Abstract
Haemophilus influenzae is both a human respiratory pathogen and pharyngeal commensal, while H. haemolyticus, the closest phylogenetic relative of H. influenzae, is arguably a strict pharyngeal commensal. A hemolytic phenotype has historically differentiated H. haemolyticus from H. influenzae, but the recent recognition of significant nonhemolytic H. haemolyticus colonization has decreased this trait's resolvability. Given this and the potential of recombination between the species, we examined the distribution of microbiologic and molecular traits between collections of H. influenzae and H. haemolyticus strains separated within a dendrogram obtained by multilocus sequence analysis (MLSA). All strains hybridizing with a probe to iga, a gene encoding an immunoglobulin A protease of H. influenzae, clustered apart from strains that did not hybridize with the probe. Other traits also segregated significantly along this division, suggesting a separation of the species. Of note, the LOS genes licA, lic2A, and lgtC of H. influenzae were approximately 2, 6, and 54 times, respectively, more prevalent in H. influenzae than in H. haemolyticus. In contrast to species separation, interspecies recombination was evidenced by the inability of single gene sequences to phylogenetically separate the species and by the “fuzzy” distribution of some species-specific traits across the species dividing line. Together, these data support the historically accurate and pragmatic division of these species while recognizing their potential for recombination. Future comparative genomic studies identifying common and distinctive genes could be useful in evaluating their role in the commensal or virulent growth, respectively, of H. influenzae.
Haemophilus influenzae is one of eight Haemophilus species that reside as commensal organisms in the pharyngeal cavity of humans. These other commensal Haemophilus species include H. parainfluenzae, H. haemolyticus, H. parahaemolyticus, H. paraphrohaemolyticus, H. segnis, H. aphrophilus, and H. paraphrophilus. H. influenzae is by far the most pathogenic member of the genus in humans. Strains possessing a type b capsule are often associated with invasive diseases, such as meningitis, sepsis, and pneumonia; and strains lacking a capsule (referred to as nontypeable [NT]) are associated with localized mucosal diseases, such as otitis media, sinusitis, and bronchitis. Other haemophili are considered to be rarely associated with disease, and H. haemolyticus has never been implicated as an infectious disease agent (1).
In contrast to other haemophili, H. haemolyticus and H. influenzae both depend on X (hemin) and V (NAD) growth factors, both lack the ability to ferment sucrose, both share similar G+C contents, and both are nearly indistinguishable by their colony and cellular morphologies (25). Initial phylogenetic studies performed by using DNA-DNA hybridization revealed that H. haemolyticus associated with H. influenzae in a cluster of the family Pasteurellaceae termed Haemophilus sensu stricto (6, 41, 47), and these results were substantiated by analysis of the 16S rRNA gene (8, 9, 43), analysis of the infB gene (20), or multilocus sequence analysis (MLSA) (42).
Despite the close relationship of the species, only the beta-hemolytic phenotype of H. haemolyticus is routinely used in the clinical setting to distinguish H. haemolyticus from H. influenzae (45, 46, 53). Beta-hemolysis, however, has recently been shown to be a poor indicator of species separation due to the finding that various H. influenzae strain collections contained significant proportions of nonhemolytic H. haemolyticus strains (38, 40, 62). Murphy and colleagues found that close to 40% of the isolates in an H. influenzae strain collection obtained from the sputum of patients with chronic obstructive pulmonary disease (COPD) were H. haemolyticus, and their identities were confirmed by phylogenetic studies and amino acid sequence analysis for differences in the P6 outer membrane protein (OMP) (40). Their study also found that more than 20% of a collection of H. influenzae strains obtained from the throats of healthy children attending day care were H. haemolyticus; and this was confirmed with a larger set of strains by Xie et al. (62), who also identified nonhemolytic H. haemolyticus strains by their inability to hybridize with a probe specific for iga, an H. influenzae gene encoding the virulence-associated immunoglobulin A (IgA) protease (28). Similar to hemolytic H. haemolyticus, those studies failed to find an association of the nonhemolytic species with disease: nonhemolytic H. haemolyticus isolates were not present among almost 300 H. influenzae isolates obtained from the middle ears of children with otitis media, were not associated with new strain acquisitions during COPD exacerbation, and were not present in nearly 600 respiratory tract isolates present in the H. influenzae multilocus sequence typing (MLST) database (40, 62).
Apart from hemolysis, however, a precise taxonomic division between H. influenzae and H. haemolyticus may be elusive. In 1976, Kilian (who was the first to note strains of H. haemolyticus that had lost their hemolytic phenotype) found that only the production of gas during glucose metabolism, H2S production, and the lack of ornithine decarboxylase (ODC) activity reasonably differentiated H. haemolyticus from H. influenzae (25). Kilian later showed that H. influenzae, in contrast to nonpathogenic Haemophilus species, expressed an IgA protease. In a phylogenetic study of the family Pasteurellaceae, Hedegaard et al. (20), however, noted two H. haemolyticus strains (strains HK855 and HK856) that had lost their hemolytic activity but that expressed the IgA protease. One strain, HK855, produced gas during glucose metabolism and produced H2S (H. haemolyticus-associated traits). Phylogenetic analysis of the infB gene revealed that HK865 was more related to H. influenzae than to H. haemolyticus but that HK855, the gas- and H2S-producing strain, was intermediately related between the species. This lack of species delineation is reminiscent of the “fuzzy species” observations noted between the pathogenic bacteria Neisseria meningitidis and the nasopharyngeal commensal N. lactamica (18). These organisms were shown to have undergone some level of interspecies recombination. Similarly, species-specific trait variations in H. influenzae and H. haemolyticus might also be explained by interspecies recombination; H. influenzae is a naturally transformable organism, and if this property exists in H. haemolyticus, the species may conceivably exchange DNA.
Given that H. haemolyticus may be a more prevalent commensal organism than was previously thought, that H. influenzae and H. haemolyticus are close phylogenetic relatives, and that the species may potentially exchange DNA through natural transformation, we have evaluated the relationships of large collections of NT H. influenzae and H. haemolyticus (hemolytic and nonhemolytic) strains by MLSA, classical taxonomic traits, and (since H. haemolyticus has not been associated with disease) the distribution of the H. influenzae iga gene and three genes (licA, lic2A, and lgtC) contributing to virulence-associated lipooligosaccharide (LOS) structures.
MATERIALS AND METHODS
Bacterial strains and culturing.
For general use, the bacteria were grown on chocolate agar plates (BD, Franklin Lakes, NJ) at 37°C with 5% CO2. For the detection of gas production, Levinthal broth (58) was supplemented with 1% glucose as described previously (25). Liquid cultures were grown without shaking.
Four H. influenzae reference strains were used in this study and represent strains whose genomes have been completely or partially sequenced: Rd (KW-20, ATCC 51907), 86-028NP (an NT nasopharyngeal strain isolated from a child with otitis media), R2846 (strain 12; an NT strain from a patient with otitis media), and R2866 (INT-1 and ATCC 51997; an NT isolate from blood). One H. haemolyticus type strain, ATCC 33390, was also used. The remaining strains were parts of various collections obtained by the laboratory of the Department of Pediatrics and Communicable Diseases, University of Michigan Medical School, or by other laboratories in previous studies (11, 16, 29, 38, 40, 50, 54) (Table 1). Briefly, the H. influenzae strains were isolated from the throats of healthy children attending day care, the throats of healthy adults in a longitudinal carriage study, or the middle ear aspirates of children with otitis media. The H. haemolyticus strains were isolated from the throats of healthy children attending day care, the adult longitudinal carriage study, or the sputum of adults enrolled in a COPD exacerbation study (Table 1). The H. haemolyticus strains from COPD patients were shown not to be new strain acquisitions that led to the exacerbation of COPD (40).
