Abstract
Sheeppox virus (SPPV) is a member of the Capripoxvirus (CaPV) genus of the Poxviridae family. Members of this genus, which also include goatpox and lumpy skin disease viruses, cause economically significant disease in sheep, goats, and cattle. A rapid diagnostic assay for CaPV would be useful for disease surveillance as well as for detection of CaPV in clinical samples and for outbreak management. Here we describe a fluorogenic probe hydrolysis (TaqMan) PCR assay designed for rapid detection of CaPV and tested on sheep experimentally infected with a virulent strain of SPPV. This assay can detect SPPV in buffy coats, nasal swabs, oral swabs, scabs, and skin lesions as well as in lung and lymph nodes collected at necropsy. This single-tube diagnostic assay can be performed in 2 h or less and can detect viral DNA in preclinical, clinical, and postmortem samples.
The Capripoxvirus (CaPV) genus of the Poxviridae family of viruses comprises sheeppox (SPPV), goatpox (GTPV), and lumpy skin disease (LSDV) viruses, which cause disease in sheep, goats, and cattle, respectively. These viruses are considered reportable agents to the World Organization for Animal Health due to their potential for significant economic impact. Members of the CaPV genus are closely related, with genomic identities ranging from 96% between viral species to 99% between isolates of the same species (27). They have complete open reading frame (ORF) colinearity (13, 26, 27) and are indistinguishable via serological methods (14-16). CaPVs tend to be host specific; however, incidences where SPPV and GTPV have crossed species, into goats and sheep, respectively, have been documented (8, 20).
Transmission of SPPV is thought to occur through exposure to aerosols or respiratory droplets produced by acutely infected animals or by direct or indirect contact with lesions or oronasal secretions (8, 17). Virus can be detected in nasal secretions of animals infected by aerosol or contact exposure (17) as well as in lesions found in the upper and lower respiratory tracts of infected animals (1c, 11). Transmission may also occur through the dermis, after contact exposure with cuts or abrasions (1c, 21, 22, 25), or via mechanical transmission by arthropod vectors (19).
Clinical signs of SPPV infection include fever, anorexia, depression, conjunctivitis, rhinitis, respiratory distress, generalized skin lesions, and enlargement of lymph nodes (9, 22). A transient viremia develops but may not be detectable throughout the entire course of infection (14). Transient viremias have also been described for cattle infected with LSDV (7, 28). Internal lesions are often seen at necropsy, especially in the lungs, although lesions in the trachea, rumen, tongue, kidney, nasal turbinates, and reproductive organs have also been reported (1c, 11, 14).
SPPV is endemic throughout much of Africa, southwest and central Asia, and the Indian subcontinent (3, 4, 23). Young animals are most susceptible, where mortality rates can be as high as 50 to 70% (10, 12). Outbreaks are controlled by ring vaccination, quarantine, and slaughter and may result in substantial economic losses due to mortality, reduced productivity, and trade restrictions (6, 24; www.fao.org). Rapid identification of CaPV is key to curtailing outbreaks before major economic damage can be inflicted.
A rapid test for CaPV that can detect virus before the onset of clinical disease would be invaluable for controlling or managing outbreaks as well as for disease surveillance. Here we describe a fluorogenic probe hydrolysis (TaqMan) PCR assay designed for rapid detection of CaPV and tested on sheep experimentally infected with a virulent strain of SPPV. This single-tube assay can be performed in 2 h or less and can detect viral DNA prior to the onset of clinical disease.
MATERIALS AND METHODS
Viruses and cell cultures.
The pathogenic field isolate SPPV strain A (SPPV-SA) was obtained from a sick sheep in the Almatinskaya region of Kazakhstan and passaged nine times in sheep at the Scientific Research Agricultural Institute, Kazakhstan (1987) (27). The pathogenic virus SPPV_RvKLP (RvKLP) was derived from SPPV-SA as previously described (2). Primary lamb testis (LT) cell cultures were obtained from the Animal and Plant Health Inspection Service (APHIS), U.S. Department of Agriculture, Plum Island Animal Disease Center, Greenport, NY.
Animal infections.
