Abstract
In staphylococci, inducible macrolide-lincosamide-streptogramin B (MLSB) resistance is conferred by the erm(C) or erm(A) gene. This phenotype is characterized by the erythromycin-clindamycin “D-zone” test. Although clindamycin appears active in vitro, exposure of MLSB-inducible Staphylococcus aureus to this antibiotic may result in the selection of clindamycin-resistant mutants, either in vitro or in vivo. We have compared the frequencies of mutation to clindamycin resistance for 28 isolates of S. aureus inducibly resistant to erythromycin and bearing the erm(C) (n = 18) or erm(A) (n = 10) gene. Seven isolates susceptible to erythromycin or bearing the msr(A) gene (efflux) were used as controls. The frequencies of mutation to clindamycin resistance for the erm(A) isolates (mean ± standard deviation, 3.4 × 10−8 ± 2.4 × 10−8) were only slightly higher than those for the controls (1.1 × 10−8 ± 6.4 × 10−9). By contrast, erm(C) isolates displayed a mean frequency of mutation to clindamycin resistance (4.7 × 10−7 ± 5.5 × 10−7) 14-fold higher than that of the S. aureus isolates with erm(A). The difference was also observed, although to a lower extent, when erm(C) and erm(A) were cloned into S. aureus RN4220. We conclude that erm(C) and erm(A) have different genetic potentials for selection of clindamycin-resistant mutants. By the disk diffusion method, erm(C) and erm(A) isolates could be distinguished on the basis of high- and low-level resistance to oleandomycin, respectively.
Clindamycin is an alternative drug for infections due to Staphylococcus aureus in case of intolerance to penicillins or resistance to methicillin. Furthermore, clindamycin represents an attractive option for several reasons. First, clindamycin is available in both intravenous and oral formulations. Second, the drug has a remarkable distribution into skin and skin structures. Third, community-acquired methicillin-resistant S. aureus (CA-MRSA), which has rapidly emerged in recent years as a cause of skin and soft-tissue infections, is frequently susceptible to several antibiotics, including clindamycin (12, 19). Finally, it has been shown that clindamycin inhibits the production of toxins and virulence factors in gram-positive organisms through inhibition of protein synthesis (7).
However, resistance to clindamycin, which is not rare, limits the use of this antibiotic in therapy. Two primary mechanisms result in resistance to macrolide antibiotics in staphylococci: macrolide efflux, controlled by the msr(A) gene, and modification of the drug-binding site on the ribosome, controlled by erm (erythromycin ribosome methylation) genes (9). The efflux mechanism yields inducible resistance to 14-membered (erythromycin, clarithromycin, roxithromycin) and 15-membered (azithromycin) macrolides and type B streptogramins but not to lincosamides (clindamycin and lincomycin). Ribosomal methylation confers cross-resistance to macrolides, lincosamides, and type B streptogramins, the so-called MLSB phenotype. In staphylococci, erm(A) or erm(C) is responsible for this cross-resistance phenotype by controlling the methylation of the 23S rRNA binding site of adenosine 2058 (A2058) (Escherichia coli numbering). Methylation results in impaired binding of the three drug classes that share this residue as a common binding site.
MLSB resistance can be expressed either constitutively or inducibly. Strains with constitutive resistance express cross-resistance to MLSB antibiotics. Strains with inducible MLSB resistance (MLSBi) display in vitro resistance to 14- and 15-membered macrolides, which are inducer antibiotics, while appearing susceptible to lincosamides and type B streptogramins, which are not inducers. In the absence of an inducer, the inactivity of the mRNA is due to the structure of its 5′ end, which comprises a leader peptide and a set of inverted repeats that form a hairpin structure which sequesters the initiation sequences (ribosome binding site and initiation codon) for the methylase by base pairing. According to the model of posttranscriptional regulation, induction arises through binding of an inducer macrolide to a ribosome during translation of the leader peptide, leading to destabilization of the hairpin structure, exposure of the initiation sequences to the ribosome, and translation of the Erm methylase (2).
Although clindamycin is not an inducer, exposure of MLSB-inducible S. aureus to this antibiotic may result in cross-MLSB resistance, either in vitro or in vivo. This is due to the selection of preexisting constitutive erm mutants (4, 6, 17). Whether in laboratory strains or in clinical isolates, deletion of the entire attenuator, point mutations, or tandem duplications in the attenuator yield constitutive expression of resistance by decreasing the stability of the hairpin structure sequestering the initiation sequences for the methylase.