TABLE 1.
Population and ecological stratification of the H. influenzae and H. haemolyticus collections
The H. influenzae and nonhemolytic H. haemolyticus strains were originally defined and tested again in this study by typical colony morphology on chocolate agar, X and V growth factor dependency, and no reaction in the porphyrin test (11, 16, 29, 38, 54). Hemolysis, used to differentiate hemolytic H. haemolyticus strains, was assessed as zones of beta-hemolysis surrounding individual colonies grown on horse blood agar (Remel, Lenexa, KS). Working stocks of each strain were made by growing the original stocks on chocolate agar (BD Biosciences, San Jose, CA) and then harvesting the overnight growth into 2 ml of sterile skim milk. Aliquots of 900 μl were then placed in duplicate 96-well plates with 1-ml wells. The plates were subsequently sealed and frozen at −80°C. Cultures were made by partially thawing one plate and pipetting 1 μl of stock onto chocolate agar. This initial growth was then subcultured as needed.
DNA template isolation and PCR.
DNA was obtained from the bacterial isolates by using a Wizard genomic DNA purification kit from Promega Co. (Madison, WI). After purification, the DNA from the strains was diluted to half the original concentration, and 1 μl was aliquoted and placed into multiple unskirted 96-well thermocycler plates from Bio-Rad Laboratories, Inc. (Hercules, CA). The plates were stored at −20°C until they were needed. PCR amplification was performed with 50-μl reaction mixtures that comprised 10 pmol of each relevant oligonucleotide primer (Table 2), 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dTTP), and 1 U of recombinant Taq DNA polymerase from Invitrogen (Carlsbad, CA). The mixture was subjected to 30 amplification cycles, including 1 min at 94°C template for denaturation, 1 min at 51 to 55°C (depending on primer specificity) for primer annealing, and 1 min at 72°C for chain extension.
TABLE 2.
Oligonucleotides used for PCR or DNA sequencing
| Gene | Primer sequencea | Position in Rd genome |
|---|---|---|
| adk | F: GGTGCACCGGGTGCAGGTAA | 376521 |
| R: CCTAAGATTTTATCTAACTC | 375899 | |
| pgi | F: GGTGAAAAAATCAATCGTAC | 1644657 |
| R: ATTGAAAGACCAATAGCTGA | 1645246 | |
| recA | F: ATGGCAACTCAAGAAGAAAA | 622556 |
| R: TTACCAAACATCACGCCTAT | 621940 | |
| infB | F: TGAAAATGAGCTTGAAGAAGCGG | 1361550 |
| R: GATAGTTGCCACAGGGCCACGACC | 1362198 | |
| 16S rRNA | F1: CCAGCAGCCGCGGTAATACG | 624342 |
| R1: ATCGGYTACCTTGTTACGACTTC | 625335 | |
| F2: GCCCGCACAAGCGGTGGAGCATGTG | 624751 | |
| R2: CTCGTAAGGGCCATGATGACTTGACG | 625039 | |
| iga | F: GTTCCACCACCTGCGCCTGCTAC | 1050386 |
| R: GTTATATTGCCCCTCGTTATTCA | 1049130 | |
| licAb | F: GTAGGATTTGTTAAAACTTGCTACAAGCC | 1608693 |
| R: GGCAATTCCTCTAACAGTTTAAATGCTGCG | 1609579 | |
| lic2Ab | F: ATATTACATAATATAGAGGAATCTAG | 571382 |
| R: CTACATAAAACGAACAATTTCTTTACC | 570690 | |
| lgtCb | F: CGGACTGTCAGTCAGACAATG | 289338 |
| R: CTCAAAATGATCATACCAAGATG | 288499 |
All oligonucleotides are based on the DNA sequences from H. influenzae strain Rd. F, forward, R, reverse.
The forward primers begin downstream of tetranucleotide repeats.
DNA sequencing.
Partial DNA sequences of the adk, pgi, recA, and infB genes were generated for MLSA according to the strategy used by Nørskov-Lauritsen et al. (42), with the exception that the oligonucleotides used for PCR amplification of these genes in the present study were based on sequences from H. influenzae strain Rd (Table 2). The oligonucleotides used for PCR amplification of the partial 16S rRNA gene sequences for the differentiation of Haemophilus species have been described previously (32) (Table 2). After the PCR amplification was confirmed by gel electrophoresis on 1% agarose, the PCR products were purified with a Qiagen (Valencia, CA) QIAquick 96 or single-sample PCR purification kit. Automated fluorescent dideoxy-DNA sequencing was done by the University of Michigan DNA sequencing core on an ABI model 3730 sequencer. Previously published sequences for type strains of the family Pasteurellaceae, including H. haemolyticus, H. parainfluenzae, H. pittmaniae, Actinobacillus lignieresii, Actinobacillus pleuropneumoniae, H. segnis, H. aphrophilus, H. paraphrophilus, Actinobacillus actinomycetemcomitans, and Pasteurella multocida have been described for adk, pgi, and recA (42) and for infB (20). The 16S rRNA gene sequences for a subset of these strains (Fig. 1) have also been published previously (8, 9). Relevant sequences from Escherichia coli K-12 and H. influenzae strains Rd, 86-028NP, R2846, and R2866 were obtained from their genome sequences (4, 10, 12, 19). Sequence editing, trimming, and concatenation were done with Lasergene software (version 7.0; DNAStar, Madison, WI), and phylogenetic analysis was done with Mega software (version 3.1) (31). Bootstrap consensus, minimum-evolution dendrograms for concatenated or individual sequences were made with 1,000 replicates.
FIG. 1.
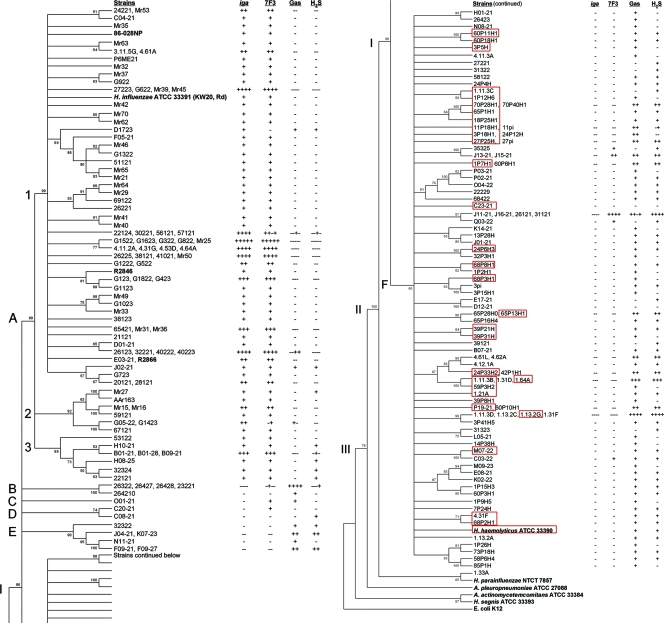
Minimum-evolution dendrogram of the Haemophilus sensu stricto cluster. The tree, rooted by E. coli strain K-12, is based on concatenated adk, pgi, recA, infB, and 16S rRNA gene sequences, with bootstrap values of ≥50% of 1,000 bootstraps indicated. Node I contains all H. influenzae and hemolytic and nonhemolytic H. haemolyticus strains within an Haemophilus sensu stricto cluster, while nodes II and III contain selected members of the family Pasteurellaceae. Strains with identical sequences are listed on the same branch. Red boxes define beta-hemolytic H. haemolyticus strains. Positive (+) and negative (−) results for the iga gene probe hybridization, P6 OMP MAb 7F3 reactivity, the production of gas during glucose metabolism, and H2S production are shown to the right of each strain. The results are also relative to the strain order on branches with multiple strains. On the basis of the stratification of iga and other results (see the text and Table 2), it is proposed that node A contains all H. influenzae strains, while nodes B to F contain all H. haemolyticus strains.