Merino lambs (3 to 4 months old) were inoculated intranasally with SPPV-SA (n = 3) or RvKLP (n = 5) at 106 PFU. Clinical signs of SPPV infection, including fever (a rectal temperature of ≥39.7°C), anorexia, lethargy, conjunctivitis, coughing, sneezing, nasal discharge, respiratory distress, recumbency, and appearance of skin lesions, were monitored daily. Nasal swabs, oral swabs, and conjunctival swabs were collected every day for the first 10 days of the experiment and every other day thereafter for the duration of the 30-day experiment. Scrapings of pox scabs were collected as they appeared. Clinical samples were collected into tubes containing Dulbecco's modified Eagle's medium (Invitrogen, San Diego, CA) with 50 μg of gentamicin/ml (Invitrogen, San Diego, CA) and stored at −70°C until use. Blood samples were collected into BD Vacutainer cell preparation tubes with sodium heparin (BD, Franklin Lakes, NJ). Buffy coats were separated by centrifugation in a swinging rotor at 1,500 rpm (800 × g), collected, spun down, resuspended in phosphate-buffered saline, and stored at 4°C until use. Lambs were euthanized when they were moribund or at the end of the 30-day experiment. Tissue samples were collected for virus isolation (VI) or DNA extraction. Tissues collected at necropsy were immediately frozen at −70°C until use.
Virus isolation.
Isolation of SPPV in clinical or necropsy samples was performed on LT cells as described in World Organization for Animal Health protocols (22). Nasal or buccal swabs, sonicated buffy coat samples, or homogenized tissue samples were filtered through a 0.45-μm Spin-X filter (Costar, Corning, NY), and 200 μl of sample was used to infect LT cells in 24-well plates in duplicate with negative controls. Cells were examined for cytopathic effect (CPE) for 14 days after infection. The contents of negative wells were collected and freeze-thawed, and the supernatant was sonicated and spun down. Samples were then used to infect fresh LT cells, which were examined for another 14 days postinfection (p.i.) for CPE. Samples that had no CPE after the second passage were considered VI negative.
DNA extraction.
DNAs were extracted from 200 μl of cell culture supernatant, transport medium containing nasal or buccal swabs, buffy coat samples, or homogenized tissue samples by using a DNeasy blood mini kit (Qiagen, Stanford, CA) and were eluted in 100 μl of AE buffer according to the manufacturer's instructions. DNA from purified virus was extracted as previously described (29).
Real-time PCR assay for SPPV.
Reagents from an EZ-RT PCR kit (Applied Biosystems, Branchburg, NJ) were used to prepare the reaction mix according to the manufacturer's guidelines. The final 25-μl assay mix contained EZ buffer solution, 5 mM manganese acetate, a 0.2 mM deoxynucleoside triphosphate mix, and 0.1 U of recombinant rTth DNA polymerase. A forward primer (5′ GGCGATGTCCATTCCCTG 3′) (200 nM), reverse primer (5′ AGCATTTCATTTCCGTGAGGA 3′) (500 nM), and fluorogenic minor groove-binding TaqMan probe (5′ CAATGGGTAAAAGATTTCTA 3′; labeled with 6-carboxyfluorescein and a nonfluorescent quencher) (200 nM) were included in each reaction mix. Sample template (2.5 μl) was added to the reaction mix in a SmartCycler (Cepheid, Sunnyvale, CA) 25-μl reaction tube. Cycling conditions consisted of an initial denaturation at 95°C for 120 seconds, followed by 45 amplification cycles (95°C for 2 s and 60°C for 60 s). The assay was run on a Cepheid SmartCycler (Cepheid, Inc., Sunnyvale, CA). Positive and negative controls were included with each set of reaction mixtures.
PCR detection.
All virus samples used to assess assay specificity were first tested using virus-specific primers to verify the presence of viral nucleic acid (not shown). Clinical samples for which there were conflicting data between VI and real-time PCR were retested by conventional PCR using the KS-1.5 and KS1.6 forward and reverse primers, as previously described (18).
Statistical analysis.