Clinical failures related to the selection of clindamycin-resistant mutants have been reported in a few cases, and there is concern over the emergence of resistance during clindamycin therapy (4, 6, 10, 16, 17). The risk for selection of resistance may depend on various factors, including the frequency of mutation to resistance, the bacterial inoculum size, and the type of infection. Since erm(A) and erm(C) attenuators display marked differences in terms of length and of leader peptides and inverted-repeat sequences, we hypothesized that the clindamycin mutation frequencies and therefore the risk of mutant selection might differ according to the gene.
In this study, we compare the clindamycin mutation frequencies of clinical isolates of S. aureus containing an erm(A) or an erm(C) gene and propose the oleandomycin test as a marker to identify the genotype of the isolate.
MATERIALS AND METHODS
Bacterial strains and antibiotic susceptibility testing.
Twenty-eight S. aureus isolates inducibly resistant to erythromycin and bearing the erm(C) (n = 18) or erm(A) (n = 10) gene were included in the study. Twelve of these isolates [five erm(A) isolates, kindly provided by Peter Appelbaum, and seven erm(C) isolates] were CA-MRSA isolates representative of European and North-American clones. Four erm(A) and seven erm(C) isolates were hospital-acquired MRSA isolates. The other isolates were susceptible to methicillin.
Three erythromycin- and clindamycin-susceptible S. aureus isolates (one clinical isolate, S. aureus RN4220, and S. aureus ATCC 29213), and four S. aureus isolates with the efflux gene msr(A) were included as controls. Escherichia coli DH10B and S. aureus RN4220 were used as recipients for cloning experiments.
Susceptibility to antibiotics was determined by the agar diffusion technique with disks (Bio-Rad, Marnes-la-Coquette, France) of erythromycin (15 μg), oleandomycin (15 μg), and clindamycin (2 μg) as recommended by the Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) (1). Briefly, a suspension in 0.9% saline equivalent to a 0.5 McFarland standard was prepared from an overnight agar medium culture plate, diluted to 1/100 (∼106 CFU/ml), and spread by swabbing onto Mueller-Hinton agar. We confirmed the inducible phenotype by the D-zone test, which consists of placing standard erythromycin and clindamycin disks in adjacent positions 15 to 26 mm apart on a Mueller-Hinton agar plate as recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) (14). Inducible resistance to clindamycin is expressed as a flattening of the clindamycin zone of inhibition adjacent to the erythromycin disk, giving a D shape to the zone of inhibited growth following 16 to 18 h of incubation at 35°C in ambient air.
Mutation frequencies.
For determination of mutation frequencies, cells from an overnight agar culture were suspended in saline to a turbidity of a 6 MacFarland standard, and ca. 108 to 5 × 109 CFU was spread onto Trypticase-soy agar plates (Bio-Rad) supplemented with 20 μg/ml of clindamycin. The bacterial inoculum was measured by using a spiral system (Interscience, Saint-Nom-la-Bretèche, France). After 48 h of incubation at 35°C, colonies were counted and the mutation frequencies were determined relative to the total count of viable organisms plated. Each experiment was repeated three or four times, and results were expressed as means and standard deviations.
The presence of hypermutable strains among the S. aureus strains studied was searched for by spreading ca. 107 to 108 CFU on Trypticase soy agar plates supplemented with rifampin (100 μg/ml) (Sigma-Aldrich, Saint Louis, MO). After 48 h of incubation at 35°C, every isolate that yielded >10 colonies on antibiotic-containing medium was considered a potential hypermutable bacterium (15). The experiment was repeated in triplicate.
The means were compared by the parametric t test and the nonparametric Mann-Whitney test. All P values were based on two-sided comparisons and were taken to be significant at ≤0.05 (5).
Molecular techniques.
DNA was extracted by the Instagen Matrix kit (Bio-Rad) as recommended by the manufacturer. The presence of the methylase genes erm(A) and erm(C) and of the efflux gene mrs(A) in the isolates studied was confirmed by PCR as described previously (11).