Classical taxonomy.
Biochemical and phenotypic analyses of H. influenzae and H. haemolyticus traits were performed as previously described by Kilian (25, 26). Briefly, the emission of H2S was assessed with lead acetate strips (Fisher Scientific, Pittsburgh, PA) placed on the lids of chocolate agar plates inoculated with individual bacterial strains. Conversion of the strips from white to black after overnight growth was considered positive. The production of gas from glucose metabolism was assessed by the collection of gas in inverted Durham tubes immersed in Levinthal broth containing 1% glucose (Fisher Scientific). H. influenzae biotype reactions have been extensively described (26, 25, 39, 60). Briefly, indole production was assessed by placing a small amount of bacteria on filter paper saturated with Kovács reagent (Remel) and observing the paper for a purple color reaction; urease activity was assessed in rapid urea broth (BD), according to the manufacturer's instructions, by mixing an inoculum loop of bacteria from the plate growth and observing the plate for a color reaction after 4 h of incubation at 37°C; and ODC activity was determined with ODC broth (Remel), according to the manufacturer's instructions.
Dot blot immunoassays for MAb 7F3 recognition of P6 OMP.
The detection of the P6 OMP by monoclonal antibody (MAb) 7F3 in dot blot immunoassays has been described previously (17). Briefly, membranes were prepared by harvesting bacteria directly from the growth on chocolate agar and placing them onto nitrocellulose membranes with a sterile pipette tip. The blots were immediately washed in phosphate-buffered saline (PBS) and then blocked in PBS containing 5% nonfat dry milk (BLOTTO [22]) for 1 h. MAb 7F3 was diluted 1:10 in BLOTTO, and the mixture was incubated overnight at room temperature. After the blots were washed three times in PBS alone, they were incubated in PBS containing goat anti-mouse IgA antibody conjugated to alkaline phosphatase (Sigma) diluted 1:5,000 for 1 h. The blots were then washed three times in PBS and exposed to nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate (Pierce, Rockford, IL) for color development.
Dot blot hybridization.
Membranes dotted with single-stranded genomic DNA obtained from crude lysates of each Haemophilus strain were made by using a modification of the method of Foxman et al. (13, 14). The bacteria were grown overnight on chocolate agar and were suspended in 4 ml of PBS to an optical density of 1.0. The bacteria were then centrifuged, and the pellets stored at −20°C until all strains were collected. The pellets were then thawed, resuspended in 8 ml of lysis solution (0.4 M NaOH and 10 mM EDTA), and heated for 30 min in a 70°C water bath. One-milliliter aliquots of each extract were placed in multiple 96-well plates with 1-ml wells. The plates were sealed and frozen at −80°C until they were needed. Dot blot membranes were made by applying 40 μl of each DNA extract to a nylon membrane in an array of 8 by 12 on a Bio-Dot microfiltration apparatus (Bio-Rad), as described previously (13, 14).
Generation of probes, hybridization, and analysis were carried out as described previously (44, 63). Oligonucleotides for the amplification of the iga, licA, lic2A, and lgtC gene probes were generated from H. influenzae strain Rd; and oligonucleotides were directed to the nonrepeat regions of each gene (Table 2). DNA was amplified by PCR as described above. The PCR products were purified from agarose gels by using the QIAquick gel extraction kit from Qiagen and labeled with the AlkPhos direct labeling and detection systems (GE Healthcare, Piscataway, NJ). The probes were hybridized to the dot blot membranes under stringent conditions and developed with an ECF detection system (GE Healthcare). The probe signal intensity was read with a Storm 860 phosphorimager and analyzed with ImageQuant (version 5.0) software (Molecular Dynamics/GE Healthcare).
Statistical analyses.
The data were entered in Excel software (Microsoft) in binary form for the presence (which was given a value of 1) or absence (which was given a value of 0) of any given trait. The prevalence ratios of the traits for H. influenzae compared to the traits for H. haemolyticus or the prevalence ratios of the traits for nonhemolytic H. haemolyticus compared to the traits for hemolytic H. haemolyticus were calculated as a ratio of the proportions of traits among each species or collection, for example, as the proportion of all H. influenzae strains with a particular trait over the proportion of all H. haemolyticus strains with the same trait. χ2 analysis was used to determine the significance of the differences of the trait associations between species or between nonhemolytic and hemolytic H. haemolyticus strains. Student's t test was used to assess significant differences in the distributions of the traits between different nodes of the same species. Statistical analyses were performed with SAS software (version 9.1).
Nucleotide sequence accession numbers.
Partial gene sequences for each of the 88 H. influenzae, 109 H. haemolyticus, and 3 H. parainfluenzae strains are available from GenBank under accession numbers EU150426 to EU150625 (for adk), EU150626 to EU150825 (for pgi), EU150826 to EU151025 (for recA), EU151026 to EU151225 (for infB), and EU151226 to EU151425 (for the 16S rRNA genes).
RESULTS
Phylogenetics of H. influenzae and H. haemolyticus within the family Pasteurellaceae.
Recently, Nørskov-Lauritsen et al. (42) defined the relationships of human-specific haemophili within the family Pasteurellaceae by performing MLSA with partial, concatenated adk, pgi, recA, and infB gene sequences. Since this approach revealed that both H. influenzae and H. haemolyticus were present in an Haemophilus sensu stricto phylogenetic cluster and were separate from other members of the family, we have used it to determine if the NT H. influenzae and H. haemolyticus (hemolytic and nonhemolytic) strains in our study also maintain the same relationships. An MLST scheme, designed to detail and track differences within the H. influenzae species, was not used. In addition, various MLST genes of H. influenzae have been shown not to be present in other members of the family Pasteurellaceae (42).
A dendrogram produced from the concatenated DNA sequences for each of the 200 strains predicted to be H. influenzae or H. haemolyticus, together with sequences from four H. influenzae type strains, one H. haemolyticus type strain, and nine other members of the family Pasteurellaceae (represented by 15 strains) (42), revealed a Haemophilus sensu stricto cluster containing the H. influenzae and H. haemolyticus type strains and all but 3 of the 200 test strains separate from other members of the family Pasteurellaceae (data not shown). The three strains not contained in the Haemophilus sensu stricto cluster were more related to H. parainfluenzae than to H. influenzae or H. haemolyticus. Since H. parainfluenzae grows independently of factor X, we reassessed the X and V growth factor requirements of these three strains and found that they grew in the presence of V factor alone, indicating that they are most likely H. parainfluenzae. Given this result, the remaining 197 strains were reassessed for their dependence on factors X and V, and all strains were found to require both factors. These results, together with the MLSA data, support the designation of these 197 strains as either H. influenzae or H. haemolyticus.