The performance of the CaPV real-time PCR assay was compared to that of virus isolation and/or conventional PCR detection in clinical and necropsy samples by using the disease/test relation calculators available at the University of Oklahoma Health Sciences Center website (http://www.fammed.ouhsc.edu/robhamm/cdmcalc.htm). The screening and diagnostic tests/validity measures option in the Describe program of WINPEPI (http://www.brixtonhealth.com) was utilized to calculate real-time PCR cutoff values.
RESULTS AND DISCUSSION
Assay design and optimization.
Complete sequences available for the genomes of eight CaPVs were used for primer and probe selection (13, 26, 27). These sequences included three strains of SPPV (SPPV-SA, SPPV_Turky, and SPPV_Niskhi), two strains of GTPV (GTPV_Pellor and GTPV_G20-LKV), and three strains of LSDV (LSDV_Neethling 2490, LSDV_Neethling Warmbaths, and LSDV_Neethling vaccine). Additionally, a partial sequence was available for the KS-1 strain of SPPV (5). Primers and probes were selected using ABI Prism Primer Express primer design software (Applied Biosystems, Foster City, CA) from central regions of the genome (ORFs 24 though 123), where the least genomic variability was observed (26, 27). Primer-probe sets were searched against the NCBI nucleotide database and selected based on 100% nucleotide identity to all members of the CaPV genus. The target region falls within ORF 068 [poly(A) polymerase (small subunit) gene]. Although this gene is present in other poxviruses, only members of the CaPV genus were detectable with the selected primer-probe system (see below).
Assay analytical sensitivity and specificity.
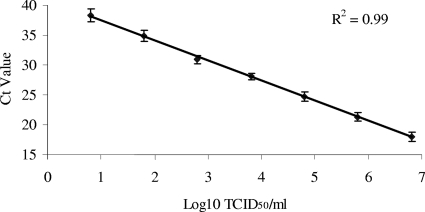
The analytical sensitivity of the CaPV real-time PCR assay was determined based on viral DNA extracted from purified SPPV-SA grown in LK cells. Virus was diluted in log10 steps in Dulbecco's modified Eagle's medium, and DNA was extracted and tested by real-time PCR to determine the limit of detection. Sensitivity was based on four independent experiments. After adjustment for extraction (100 μl) and sample (2.5 μl) volume, the sensitivity of the assay ranged between 1.6 and 15.8 50% tissue culture infective doses of SPPV. The efficiency of amplification [10(−1/slope)] was 1.99, with an R2 value of 0.99 (Fig. 1), indicating an approximate doubling of product after each cycle. In four independent experiments, the mean standard deviation was determined to be 1.0 cycle threshold (CT) among all dilutions.
FIG. 1.
Sensitivity of the CaPV real-time PCR assay, calculated using SPPV samples obtained from infected cell culture supernatants and run in quadruplicate. An inverse linear relationship exists between CT values and log10 50% tissue culture infective dose (TCID50)/ml of virus in the samples. The efficiency of amplification was 1.99, with an R2 value of 0.99, indicating an approximate doubling of product after each cycle.
The specificity of the CaPV real-time assay was determined for members of the CaPV genus. DNAs were extracted from five strains of SPPV, two strains of GTPV, and four strains of LSDV and tested by real-time PCR (Table 1). All samples were positive, with CT values ranging from 16 to 23. Members of the Orthopoxvirus, Parapoxvirus, and Suipoxvirus genera of the Poxviridae tested negative by the CaPV real-time PCR assay (Table 1).
TABLE 1.
