifiable by the ASPE assay. Using ITS sequencing, one was identified as Acinetobacter sp. strain ADP1, and the other two isolates showed an ITS similarity to the 13 reference species of 81 to 85%. In summary, 79 of the 88 clinical isolates could be assigned to species of the ACB complex using the ASPE assay, with an identification rate of 90%.
Antimicrobial susceptibilities.
The antimicrobial susceptibility test showed that 56 clinical A. baumannii isolates were resistant to most antimicrobial agents, including ciprofloxacin, meropenem, ampicillin-sulbactam, aztreonam, gentamicin, cefazidime, and cefepime, with resistance rates of 54 to 95%, but only 32% were resistant to imipenem (Table 3). Fewer genospecies 3 and 13TU isolates than other isolates were susceptible to imipenem, meropenem, aztreonam, gentamicin, cefazidime, and cefepime, but more were susceptible to ciprofloxacin and ampicillin-sulbactam. Thus, the antimicrobial agent to which A. baumannii isolates were most susceptible was imipenem, but the agents to which genospecies 3 and 13TU isolates were most susceptible were ciprofloxacin and ampicillin-sulbactam.
DISCUSSION
Although other molecular methods for Acinetobacter species identification are used, they are not ideal for rapid or large-scale applications. The microsphere-based array platform can be finished within 8.5 h (20) and is very suitable for high-throughput applications. The ASPE assay is based on the design of a single-nucleotide polymorphism (SNP)-specific nucleotide at the 3′ end of each extension primer and can readily discriminate any SNP or mutant. The utilization of the ZipCode/cZipCode hybridization enabled universal hybridization with microspheres (29, 36). The cZipCode-coupled beads also can be used in other DNA-based tests, which will economize bead usage.
In this study, the results showed that the microsphere-based array platform can be applied to the multiplexed identification of 13 reference Acinetobacter spp. For the identification of 92 clinical isolates, 88 isolates belonging to the ACB complex have been discriminated, of which 56 A. baumannii isolates were accurately identified by the ASPE assay. The results show that the most common clinical Acinetobacter species isolates in Taiwan are A. baumannii isolates. However, discrepant identification results between the ASPE assay and ITS sequencing were found for nine isolates belonging to genospecies 3 and 13TU. It was presumed that the interspecies diversity between genospecies 3 and 13TU clinical strains was low.
To assess the specificity of the ASPE assay, 14 different non-Acinetobacter species nosocomial bacteria were subjected to the ASPE assay with all 13 species-specific ASPE primers and UniA. The data (see Fig. S1 in the supplemental material) clearly demonstrated that all ASPE primers were highly specific for their targeted genus and species. The high specificity of the ASPE assay may be advantageous in nosocomial pathogen detection. The sensitivity of the ASPE assay is fewer than 100 CFU per ml of blood, which suggests that the ASPE assay can detect target species in human blood. In addition, the result of the simulated multiple Acinetobacter species infections suggested that multiple species could be discriminated by the ASPE assay despite being present in the mixture at different ratios. Polymicrobial bacteremia was identified in 5 to 22% of bacteremia cases (25, 33), and blood cultures may be inefficient in detecting polymicrobial bacteremia (27). Therefore, these data suggested that the ASPE assay is useful for clinical application.
Further investigations are required to define the clinical significance of Acinetobacter spp. other than A. baumannii (2). In this study, a combination of the identified results and antimicrobial susceptibilities of the clinical isolates showed that most A. baumannii isolates were resistant to most antimicrobial agents other than imipenem, but the majority of genospecies 3 and 13TU isolates were susceptible to ciprofloxacin and ampicillin-sulbactam. There are a few reports describing the significant differences of antimicrobial susceptibility among members of the ACB complex. The antimicrobial patterns described in Korean reports by Lim et al. (21) and Lee et al. (19) are similar to those of our findings: almost all A. baumannii isolates were highly resistant to most antimicrobial agents except carbapenems, while Acinetobacter genospecies 3 and 13TU isolates mostly were susceptible to ciprofloxacin and ampicillin-sulbactam. In Hong Kong, Houang et al. also reported significant differences in the antimicrobial susceptibilities of isolates of A. baumannii, genospecies 3, and genospecies 13TU and suggested that the delineation of genospecies is important in surveillance studies of antimicrobial susceptibilities (10). During the study period, tigecycline and colistin, the two antimicrobial agents with encouraging activity against MDR Acinetobacter spp., were not available in Taiwan. In many countries, carbapenems (e.g., imipenem and meropenem) have been the drugs of choice against Acinetobacter infections and have retained better activity than other antimicrobial agents (2). However, carbapenem-resistant Acinetobacter spp. have been reported worldwide (1) and are rapidly increasing in prevalence, from 5.88% in 1993 to 21.5% in 2000 in Taiwan (12). Our results suggested that accurately differentiating genospecies 3 and 13TU from A. baumannii isolates is significant because of their differences in antimicrobial susceptibility. Ciprofloxacin or ampicillin-sulbactam might be a better choice than carbapenems for the treatment of the Acinetobacter genospecies 3 and 13TU infections. A recent clinical report in Taiwan suggested that combined carbapenem and ampicillin-sulbactam regimens were associated with a better outcome than the combination of carbapenem and amikacin or carbapenem alone (17). These conclusions might provide clinicians with information for the treatment of Acinetobacter infections.