Because 16S rRNA gene sequences were previously used to define members of the family Pasteurellaceae (43) and to separate the nonhemolytic form of H. haemolyticus from H. influenzae (40), partial sequences of the 16S rRNA gene were obtained from the H. influenzae and the H. haemolyticus test strains described above and concatenated with the four housekeeping gene sequences. A dendrogram created from the extended sequences of these strains, together with those of the H. influenzae and H. haemolyticus type strains and four members of the family Pasteurellaceae for which published 16S rRNA gene sequences were available, revealed that all H. influenzae and H. haemolyticus strains remained in an Haemophilus sensu stricto cluster similar to that described with the four housekeeping genes alone (Fig. 1 and 2A). Within the cluster, some branches were found to contain groups of two to five strains with identical sequences. Of the 197 strains in the collection, 88 (45%) had sequences identical to those of 1 or more strains; and the remaining strains, which had unique sequences, were found on separate branches (Fig. 1). The Haemophilus sensu stricto cluster emerged from node I of the dendrogram, separate from nodes II and III, which contained four other members of the family Pasteurellaceae (Fig. 1). The Haemophilus sensu stricto cluster at node I was composed of six subnodes, designated A to F; node A contained all four H. influenzae type strains together with 88 of the 197 test strains, and node F contained the H. haemolyticus type strain, ATCC 33390, together with 95 test strains. The 14 remaining strains were present on four branches that emerged from nodes B to E (Fig. 1).
FIG. 2.
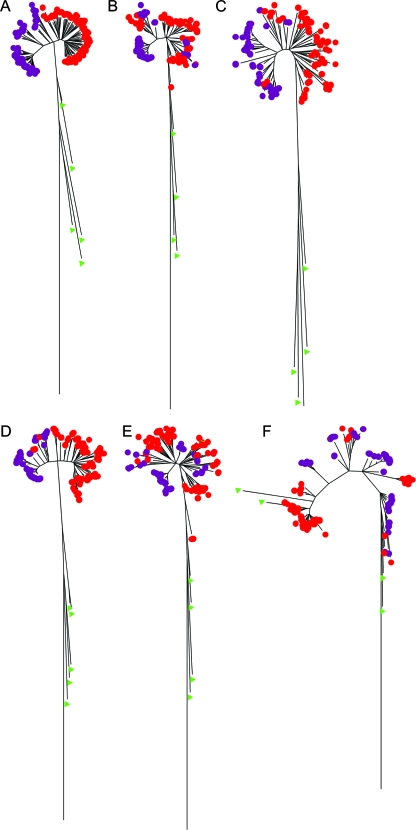
Multiple and single DNA sequence segregation of H. influenzae and H. haemolyticus strains. Radiation tree views of the (A) five concatenated sequences and individual (B) adk, (C) pgi, (D) recA, (E) infB, and (F) 16S rRNA gene sequences. H. influenzae (purple dots) and H. haemolyticus (red dots) strains are shown together with other members of the family Pasteurellaceae (green triangles).
Phylogenetic distribution of iga gene probe hybridization and P6 MAb reactivity between Haemophilus species.
In a previous study, H. influenzae strains were defined as factor X- and V-dependent, nonhemolytic strains that both hybridized to an iga gene probe and reacted with P6 OMP MAb 7F3, an antibody previously shown to react with the conserved P6 OMP of H. influenzae but not with that of H. haemolyticus (40, 62). These strains were distinguished from nonhemolytic H. haemolyticus that lacked at least one of these traits (62). Therefore, the 197 strains belonging to the Haemophilus sensu stricto collection described above were also examined for their ability to hybridize with an iga gene probe by dot blot hybridization. As expected, the four H. influenzae reference strains hybridized with the probe, while the H. haemolyticus type strain, ATCC 33390, did not. All 88 strains in clades emerging from node A of the MLSA-based dendrogram hybridized with the iga gene probe, while the 109 strains in clades and branches emerging from nodes B to F did not hybridize to the probe (Fig. 1). Next, a dot blot immunoassay was performed for all strains with MAb 7F3. The four H. influenzae type strains, but not the H. haemolyticus type strain, reacted with MAb 7F3. All but 3 of the 88 strains (97%) that hybridized to the iga gene probe also reacted with MAb 7F3. In contrast, the antibody reacted with only 13 of the 109 strains (12%) that did not hybridize to the iga gene probe (Fig. 1; Table 3). In contrast to the clear segregation of the iga gene probe hybridization in the MLSA-based dendrogram, strains with discordant results for MAb 7F3 reactivity were distributed throughout the dendrogram. Thus, the species designations of the strains in this study were made primarily by MLSA and iga gene probe hybridization. In this context, the data suggest that node A of the MLSA-based dendrogram contains 88 H. influenzae strains that hybridize with an iga gene probe and that predominantly react with MAb 7F3, while nodes B to F of the dendrogram contain 109 H. haemolyticus strains (hemolytic or nonhemolytic) that lack iga hybridization and that mostly lack MAb 7F3 reactivity.
TABLE 3.
Distribution of taxonomic traits among H. influenzae and H. haemolyticus isolates
| Trait | No. (%) of isolatesa
|
PRb | Result of χ2 analysis (P valued) | |
|---|---|---|---|---|
| H. influenzae (n = 88) | H. haemolyticus (n = 109) | |||
| Molecular-based traits | ||||
| iga gene probe hybridization | 88 (100) | 0 (0.0) | Referent | |
| MAb 7F3 reactivity | 85 (96.6) | 13 (11.9) | 8.10 | <0.0001 |
| Classical traits | ||||
| H2S production | 11 (12.5) | 75 (68.8) | 0.18 | <0.0001 |
| Gas production | 5 (5.7) | 96 (88.1) | 0.06 | <0.0001 |
| Indole production | 73 (83.0) | 68 (62.4) | 1.33 | 0.0015 |
| Urease activity | 74 (84.1) | 77 (70.6) | 1.19 | 0.0265 |
| ODC activity | 24 (27.3) | 8 (7.3) | 3.72 | 0.0002 |
| Biotype I | 18 (20.5) | 4 (3.7) | 5.57 | 0.0002 |
| Biotype II | 44 (50.0) | 43 (39.4) | 1.27 | 0.1382 |
| Biotype III | 12 (13.6) | 28 (25.7) | 0.53 | 0.0366 |
| Biotype IV | 0 (0)c | 2 (1.8) | 0.31 | 0.2015 |
| Biotype V | 5 (5.7) | 2 (1.8) | 3.10 | 0.1470 |
| Biotype VI | 1 (1.1) | 0 (0)c | 2.48 | 0.2645 |
| Biotype VII | 6 (6.8) | 19 (17.4) | 0.39 | 0.0261 |
| Biotype VIII | 2 (2.3) | 11 (10.1) | 0.23 | 0.0280 |
| LOS gene virulence traits | ||||
| licA | 84 (95.5) | 46 (42.2) | 2.26 | <0.0001 |
| lic2A | 80 (90.0) | 17 (15.6) | 5.83 | <0.0001 |
| lgtC | 88 (100.0) | 2 (1.8) | 54.50 | <0.0001 |
The species designation is based on the stratification of strains in the MLSA dendrogram of Fig. 1 and on the presence or absence of hybridization with an iga gene probe.
PR, prevalence ratio. The prevalence ratios were calculated for H. influenzae by using H. haemolyticus as the referent group.