CaPVs and other poxviruses tested by the CaPV real-time PCR assaya
| Genus | Virus | Strain | Species of origin | 6-Carboxyfluorescein CT | Reference or source |
|---|---|---|---|---|---|
| Capripoxvirus | SPPV | Almatinskaya (SA) | Sheep | 16.27 | 27 |
| SPPV | Turkey | Sheep | 19.88 | 27 | |
| SPPV | Niskhi | Sheep | 22.32 | 27 | |
| SPPV | Kenya | Sheep | 17.28 | USDA, FADDL | |
| SPPV | Romania | Sheep | 19.6 | USDA, FADDL | |
| GTPV | Pellor | Goat | 22.48 | 27 | |
| GTPV | Israel | Goat | 18.26 | USDA, FADDL | |
| LSDV | Makhana | Cow | 24.4 | USDA, FADDL | |
| LSDV | Neethling | Cow | 23.28 | 26 | |
| LSDV | Ismalia | Cow | 16.22 | USDA, FADDL | |
| LSDV | Cameroon | Cow | 18.58 | USDA, FADDL | |
| Parapoxvirus | Orf virus | Oregon | Goat | 0 | USDA, FADDL |
| Orf virus | Virginia | Goat | 0 | USDA, FADDL | |
| Orf virus | Indiana | Goat | 0 | USDA, FADDL | |
| Bovine papular stomatitis virus | Kansas | Cow | 0 | USDA, FADDL | |
| Bovine papular stomatitis virus | California | Cow | 0 | USDA, FADDL | |
| Orthopoxvirus | Vaccinia virus | Western Reserve | NA | 0 | |
| Camelpox virus | Mangistauskiy | Camel | 0 | 1b | |
| Horsepox virus | Mongolia | Horse | 0 | 26 | |
| Suipoxvirus | Swinepox virus | Nebraska | Pig | 0 | 1a |
Samples were first tested by PCR or reverse transcription-PCR, using virus-specific primers, and then by real-time PCR using the CaPV real-time assay. Positive (SA) and no-template controls were included with each set of reactions.
Agents included in differential diagnosis of CaPV as well as samples of other viral vesicular diseases affecting sheep, goats, or cattle were obtained from the Foreign Animal Disease Diagnostic Laboratory (FADDL), APHIS (U.S. Department of Agriculture, Plum Island Animal Disease Center, Greenport, NY), and tested using virus-specific primers in order to verify the presence of viral nucleic acid. Samples were then examined using the CaPV real-time assay. Bovine herpesvirus 2, bluetongue virus, rinderpest virus, peste des petits ruminants virus, foot-and-mouth disease virus, and vesicular stomatitis virus all tested negative by the CaPV real-time PCR assay (not shown).
Determination of cycle cutoff values.
The optimum cutoff value for the real-time PCR assay was determined using clinical samples (buffy coats and nasal swabs) collected from eight SPPV-inoculated animals and tested by real-time PCR and VI, with conventional PCR used as a confirmatory test (18). The Describe program of the WINPEPI statistical analysis program was used to calculate an optimum cutoff value of 45.7 cycles. Further analysis showed that while increasing the cutoff value to 50 cycles resulted in a sensitivity of 100% for both sample types, there was a resulting loss of specificity (90% buffy coats and 80% nasal swabs) due to the appearance of false-positive results. All subsequent results were based on a cutoff value of 45 cycles.
Assay clinical sensitivity and specificity.
Infected lambs developed signs of clinical disease, including fever, loss of appetite, depression, conjunctivitis, and nasal discharge. All lambs developed fever by day 5 p.i. Lesions developed by day 7 and began to scab over by day 14 p.i. All lambs developed neutralizing antibodies by day 14, and all but one lamb were euthanized by day 21 or when they were found moribund. There was no significant difference in disease onset or severity between animals inoculated with the SPPV-SA and RvKLP viruses.
Oral swabs, conjunctival swabs, nasal swabs, buffy coats, and lesion scabs were evaluated as clinical samples for early SPPV detection. Oral and conjunctival swabs were not considered good candidates for early detection of SPPV, since positive results were not obtained until after the onset of clinical signs (not shown). Nasal swabs were found to be positive by real-time PCR starting on day 2 p.i., with all samples being positive by day 4 (Fig. 2). The real-time assay was able to detect viral DNA in nasal swabs 1 to 5 days prior to the onset of clinical disease, as defined by fever (temperature of ≤39.7°C) and the presence of skin lesions, and up to 2 days before VI-positive results were seen. Virus was detected in nasal secretions of infected animals until the day of death or day 20 for the one surviving animal. The clinical sensitivity for the real-time assay on nasal swabs was 100%, with a specificity of 95.2% (Table 2). In buffy coat samples, virus was first detected by the CaPV real-time assay on day 3, 1 to 3 days prior to the onset of clinical disease. However, comparison to VI results showed a few instances (4/42 samples) where samples were positive by VI but negative by real-time PCR. The VI results were confirmed by conventional PCR, indicating that for buffy coat samples, VI was more sensitive than real-time PCR (Table 2). This may reflect the presence of inhibitors in blood or the relatively low viremias associated with SPPV infections. Virus was not detected in buffy coat samples by real-time PCR or VI after day 12, even though virus was present in nasal secretions of infected animals until day 20.