Sequences upstream from the methylase genes of 18 clindamycin-resistant mutants, 9 derived from S. aureus strain 23 [a clinical isolate containing the erm(A) gene] and 9 derived from S. aureus strain 9 [a clinical isolate containing the erm(C) gene] were amplified by PCR with the oligonucleotides shown in Table 1. The PCR consisted of 30 cycles of denaturation (95°C, 30 s), annealing [50°C, 30 s, for erm(A) and 48°C, 30 s, for erm(C)], and extension (72°C, 1 min). Amplified DNA strands were sequenced in both directions. DNA analyses and nucleotide comparisons were carried out using the National Center for Biotechnology Information server at http://www.ncbi.nlm.nih.gov or the MultAlin server at http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html.
TABLE 1.
Primers used in this study
| Gene | Primer designation | Primer sequence (5′ to 3′)a | Product size (bp) |
|---|---|---|---|
| For cloning into S. aureus RN4220 | |||
| erm(A) | erm(A)BH | + CAGTGGATCCTCTTATCAAG | 1,107 |
| erm(A)XB | − CTTGTGATCTAGAGAACGC | ||
| erm(C) | erm(C)BH | + TTAGATGGATCCCTCATATC | 999 |
| erm(C) XB | − TGCATCTCTAGACTTACTTATT | ||
| For sequencing of the translational attenuator | |||
| erm(A) | erm(A)1LP | + TTTTTAGTAAAGACAGTGGC | 481 |
| erm(A)2LP | − GTCCTTTTCCTGATCCGA | ||
| erm(C) | erm(C)1LP | + TACAAGAAAAAAGAAATTAG | 327 |
| erm(C)2LP | − ATCTATATTATGTTTTGAAG |
+, sense primer; −, antisense primer.
Cloning experiments.
The entire erm(A) and erm(C) genes were amplified from two clinical isolates with the specific primers shown in Table 1 and were cloned into the multicopy shuttle vector pAT28 (spectinomycin resistance) using the BamHI and XbaI restriction sites (18). The plasmid constructs were introduced into E. coli DH10B by electroporation and were subsequently extracted and electroporated into S. aureus RN4220. Staphylococcal transformants were selected on Trypticase soy agar plates containing 60 μg/ml of spectinomycin and 50 μg/ml of erythromycin. The MLSB resistance phenotype of two transformants, called S. aureus RN4220erm(A) and RN4220erm(C), was verified by the D-zone test, and mutation frequencies on clindamycin were determined as mentioned above.
RESULTS
Phenotypes and mutation frequencies.
The inducible MLSB phenotype was confirmed for the erm(A) and erm(C) isolates by the D-zone test. Notably, no inhibition zone was observed with the oleanomycin disk for erm(C) isolates, whereas erm(A) isolates displayed zone diameters greater than 15 mm (range, 16 mm to 21 mm).
No clindamycin-resistant mutant could be selected for S. aureus ATCC 29213, S. aureus RN4220, or one erm(A) and three msr(A) isolates by using inocula between 1.2 × 108 and 2.5 × 109 CFU. For those strains that did not yield mutants, the inverse of the inoculum value was taken as the frequency of mutation for the purpose of calculations.
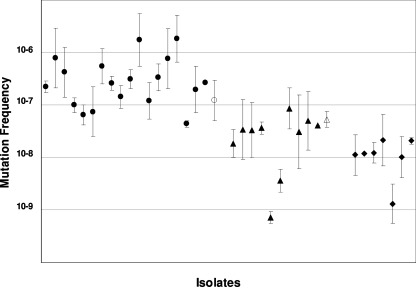
The frequencies of mutation to clindamycin resistance for the erm(A) isolates (range, 7.3 × 10−10 to 1.8 × 10−8; mean ± standard deviation, 3.4 × 10−8 ± 2.4 × 10−8) were slightly higher than those for the controls (range, 1.3 × 10−9 to 1 × 10−8; mean ± standard deviation, 1.1 × 10−8 ± 6.4 × 10−9) (Fig. 1). However, the difference was significant by the t test (P = 0.046) but not by the Mann-Whitney test (P = 0.055).
FIG. 1.
Frequencies of mutation to clindamycin resistance of S. aureus strains, including 18 with erm(C) (solid circles), 10 with erm(A) (solid triangles), and 7 controls (solid diamonds). Open circle and triangle represent RN4220erm(C) and RN4220erm(A), respectively. Approximately 108 to 5 × 109 CFU of cells was spread onto agar plates containing 20 μg/ml of clindamycin. After 48 h of incubation at 35°C, colonies were counted, and the mutation frequencies were determined relative to the total count of viable organisms plated. Each data point represents the mean mutation frequency calculated from three experiments for one strain.