Logit, 0.5 was used in place of 0 for prevalence ratio and statistical calculations.
A P value of <0.05 is considered statistically significant.
Distribution of classically defined taxonomic traits between the Haemophilus species.
On the basis of this stratification of species, we estimated the distribution of classically defined taxonomic traits such as the production of gas during glucose metabolism, H2S emission, and the lack of ODC activity (traits previously shown to be associated with H. haemolyticus [25]). In addition, we examined indole production and urease activity since these markers (together with ODC) have been routinely used for H. influenzae biotyping and may therefore be differentially distributed among the species.
As expected, the H. haemolyticus type strain, ATCC 33390, was found to produce gas during glucose metabolism, emit H2S, and lack ODC activity, while the four H. influenzae type strains expressed the opposite phenotypes (with the exception of strain 86-028NP, which lacked ODC activity). Gas production and H2S emission were 69 and 88% prevalent, respectively, among the phylogenetically defined H. haemolyticus strains and 12 and 6% prevalent, respectively, among the phylogenetically defined H. influenzae strains (Table 3 and Fig. 1), suggesting that these traits were 17 and 6 times more prevalent, respectively (P < 0.05 for both traits), in H. haemolyticus than in H. influenzae (the prevalence ratios are the inverse of those shown in Table 3). In contrast, ODC activity was about four times more prevalent (P < 0.05) among the H. influenzae strains than among the H. haemolyticus strains. Indole production and urease activity were significantly different (P < 0.05), but they were only 1.3 and 1.2 times more prevalent, respectively, in H. influenzae strains than in H. haemolyticus strains (Table 3). Because H. influenzae and H. haemolyticus are such close phylogenetic relatives, we unconventionally assigned H. influenzae biotype profiles (biotypes I to VIII) to H. haemolyticus strains and compared the prevalence of the profiles between the species. Biotype I was almost six times more prevalent among the H. influenzae strains and biotype III was about two times more prevalent among the H. haemolyticus strains (P < 0.05 for both traits). The overall prevalence rates of these and other biotypes, however, were not high enough to be useful for species differentiation (Table 3). Together, the results of the classical taxonomic traits correlate well with those of a previous taxonomic study by Kilian that used nine H. haemolyticus strains (25), but none of the traits were able to completely differentiate H. influenzae from H. haemolyticus in our larger strain collections.
Distribution of LOS genes with tetranucleotide repeats among H. influenzae and H. haemolyticus strains.
Because the presence or absence of the virulence-associated iga gene appears to corroborate the data obtained by MLSA in segregating H. influenzae from H. haemolyticus, we speculated that other virulence-associated genes might separate the species in a similar manner. LOS-modifying genes in H. influenzae that contain tetranucleotide repeats undergo slipped-strand mispairing, resulting in on-off phase variation in gene expression. These repeats allow the organism to modify its LOS structures in response to host immune pressure, and therefore, the loci are strongly suspected of facilitating virulence (2, 37). Because H. haemolyticus has never been implicated in disease, we hypothesized that LOS genes with tetranucleotide repeats might be lower in prevalence or altogether absent in H. haemolyticus strains compared to their abundance in H. influenzae strains.
Dot blot hybridization was performed with probes specific for licA, lic2A, and lgtC, three LOS genes that contain tetranucleotide repeats. All three genes hybridized to the four H. influenzae type strains but not to the H. haemolyticus type strain (data not shown). The licA, lic2A, and lgtC gene probes hybridized to 96, 90, and 100% of the H. influenzae strains, respectively, and to 42, 16, and 2% of the H. haemolyticus strains, respectively (Table 3). The prevalence ratios calculated from these values showed that the licA, lic2A, and lgtC genes were about 2, 6, and 54 times more prevalent, respectively (P < 0.05 for all associations), in H. influenzae than in H. haemolyticus (Table 3). Of the three LOS contingency genes, these data suggest that only lgtC segregates between the species, in similarity to the iga gene results. Although the prevalence rates of the licA and lic2A genes were high in the H. influenzae strains, they were variably present among the H. haemolyticus strains. In this regard, the relatively high prevalence of the licA gene in H. haemolyticus (42%) may suggest that it facilitates the normal commensal growth of H. haemolyticus.
Recombination between species.
The H. influenzae and H. haemolyticus species designations in this study are supported by a combination of MLSA, molecular, and traditional taxonomic traits. These bacteria, however, may undergo interspecies recombination through natural transformation, a feature that has been well documented between other recombinogenic pathogens and their commensal relatives (18, 23).
To estimate interspecies recombination in H. influenzae and H. haemolyticus, we examined the utility of individual rather than concatenated gene sequences for segregation of the species. Compared to the MLSA-based dendrogram containing the concatenated sequences (shown as a radial view in Fig. 2A), dendrograms constructed from individual adk, pgi, recA, infB, and 16S rRNA gene sequences (Fig. 2B to F, respectively) failed to completely separate H. influenzae from H. haemolyticus. In addition, only the recA sequence was able to maintain a complete Haemophilus sensu stricto cluster separate from four other members (five strains) of the family Pasteurellaceae used in the trees. The 16S rRNA gene sequences poorly defined the cluster; and H. parainfluenzae strain 1.33A did not segregate from the H. influenzae and H. haemolyticus strains with individual adk, pgi, or infB sequences.
Next, we investigated the potential for the unequal distribution of species-specific traits among the MLSA-based nodes separating the species. For instance, strains that phylogenetically border the species dividing line in the MLSA-based dendrogram (Fig. 1) might be expected to exhibit more interspecies recombination than strains that are more distal to that division. As mentioned, node A contains all H. influenzae strains and is separate from nodes B to F, which contain all H. haemolyticus strains. In addition, node A is composed of three clades emerging at subnodes A1 (72 strains), A2 (8 strains), and A3 (8 strains). In the H. haemolyticus strains of nodes B to F, 14 strains were found in nodes B to E, while 95 strains were more distally positioned in the clade emerging from node F. Therefore, the trait prevalence was compared between the H. influenzae strains at node A1 and the combined strains (to increase the sample size) at nodes A2 and A3. Similarly, the trait prevalence was compared between the combined H. haemolyticus strains at nodes B to E and the H. haemolyticus strains at node F.
H2S production (an H. haemolyticus-associated trait) was found to be significantly less prevalent among the H. influenzae strains at node A1 (6%) than among the H. influenzae strains at nodes A2 and A3 (44%), while indole production, the licA gene, and the lic2A gene (all H. influenzae-associated traits) were significantly more prevalent in strains at node A1 (88, 100, and 99% of strains, respectively) than in the combined strains at nodes A2 and A3 (63, 75, and 56%, respectively) (P < 0.05 for all associations) (Table 4). Although the differences were not statistically different, H2S production was less prevalent in the H. haemolyticus strains at nodes B and E (50%) than in the H. haemolyticus strains at node F (72%); and indole production, the licA gene, and the lic2A gene were more common in the strains at nodes B to E (71, 57, and 21%, respectively) than in the strains at node F (61, 40, and 15%, respectively). In addition, the strains at nodes B to E were twice as likely as the strains at node F to react with MAb 7F3 (an H. influenzae-associated trait) (Table 4). In contrast to the results presented above, however, the possession of an lgtC gene and gas production appeared to be equally distributed between clades within the same species, and the urease and ODC activities did not exhibit the trends described above (Table 4). Together, the results of these phylogenetic and taxonomic studies suggest the possibility of recombination between H. influenzae and H. haemolyticus.