In conclusion, the microsphere-based array is rapid and reliable and has a multiplex capability for the identification of Acinetobacter spp. This method may be of help in clinical applications. Furthermore, distinct resistance patterns among Acinetobacter spp. also were observed, thus highlighting the importance of accurate species identification.
Supplementary Material
Acknowledgments
This work was supported by grants DOH96-DC-2017 and DOH96-DC-1010 from the Centers for Disease Control, Department of Health, Taiwan.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Afzal-Shah, M., and D. M. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J. Antimicrob. Chemother. 41576-577. [DOI] [PubMed] [Google Scholar]
- 2.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 431632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., M. A. Iannone, M. S. Li, J. D. Taylor, P. Rivers, A. J. Nelsen, K. A. Slentz-Kesler, A. Roses, and M. P. Weiner. 2000. A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res. 10549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Colinas, R. J., R. Bellisario, and K. A. Pass. 2000. Multiplexed genotyping of beta-globin variants from PCR-amplified newborn blood spot DNA by hybridization with allele-specific oligodeoxynucleotides coupled to an array of fluorescent microspheres. Clin. Chem. 46996-998. [PubMed] [Google Scholar]
- 7.Cornaglia, G., M. L. Riccio, A. Mazzariol, L. Lauretti, R. Fontana, and G. M. Rossolini. 1999. Appearance of IMP-1 metallo-beta-lactamase in Europe. Lancet 353899-900. [DOI] [PubMed] [Google Scholar]
- 8.Gerner-Smidt, P. 1992. Ribotyping of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Clin. Microbiol. 302680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerner-Smidt, P., I. Tjernberg, and J. Ursing. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houang, E. T., Y. W. Chu, K. Y. Chu, K. C. Ng, C. M. Leung, and A. F. Cheng. 2003. Significance of genomic DNA group delineation in comparative studies of antimicrobial susceptibility of Acinetobacter spp. Antimicrob. Agents Chemother. 471472-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsueh, P. R., W. H. Chen, and K. T. Luh. 2005. Relationships between antimicrobial use and antimicrobial resistance in gram-negative bacteria causing nosocomial infections from 1991-2003 at a university hospital in Taiwan. Int. J. Antimicrob. Agents 26463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsueh, P. R., L. J. Teng, C. Y. Chen, W. H. Chen, C. J. Yu, S. W. Ho, and K. T. Luh. 2002. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 8827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iannone, M. A., J. D. Taylor, J. Chen, M. S. Li, P. Rivers, K. A. Slentz-Kesler, and M. P. Weiner. 2000. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39131-140. [PubMed] [Google Scholar]
- 14.Janssen, P., K. Maquelin, R. Coopman, I. Tjernberg, P. Bouvet, K. Kersters, and L. Dijkshoorn. 1997. Discrimination of Acinetobacter genospecies by AFLP fingerprinting. Int. J. Syst. Bacteriol. 471179-1187. [DOI] [PubMed] [Google Scholar]
- 15.Jeon, B. C., S. H. Jeong, I. K. Bae, S. B. Kwon, K. Lee, D. Young, J. H. Lee, J. S. Song, and S. H. Lee. 2005. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 β-lactamase in Korea. J. Clin. Microbiol. 432241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krawczyk, B., K. Lewandowski, and J. Kur. 2002. Comparative studies of the Acinetobacter genus and the species identification method based on the recA sequences. Mol. Cell Probes 161-11. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, L. C., C. C. Lai, C. H. Liao, C. K. Hsu, Y. L. Chang, C. Y. Chang, and P. R. Hsueh. 2007. Multidrug-resistant Acinetobacter baumannii bacteraemia: clinical features, antimicrobial therapy and outcome. Clin. Microbiol. Infect. 13196-198. [DOI] [PubMed] [Google Scholar]