By contrast, mutants were readily obtained for all the S. aureus isolates with erm(C). On average, erm(C) isolates displayed a 14-fold higher frequency of mutation to clindamycin resistance (range, 1.7 × 10−6 to 4.4 × 10−8; mean ± standard deviation, 4.7 × 10−7 ± 5.5 × 10−7) than the S. aureus isolates with erm(A) (Fig. 1). These differences were observed whether the isolates were susceptible or resistant to methicillin. The means of mutation frequencies in the erm(C) group differed significantly from those in the erm(A) and control groups by the Mann-Whitney test (P < 0.001 and P = 0.002, respectively) and by the t test (P = 0.019 and P = 0.040, respectively). Therefore, clinical strains of S. aureus with the erm(C) gene are the most likely to readily develop constitutive resistance to clindamycin.
We could not exclude the possibility that the higher mutation frequencies observed for the erm(C) isolates might be related to a hypermutable background of the isolates. Therefore, we screened for a putative hypermutable status of those isolates. No potential hypermutable bacteria were detected, and no difference could be observed between erm(A) and erm(C) isolates. In addition, no clear difference between mutation rates for hospital-acquired MRSA versus CA-MRSA erm(C) and erm(A) isolates was observed.
The introduction of erm(C) and erm(A) genes into S. aureus RN4220 resulted in differences in frequencies of mutation to clindamycin resistance with, again, a higher frequency for erm(C) [1.2 × 10−7 ± 2.1 × 10−7 for erm(C) versus 5.3 × 10−8 ± 3.6 × 10−8 for erm(A)]. This observation confirmed the data for clinical isolates, although the difference was less striking.
Methylase attenuator mutations.
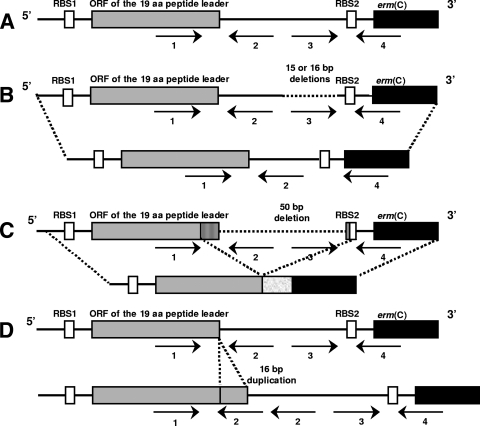
The sequence of the attenuator was compared between MLSBi parents and derivatives. Structural alterations in the attenuators of four of nine clindamycin-resistant derivatives of S. aureus strain 9 [erm(C)] were observed. Two deletions of 15 and 16 bp, one deletion of 50 bp, and one tandem duplication of a 16-bp sequence resulted in marked alterations in a series of inverted repeats involved in the control of methylase expression (Fig. 2). In all cases, alterations of the attenuator would potentially lead to constitutive expression of the methylase, whether under the control of the ribosome binding site of the leader peptide or of its own ribosome binding site. In particular, the 50-bp deletion would delete the stop codon of the leader peptide, creating a translational fusion of the peptide with the methylase.
FIG. 2.
Schematic presentation of the regulatory regions of the inducibly expressed erm(C) gene from S. aureus 9 (A) and the constitutively expressed erm(C) genes selected in this study (B, C, and D). RBS1 and RBS2, ribosome binding sites of the leader peptide and the erm(C) gene, respectively. Arrows indicate the inverted repeated sequences IR1 to IR4. (A) Wild-type erm(C) gene; (B) derivatives with 15- and 16-bp deletions in IR3; (C) derivative with a 50-bp deletion (deletion of the stop codon of the leader peptide generates a translational fusion with the methylase); (D) derivative with tandem duplication of a 16-bp sequence (insertion of a new IR2).
By contrast, none of the nine erm(A) mutants analyzed contained any alteration of the regulatory region. This unexpected result might be related to the use of a relatively low inoculum (approximately 109 CFU), which might be insufficient to select mutants with alterations of the erm(A) attenuator in S. aureus 23 but enough to select ribosomal mutants.