TABLE 4.
Trends of species-specific traits within a species
| Trait | No. (%) of strainsa
|
|||
|---|---|---|---|---|
|
H. influenzae nodes
|
H. haemolyticus nodes
|
|||
| A1 (n = 72) | A2 and A3 (n = 16) | B to E (n = 14) | F (n = 95) | |
| H2S production | 4 (5.6) | 7 (43.8)c | 7 (50.0) | 68 (71.6) |
| Indole production | 63 (87.5) | 10 (62.5)c | 10 (71.4) | 58 (61.1) |
| licA | 72 (100) | 12 (75.0)c | 8 (57.1) | 38 (40.0) |
| lic2A | 71 (98.6) | 9 (56.3)c | 3 (21.4) | 14 (14.7) |
| P6 positive | 70 (97.2) | 15 (93.7) | 3 (21.4) | 10 (10.5) |
| lgtC | 72 (100) | 16 (100) | 0 (0)b | 2 (2.1) |
| Gas production | 4 (5.6) | 1 (6.3) | 12 (85.7) | 84 (88.4) |
| Urease activity | 59 (81.9) | 15 (93.8) | 6 (42.8) | 71 (74.7)c |
| ODC activity | 16 (22.2) | 8 (50.0)c | 1 (7.1) | 7 (7.4) |
The nodes and the number of strains in each node are relative to those in the dendrogram in Fig. 1.
Logit, 0.5 was used instead of 0 for statistical calculations.
Nodes possessing a statistically significant difference (P < 0.05) for the trait on the basis of Student's t test.
Distribution of traits between hemolytic and nonhemolytic H. haemolyticus strains.
The relationships between hemolytic and nonhemolytic H. haemolyticus strains are poorly understood. In the MLSA-based dendrogram (Fig. 1), 33 hemolytic H. haemolyticus strains were interdispersed among 62 nonhemolytic H. haemolyticus strains emerging from node F, suggesting that hemolytic and nonhemolytic H. haemolyticus strains do not cluster as a separate subspecies. No hemolytic strains were present in the 14 strains emerging from nodes B to E that bordered the species dividing line. Stratification of the molecular and classical taxonomic traits described above between hemolytic and nonhemolytic H. haemolyticus strains from all nodes revealed that the prevalence of H2S emission, gas formation during glucose metabolism, ODC activity, and the possession of an lgtC gene were not significantly different (P > 0.05) between hemolytic and nonhemolytic H. haemolyticus strains (Table 5). Although urease activity was significantly more prevalent in hemolytic strains than in nonhemolytic strains, reactivity with MAb 7F3 and possession of the licA and lic2A genes were all significantly more prevalent (P < 0.05 for all associations) in nonhemolytic strains than in hemolytic strains (Table 5). Of the 13 total H. haemolyticus strains that reacted with MAb 7F3, none were hemolytic, and of the 17 total H. haemolyticus strains that hybridized with the lic2A probe, only one was hemolytic. Similarly, the licA gene was almost twice as prevalent in nonhemolytic H. haemolyticus strains than in hemolytic H. haemolyticus strains. To decrease the potential for interspecies recombination to contribute to these results, the prevalence of the traits was recalculated for the H. haemolyticus strains emerging only from node F. With the exception of urease activity (which lost its statistical significance), the associations remained very similar to those obtained when all H. haemolyticus strains were used (data not shown). Although hemolytic and nonhemolytic H. haemolyticus strains appear to be phylogenetically inseparable, the results suggest the possibility that some genes and traits common to H. influenzae exist more frequently in nonhemolytic than hemolytic H. haemolyticus strains.
TABLE 5.
Distribution of taxonomic traits between hemolytic and nonhemolytic H. haemolyticus strains
| Trait | No. (%) of strains
|
PRa | Results of χ2 analysis (P valuec) | |
|---|---|---|---|---|
| Hemolytic (n = 33) | Nonhemolytic (n = 76) | |||
| Molecular-based traits | ||||
| Hemolysis | 33 (100) | 0 (0) | Referent | |
| MAb 7F3 | 0 (0)b | 13 (17.1) | 11.3 | 0.0114 |
| Traditional traits | ||||
| H2S production | 24 (72.7) | 51 (67.1) | 0.92 | 0.5605 |
| Gas production | 31 (93.9) | 65 (85.5) | 0.88 | 0.2131 |
| Indole production | 20 (60.6) | 48 (63.2) | 1.04 | 0.8005 |
| Urease activity | 28 (84.8) | 49 (64.5) | 0.76 | 0.0319 |
| ODC activity | 2 (6.1) | 6 (7.9) | 1.30 | 0.7358 |
| LOS gene virulence traits | ||||
| licA | 8 (24.2) | 38 (50.0) | 2.06 | 0.0124 |
| lic2A | 1 (3.0) | 16 (21.1) | 6.95 | 0.0172 |
| lgtC | 0 (0)b | 2 (2.6) | 1.74 | 0.3469 |
PR, prevalence ratio. Prevalence ratios were calculated for nonhemolytic strains by using the hemolytic strains as the referent group.
Logit, 0.5 was used instead of 0 for prevalence ratio and statistical calculations.
A P value of <0.05 is considered statistically significant.
DISCUSSION
Since its first description as “Bacillus X” in 1919, H. haemolyticus has been differentiated from H. influenzae predominantly by its ability to produce hemolysis on blood agar plates (46). Shortly after this description, arguments were made that H. haemolyticus was a hemolytic variant of H. influenzae, since the two species appeared to be closely related by their X and V growth factor dependencies and other criteria (36, 57, 59). In 1953, however, Margaret Pittman (45) empirically proposed that a separate H. haemolyticus species designation should continue, according to the 5th edition of Bergey's Manual of Determinative Bacteriology, published in 1939 (3). Numerous phylogenetic studies, including the current study, clearly validate separation of the species (8, 9, 20, 25, 42, 47). In addition, the same studies have substantiated that H. influenzae and H. haemolyticus are perhaps the closest phylogenetic relatives of each other.
The recent edition of Bergey's Manual of Systematic Bacteriology suggests the possible existence of novel species that are intermediately related to H. influenzae and H. haemolyticus (27, 43). These species include H. intermedius subsp. gazogenes (a nonhemolytic taxon noted for its ability to produce gas in glucose broth, a distinctive feature of the nonhemolytic H. haemolyticus strains in this study) and H. intermedius subsp. intermedius (a species that grows independently of factor X). These proposals of novel species, however, are based only on a few strains that have been segregated with single DNA sequences (e.g., by use of the 16S rRNA gene alone). The increased recognition of recombination within and between species, as evidenced by the tremendous diversity in the genome sequences of recombinogenic bacteria (including H. influenzae [10, 19]), has prompted the recommendation that the phylogenetic designation of species should include analyses of multiple strains of each species together with MLSA (7, 15, 52). The difficulty of separating individual strains by the analysis of individual genes is seen in the current study: H. parainfluenzae strain 1.33A segregated from the Haemophilus sensu stricto phylogenetic cluster in a dendrogram containing five concatenated gene sequences but failed to segregate from H. influenzae and H. haemolyticus in trees created from most individual DNA sequences. Thus, this strain, which is dependent only on factor V, may have been classified as H. intermedius subsp. intermedius if it had been assessed by the use of only one DNA sequence. Although it is not the focus of the current study, these observations also highlight the overall poor understanding of the interspecies relationships of H. parainfluenzae with H. haemolyticus and H. influenzae.