- 18.La Scola, B., V. A. Gundi, A. Khamis, and D. Raoult. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. H., C. H. Choi, H. Y. Kang, J. Y. Lee, J. Kim, Y. C. Lee, S. Y. Seol, D. T. Cho, K. W. Kim, D. Y. Song, and J. C. Lee. 2007. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genospecies 13TU. J. Antimicrob. Chemother. 59633-639. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. H., D. R. Walker, P. B. Cregan, and H. R. Boerma. 2004. Comparison of four flow cytometric SNP detection assays and their use in plant improvement. Theor. Appl. Genet. 110167-174. [DOI] [PubMed] [Google Scholar]
- 21.Lim, Y. M., K. S. Shin, and J. Kim. 2007. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genospecies of Acinetobacter isolates. J. Clin. Microbiol. 45902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naas, T., B. Coignard, A. Carbonne, K. Blanckaert, O. Bajolet, C. Bernet, X. Verdeil, P. Astagneau, J. C. Desenclos, and P. Nordmann. 2006. VEB-1 extended-spectrum beta-lactamase-producing Acinetobacter baumannii, France. Emerg. Infect. Dis. 121214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page, B. T., and C. P. Kurtzman. 2005. Rapid identification of Candida species and other clinically important yeast species by flow cytometry. J. Clin. Microbiol. 434507-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quale, J., S. Bratu, D. Landman, and R. Heddurshetti. 2003. Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin. Infect. Dis. 37214-220. [DOI] [PubMed] [Google Scholar]
- 25.Rello, J., E. Quintana, B. Mirelis, M. Gurgui, A. Net, and G. Prats. 1993. Polymicrobial bacteremia in critically ill patients. Intensive Care Med. 1922-25. [DOI] [PubMed] [Google Scholar]
- 26.Seifert, H., R. Baginski, A. Schulze, and G. Pulverer. 1993. Antimicrobial susceptibility of Acinetobacter species. Antimicrob. Agents Chemother. 37750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stetz, E. M., and W. J. Martin. 1985. Repeat subculture of known positive blood cultures: costly and ineffective in detecting polymicrobial bacteremias. Diagn. Microbiol. Infect. Dis. 3113-118. [DOI] [PubMed] [Google Scholar]
- 28.Tjernberg, I. 1990. Antimicrobial susceptibility of Acinetobacter strains identified by DNA-DNA hybridization. APMIS 98320-326. [PubMed] [Google Scholar]
- 29.Ugozzoli, L. A. 2004. Multiplex assays with fluorescent microbead readout: a powerful tool for mutation detection. Clin. Chem. 501963-1965. [DOI] [PubMed] [Google Scholar]
- 30.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. de Vos, G. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genospecies by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 3311-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Looveren, M., and H. Goossens. 2004. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10684-704. [DOI] [PubMed] [Google Scholar]
- 32.Wang, J. T., L. C. McDonald, S. C. Chang, and M. Ho. 2002. Community-acquired Acinetobacter baumannii bacteremia in adult patients in Taiwan. J. Clin. Microbiol. 401526-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein, M. P., J. R. Murphy, L. B. Reller, and K. A. Lichtenstein. 1983. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Rev. Infect. Dis. 554-70. [DOI] [PubMed] [Google Scholar]
- 34.Wu, C. J., H. C. Lee, N. Y. Lee, H. I. Shih, N. Y. Ko, L. R. Wang, and W. C. Ko. 2006. Predominance of gram-negative bacilli and increasing antimicrobial resistance in nosocomial bloodstream infections at a university hospital in southern Taiwan, 1996-2003. J. Microbiol. Immunol. Infect. 39135-143. [PubMed] [Google Scholar]
- 35.Yao, S. M., Y. C. Lin, C. Y. Chou, Y. Y. Chen, M. J. Hsiao, H. Y. Chen, J. J. Yan, H. P. Su, and S. Y. Li. 2005. Antigenic divergence of Bordetella pertussis isolates in Taiwan. J. Clin. Microbiol. 435457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, Y., D. W. Hein, M. A. Doll, K. K. Reynolds, N. Abudu, R. Valdes, Jr., and M. W. Linder. 2006. Simultaneous determination of 7 N-acetyltransferase-2 single-nucleotide variations by allele-specific primer extension assay. Clin. Chem. 521033-1039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.