DISCUSSION
Clindamycin is widely used to treat serious staphylococcal infections, in particular for children, due to limited alternative therapy. However, recent reports indicate that failure may occur in the case of inducible MLSB resistance in spite of in vitro susceptibility to clindamycin (4, 6, 10, 16, 17). In most cases, failure was related to the selection of MLSB constitutive mutants resistant to clindamycin. Although clinical data are limited and sometimes conflicting, concern over the emergence of resistance during clindamycin therapy has led the CLSI to recommend the use of a simple laboratory test, the erythromycin-clindamycin D-zone test, to differentiate isolates that have a high genetic potential to become resistant during clindamycin therapy (those with erm genes) from isolates that have a low genetic potential to become resistant [those with the msr(A) gene]. In the study by Lina et al., 70% of MLSBi S. aureus isolates contained erm(C) and 30% contained erm(A) (11). In a recent study, among 402 S. aureus isolates, the overall prevalence of MLSBi was 52%, with 50% of MRSA and 60% of methicillin-susceptible S. aureus isolates exhibiting MLSBi (12). CA-MRSA strains had a lower prevalence of MLSBi than hospital-associated MRSA strain (33% versus 55%). Therefore, the MLSBi phenotype is common in staphylococci, although less prevalent in CA-MRSA strains.
In this study, we were able to select mutants resistant to clindamycin from most isolates, including those that were susceptible to erythromycin. Generally, frequencies of mutation to clindamycin were around 10−8 or below for susceptible isolates. Although we did not study the mechanisms of clindamycin resistance, mutations in ribosomal structures such as the rrl gene (23S rRNA) or ribosomal proteins were probably involved. In agreement with this idea, a previous study has shown that selection of ribosomal mutants by erythromycin occurred in erythromycin-susceptible and nonhypermutable S. aureus strains at frequencies similar to those determined in our study for clindamycin resistance (15). Mutations to clindamycin resistance were within the same range for isolates with the msr(A) gene, confirming that this type of isolate was not particularly at risk for selection of resistant mutants, similarly to wild-type isolates.
The erm(A) isolates had mutation frequencies between 10−7 and 10−8, slightly higher than those for the msr(A) or susceptible isolates. By contrast, mutation frequencies were generally between 10−6 and 10−7 for erm(C) isolates; thus, the mean risk of mutation was 14 times greater than that for the erm(A) isolates. The behavior of two strains of S. aureus RN4220 that differed only in carrying the erm(A) or the erm(C) gene confirmed this observation. However, the mutation frequencies for the constructs S. aureus RN4220erm(A) and RN4220erm(C) were only slightly different. This could be due to the cloning of erm(A) on a multicopy plasmid, whereas under natural conditions the gene is usually borne by transposon Tn554 and is present in a single copy or a few copies in the chromosome of S. aureus (13). By contrast, erm(C) is often borne by small multicopy plasmids in clinical isolates (8).
The reason for the differences in mutation frequencies between erm(A) and erm(C) might be explained by differences in the respective structures of attenuators. The regulatory region of erm(C) comprises one leader peptide and four inverted repeats. erm(A) has a longer regulatory region, with a much more complex structure that includes two leader peptides and six inverted repeats (13). Possibly, due to the complexity of the erm(A) attenuator, there are fewer opportunities for mutations spontaneously occurring in the regulatory region to generate constitutive expression of MLSB resistance than there are for the erm(C) attenuator. Alternatively, the particular hairpin structure of the erm(C) attenuator might favor errors of the DNA polymerase, particularly deletions, more than the erm(A) attenuator would. This would explain why we did not observe attenuator alterations in the nine mutants selected from S. aureus 23 [erm(A)] at the inoculum that we used. By contrast, in four of nine mutants from S. aureus 9, deletions or duplications affected the regulatory region of the erm(C) gene.