The hemolytic properties of H. haemolyticus are largely uncharacterized. In 1920, Stillman and Bourn (53) noted that the hemolytic activity of Haemophilus species isolated from the throat (which were referred to as Bacillus X but which were most likely H. parahaemolyticus or H. haemolyticus) was unfilterable and stable on ice but was heat labile at 56°C. In addition, strain-to-strain variations in the hemolytic phenotype were noted. As mentioned above, some H. haemolyticus strains, including the type strain, ATCC 33390, are known to have lost their beta-hemolytic phenotype on passage. Interestingly, we have found strain ATCC 33390 to be consistently hemolytic on 5% horse blood agar. Similar to what was reported by Stillman and Bourn (53), the hemolytic activity of this strain was unfilterable and heat labile at 56°C. In addition, the zonal size and clarity of beta-hemolysis among hemolytic H. haemolyticus strains in our collection were highly variable, but beta-hemolysis was consistently present (K. W. McCrea, J. Xie, C. F. Marrs, and J. R. Gilsdorf, unpublished data). The regulation of hemolytic activity in H. haemolyticus is unknown. The hemolytic phenotype in other bacteria is subject to variations in oxygen tension, the concentrations of specific ions, the type of basal medium used, and the type of animal blood used for analysis (24, 48, 49). Regardless of the mechanism, however, the observations of hemolysis in this study strongly support the conclusion of Kilian in 1976 that hemolysis is not a reliable trait to be used for the differentiation of some Haemophilus species (24, 25).
As mentioned above, neither hemolytic nor nonhemolytic H. haemolyticus strains have been associated with disease (40, 62). Therefore, distinguishing H. influenzae from other bacterial species infecting normally sterile sites (i.e., the middle ear or blood) can be done reliably by determining X and V growth factor dependence. The differentiation of H. influenzae from H. haemolyticus in culture specimens potentially contaminated by the pharyngeal microbiome may still rely, in part, on determining the absence of beta-hemolysis, since the prevalence of hemolytic H. haemolyticus strains in specimen collections is highly variable and ranges from 0 to 51% (5, 30, 40, 51). In contrast, a beta-hemolytic phenotype would not be useful in differentiating nonhemolytic H. haemolyticus strains, which are now know to be significantly prevalent in the pharyngeal cavities of healthy children and of adults with COPD (40, 62). A rapid, clinically useful marker for species differentiation is currently not available. In our study, dot blot hybridization with iga and lgtC gene probes specifically differentiated H. haemolyticus from H. influenzae, but further work is necessary to refine their clinical applicability.
Although H. influenzae and H. haemolyticus are closely related commensal organisms of the human pharynx and both are subject to the same host or environmental factors facilitating infection, only H. influenzae has the genetic constitution necessary to cause disease. Thus, comparative genomic studies at the species population level should be able to differentiate unique genotypes of H. influenzae that are associated with disease from common genotypes of both species that are associated with commensal growth. The data presented in the current study begin to examine this. Significant differences in the prevalence of H. influenzae LOS genes associated with virulence were found between the species. The lic2A and lgtC genes were nearly ubiquitous in H. influenzae (≥90%) but were only 16 and 2% prevalent, respectively, in H. haemolyticus. Interestingly, the combined expression of these genes in H. influenzae is necessary to form a LOS digalactoside structure (34, 35) that antigenically mimics the pK blood group of humans and increases the serum resistance of the organism by inhibiting the deposition of the C4b released from the classical complement pathway (21, 61). Of the 17 H. haemolyticus strains in this study that possessed a lic2A gene, none possessed the lgtC gene (unpublished data), suggesting that the digalactoside structure is not made in H. haemolyticus. In contrast to the absence of genes facilitating the formation of the digalactoside structure in H. haemolyticus, the licA gene of H. influenzae was found in close to half (42%) of the H. haemolyticus strains. In H. influenzae, the licA gene is part of the lic-1 locus (which contains the licA, licB, licC, and licD genes), required for modifying LOS with phosphorylcholine (or ChoP). ChoP structurally mimics phosphatidylcholine, an abundant component in human cell membranes, and in H. influenzae ChoP has been shown to provide a defense against host-derived antimicrobial peptides (33) and to mediate bacterium adherence to and invasion of host cells through the platelet-activating factor receptor (55). In addition, ChoP has been shown to enhance the ability of H. influenzae to colonize the middle ear and induce otitis media in the chinchilla model of infection (56). The presence of the licA gene in the H. haemolyticus strains of this study suggests that they may contain a lic-1 locus capable of expressing ChoP. If this were found to be the case, the relatively high prevalence of strains with this locus would suggest that ChoP facilitates normal commensal growth, since H. haemolyticus has not been implicated as a disease agent. Further studies, however, are necessary to demonstrate ChoP expression, its association with LOS, and its function in H. haemolyticus compared with the function of ChoP expression in H. influenzae.
Although this study has focused on the phylogenetic and taxonomic relationships of H. influenzae and H. haemolyticus, the question of how these organisms differ in regard to their virulence and commensalism is more prominent. Although H. influenzae is a serious pathogen, it is by far more devoted to commensal growth rather than pathogenic growth in humans. Yet, little is clearly understood about how genetic factors facilitate the different growth environments. Comprehensive taxonomic studies between H. influenzae and H. haemolyticus by the use of modern bioinformatic tools should be able to generate genotypic and phenotypic blueprints contrasting species similarities that facilitate commensal growth from species differences that facilitate virulent growth in H. influenzae.
Acknowledgments
This work was supported, in part, by Public Health Service grants R03 DC006585-01 to K.W.M. and R01 DC05840 to J.R.G. from the National Institute on Deafness and Other Communication Disorders.
We thank Patricia Juliao for assistance with statistical analysis.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Albritton, W. L. 1982. Infections due to Haemophilus species other than H. influenzae. Annu. Rev. Microbiol. 36199-216. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergey, D. H., R. S. Breed, E. G. D. Murray, and A. P. Hitchens. 1939. Genus IV. Haemophilus Winslow et al., 5th ed. The Williams & Wilkins Co., Baltimore, MD.