More clinical isolates should be studied to reinforce the validity of our results. In addition, the clinical relevance of the difference observed in vitro between erm(C) and erm(A) in the selection of clindamycin resistance remains to be fully established. Since the type of erm gene was not reported for the published cases of clindamycin failure, we do not know if erm(C) was mostly involved, as expected from our results. However, the frequencies of mutation to clindamycin resistance, which are between 10−6 and 10−7 for the inducible erm(C) staphylococci, can be considered relevant for clindamycin therapy of infections with bacterial inocula usually exceeding 107 CFU, such as abscesses requiring drainage, mediastinitis, and some lower respiratory tract infections. In these cases, clindamycin therapy has a high probability of failure and should be avoided. The lower frequencies of mutation observed for inducible erm(A) staphylococci do not mean that the use of clindamycin is safe in those cases, and if clindamycin is used for treatment of infections due to such isolates, close follow-up and monitoring for failure or relapse are needed.
Further clinical studies are required to establish whether, for a given MLSBi isolate, the identification of the type of the erm gene has any importance for decision making if clindamycin therapy is considered. Research protocols should include distinction between erm(A) and erm(C) by genotypic techniques, such as PCR, and the oleandomycin test as a phenotypic marker. In a previous publication, Di Modugno et al. showed that oleandomycin MICs for staphylococci with inducible erm(C) genes were greater than 32 μg/ml, whereas those for staphylococci with inducible erm(A) genes were 2 to 8 μg/ml, and they proposed to use high-level resistance to oleandomycin as a marker of inducible erm(C) genes (3). In the same study, similar differences in clarithromycin MICs were also observed, and these could also be considered as a suitable marker. We confirmed the conclusions of this study, and we propose the use of a disk of oleandomycin (15 μg) to distinguish staphylococci with inducible erm(C) genes (no inhibition zone) from those with inducible erm(A) genes (zone diameters greater than 15 mm by the CA-SFM technique).
In conclusion, the D-zone test is useful for identifying the non-MLSBi S. aureus isolates against which clindamycin can be used safely in therapy. When isolates display the MLSBi phenotype, the oleandomycin test may provide additional information for evaluation of the potential therapeutic use of clindamycin. In the future, the development of genetic techniques for routine identification of resistance genes will be helpful in guiding therapy more accurately.
Acknowledgments
We thank Peter Appelbaum for the gift of strains.
This work was supported in part by a grant from the Fondation pour la Recherche Médicale.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2007. Communiqué 2007. Société Française de Microbiologie, Paris, France. http://www.sfm.asso.fr/.
- 2.Depardieu, F., I. Podglajen, R. Leclercq, E. Collatz, and P. Courvalin. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 2079-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Modugno, V., M. Guerrini, S. Shah, and J. Hamilton-Miller. 2002. Low level resistance to oleandomycin as a marker of ermA in staphylococci. J. Antimicrob. Chemother. 49425-427. [DOI] [PubMed] [Google Scholar]
- 4.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48315-316. [DOI] [PubMed] [Google Scholar]
- 5.Fleiss, J. L. 1981. Statistical methods for rates and populations, 2nd ed. Wiley, New York, NY.
- 6.Frank, A. L., J. F. Marcinak, P. D. Mangat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21530-534. [DOI] [PubMed] [Google Scholar]
- 7.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 692996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol. 150804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34482-492. [DOI] [PubMed] [Google Scholar]
- 10.Lewis, J. S., II, and J. H. Jorgensen. 2005. Inducible clindamycin resistance in staphylococci: should clinicians and microbiologists be concerned? Clin. Infect. Dis. 40280-285. [DOI] [PubMed] [Google Scholar]
- 11.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 431062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcinak, J. F., and A. L. Frank. 2006. Epidemiology and treatment of community-associated methicillin-resistant Staphylococcus aureus in children. Expert Rev. Anti-Infect. Ther. 491-100. [DOI] [PubMed] [Google Scholar]
- 13.Murphy, E. 1985. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J. Bacteriol. 162633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NCCLS/CLSI. 2004. Performance standards for antimicrobial susceptibility testing: 12th informational supplement. NCCLS document M100-S14. NCCLS/CLSI, Wayne, PA.
- 15.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclercq. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 1871709-1716. [DOI] [PubMed] [Google Scholar]
- 16.Rao, G. G. 2000. Should clindamycin be used in treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob. Chemother. 45715. [DOI] [PubMed] [Google Scholar]
- 17.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 371257-1260. [DOI] [PubMed] [Google Scholar]
- 18.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 184296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tristan, A., M. Bes, H. Meugnier, G. Lina, B. Bozdogan, P. Courvalin, M. E. Reverdy, M. C. Enright, F. Vandenesch, and J. Etienne. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]