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Branson, D. 1968. Bacteriology and clinical significance of hemolytic Haemophilus in the throat. Appl. Microbiol. 16256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casin, I., F. Grimont, and P. A. Grimont. 1986. Deoxyribonucleic acid relatedness between Haemophilus aegyptius and Haemophilus influenzae. Ann. Inst. Pasteur Microbiol. 137B155-163. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, H., M. Bisgaard, W. Frederiksen, R. Mutters, P. Kuhnert, and J. E. Olsen. 2001. Is characterization of a single isolate sufficient for valid publication of a new genus or species? Proposal to modify recommendation 30b of the Bacteriological Code (1990 Revision). Int. J. Syst. Evol. Microbiol. 512221-2225. [DOI] [PubMed] [Google Scholar]
- 8.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 1742002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1993. Phylogeny of the Pasteurellaceae as determined by comparison of 16S ribosomal ribonucleic acid sequences. Zentralbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 27935-44. [DOI] [PubMed] [Google Scholar]
- 10.Erwin, A. L., and A. L. Smith. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 15355-362. [DOI] [PubMed] [Google Scholar]
- 11.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 2341-46. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 13.Foxman, B., L. Zhang, K. Palin, P. Tallman, and C. F. Marrs. 1995. Bacterial virulence characteristics of Escherichia coli isolates from first-time urinary tract infection. J. Infect. Dis. 1711514-1521. [DOI] [PubMed] [Google Scholar]
- 14.Foxman, B., L. Zhang, P. Tallman, K. Palin, C. Rode, C. Bloch, B. Gillespie, and C. F. Marrs. 1995. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J. Infect. Dis. 1721536-1541. [DOI] [PubMed] [Google Scholar]
- 15.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3733-739. [DOI] [PubMed] [Google Scholar]
- 16.Gilsdorf, J. R., H. Y. Chang, K. W. McCrea, and L. O. Bakaletz. 1992. Comparison of hemagglutinating pili of Haemophilus influenzae type b with similar structures of nontypeable H. influenzae. Infect. Immun. 60374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilsdorf, J. R., K. McCrea, and L. Forney. 1990. Conserved and nonconserved epitopes among Haemophilus influenzae type b pili. Infect. Immun. 582252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanage, W. P., C. Fraser, and B. G. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 1874627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedegaard, J., H. Okkels, B. Bruun, M. Kilian, K. K. Mortensen, and N. Norskov-Lauritsen. 2001. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology 1472599-2609. [DOI] [PubMed] [Google Scholar]
- 21.Ho, D. K., S. Ram, K. L. Nelson, P. J. Bonthuis, and A. L. Smith. 2007. lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J. Immunol. 1781002-1012. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. A., J. W. Gautsch, J. R. Sportsman, and J. Elder. 1984. Improved technique utilizing nonfat dry milk for analysis of proteins and nucleic acids transfer to nitrocellulose. Gene Anal. Tech. 13-8. [Google Scholar]
- 23.Kalia, A., M. C. Enright, B. G. Spratt, and D. E. Bessen. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 694858-4869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Kilian, M. 1976. The haemolytic activity of Haemophilus species. Acta Pathol. Microbiol. Scand. Sect. B 84339-341. [DOI] [PubMed] [Google Scholar]
- 25.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 939-62. [DOI] [PubMed] [Google Scholar]
- 26.Kilian, M. 1991. Haemophilus, p. 463-470. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, DC.
- 27.Kilian, M. 2005. Genus III. Haemophilus Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 561AL, p. 883-904. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, NY.
- 28.Kilian, M., J. Mestecky, and R. E. Schrohenloher. 1979. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuklinska, D., and M. Kilian. 1984. Relative proportions of Haemophilus species in the throat of healthy children and adults. Eur. J. Clin. Microbiol. 3249-252. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignments. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 32.Lu, J. J., C. L. Perng, S. Y. Lee, and C. C. Wan. 2000. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol. 382076-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 681664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masoud, H., A. Martin, P. Thibault, E. R. Moxon, and J. C. Richards. 2003. Structure of extended lipopolysaccharide glycoforms containing two globotriose units in Haemophilus influenzae serotype b strain RM7004. Biochemistry 424463-4475. [DOI] [PubMed] [Google Scholar]
- 35.Masoud, H., E. R. Moxon, A. Martin, D. Krajcarski, and J. C. Richards. 1997. Structure of the variable and conserved lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae serotype b strain Eagan. Biochemistry 362091-2103. [DOI] [PubMed] [Google Scholar]
- 36.Miles, A. A., and J. Gray. 1938. Haemophilus para-influenzae endocarditis. J. Path. Bacteriol. 47257. [Google Scholar]
- 37.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 424-33. [DOI] [PubMed] [Google Scholar]
- 38.Mukundan, D., Z. Ecevit, M. Patel, C. F. Marrs, and J. R. Gilsdorf. 2007. Pharyngeal colonization characteristics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J. Clin. Microbiol. 453207-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, P. G., I. Craig, A. C. Lafong, and E. T. Smyth. 1990. Evaluation of two rapid methods for identifying and biotyping Haemophilus influenzae. J. Clin. Pathol. 43581-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 19581-89. [DOI] [PubMed] [Google Scholar]
- 41.Mutters, R., W. Mannheim, and M. Bisgaard. 1989. Taxonomy of the group, p. 3-34. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, United Kingdom.
- 42.Nørskov-Lauritsen, N., B. Bruun, and M. Kilian. 2005. Multilocus sequence phylogenetic study of the genus Haemophilus with description of Haemophilus pittmaniae sp. nov. Int. J. Syst. Evol. Microbiol. 55449-456. [DOI] [PubMed] [Google Scholar]
- 43.Olsen, I., F. E. Dewhirst, B. J. Paster, and H. Busse. 2005. Family I. Pasteurellaceae Pohl 1981b, 382VP (Effective publication: Pohl 1979, 81), p. 851-856. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, NY.
- 44.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 703551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pittman, M. 1953. A classification of the hemolytic bacteria of the genus Haemophilus: Haemophilus haemolyticus Bergey et al. and Haemophilus parahaemolyticus nov. spec. J. Bacteriol. 65750-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchett, I. W., and E. G. Stillman. 1919. The occurrence of Bacillus influenzae in throats and saliva. J. Exp. Med. 29259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quentin, R., C. Martin, J. M. Musser, N. Pasquier-Picard, and A. Goudeau. 1993. Genetic characterization of a cryptic genospecies of Haemophilus causing urogenital and neonatal infections. J. Clin. Microbiol. 311111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowe, G. E., and R. A. Welch. 1994. Assays of hemolytic toxins. Methods Enzymol. 235657-667. [DOI] [PubMed] [Google Scholar]
- 49.Ruoff, K. L., R. A. Whiley, and D. Beighton. 2003. Streptococcus, p. 405-421. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 50.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347465-471. [DOI] [PubMed] [Google Scholar]
- 51.Sims, W. 1970. Oral haemophili. J. Med. Microbiol. 3615-625. [DOI] [PubMed] [Google Scholar]
- 52.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 521043-1047. [DOI] [PubMed] [Google Scholar]
- 53.Stillman, E. G., and J. M. Bourn. 1920. Biological study of the hemophilic bacilli. J. Exp. Med. 32665-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St. Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swords, W. E., B. A. Buscher, K. Ver Steeg Ii, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 3713-27. [DOI] [PubMed] [Google Scholar]
- 56.Tong, H. H., L. E. Blue, M. A. James, Y. P. Chen, and T. F. DeMaria. 2000. Evaluation of phase variation of nontypeable Haemophilus influenzae lipooligosaccharide during nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 684593-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Topley, W. W. C. 1946. Topley and Wilson's principles of bacteriology and immunity, vol. 1, 3rd ed. The Williams & Wilkins Co., Baltimore, MD.
- 58.Turk, D. C., and J. R. May. 1967. Haemophilus influenzae; its clinical importance. English University Press, London, United Kingdom.
- 59.Valentine, F. C. O., and T. M. Rivers. 1927. Further observations concerning growth requirements of hemophilic bacilli. J. Exp. Med. 45993-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vracko, R., and J. C. Sherris. 1963. Indole-spot test in bacteriology. Tech. Bull. Regist. Med. Technol. 3347-50. [PubMed] [Google Scholar]
- 61.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30767-775. [DOI] [PubMed] [Google Scholar]
- 62.Xie, J., P. C. Juliao, J. R. Gilsdorf, D. Ghosh, M. Patel, and C. F. Marrs. 2006. Identification of new genetic regions more prevalent in nontypeable Haemophilus influenzae otitis media strains than in throat strains. J. Clin. Microbiol. 444316-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44225-233. [DOI] [PubMed] [Google Scholar]