FIG. 2.
Detection of SPPV in nasal secretions of lambs at 0 to 8 days postinoculation. Bars represent body temperature (°C), and the solid horizontal line indicates the cutoff temperature of 39.7°C, above which lambs were considered febrile. Arrows indicate the first day that clinical signs were evident. Empty bars represent samples that were negative both by real-time PCR and VI. Hatched bars represent samples that were positive by real-time PCR but negative by VI. Filled bars represent samples that were positive by both real-time PCR and VI.
TABLE 2.
Performance of CaPV real-time PCR assay compared with VI or PCR detection with buffy coats, nasal swabs, and lesion biopsies/scabs
| Sample type and parameter | % of samples with evaluated result (no. of samples with evaluated result/total no. of samples tested) | 95% Confidence interval |
|---|---|---|
| Nasal swab | ||
| Sensitivitya | 100.0 (51/51) | 100.0-100.0 |
| Specificityb | 95.2 (20/21) | 86.1-104.3 |
| False-positive ratec | 4.8 (1/21) | 0.0-13.9 |
| False-negative rated | 0.0 (0/51) | 0.0-0.0 |
| Positive predictive valuee | 98.1 (51/52) | 94.3-101.8 |
| Negative predictive valuef | 100.0 (20/20) | 100.0-100.0 |
| Overall accuracyg | 98.6 (71/72) | 95.9-101.3 |
| Buffy coat | ||
| Sensitivitya | 90.5 (38/42) | 81.6-99.4 |
| Specificityb | 96.7 (29/30) | 90.2-103.1 |
| False-positive ratec | 3.3 (1/30) | 0.0-9.8 |
| False-negative rated | 9.5 (4/42) | 0.6-19.5 |
| Positive predictive valuee | 97.4 (38/39) | 92.5-102.4 |
| Negative predictive valuef | 87.9 (29/33) | 76.7-99.0 |
| Overall accuracyg | 93.1 (67/72) | 87.2-98.9 |
| Skin lesion/scab | ||
| Sensitivitya | 95.5 (21/22) | 86.8-104.2 |
| Specificityb | 100.0 (17/17) | 100.0-100.0 |
| False-positive ratec | 0.0 (0/17) | 0.0-0.0 |
| False-negative rated | 4.6 (1/22) | 4.2-22.8 |
| Positive predictive valuee | 100.0 (21/21) | 100.0-100.0 |
| Negative predictive valuef | 94.4 (17/18) | 83.9-105.0 |
| Overall accuracyg | 97.4 (25/26) | 92.5-102.4 |
Number of PCR-positive samples/number of VI-positive samples.
Number of PCR-negative samples/number of VI-negative samples.
Number of PCR-positive samples confirmed to be negative/number of truly positive samples.
Number of PCR-negative samples confirmed to be negative/number of truly positive samples.
Number of samples that were PCR positive and had SPPV.
Number of samples that were PCR negative and did not have SPPV.
Total number of truly positive and truly negative samples/total number of samples.
Scabs from pox lesions collected throughout the course of the experiment and skin lesion samples collected at necropsy were tested by VI and real-time PCR. Skin samples taken from noninfected lambs and scabs collected from sheep, goats, and cows infected with parapoxvirus (provided by APHIS/FADDL) were used as negative controls. The results showed a clinical sensitivity of 95.5% and a specificity of 100% (Table 2). We tested the ability of our assay to detect SPPV in various tissues obtained from acutely infected sheep during postmortem examination. The CaPV real-time assay was able to detect virus in the lungs, perilumbar lymph nodes, and pharyngeal tonsils of all eight lambs tested.
The clinical sensitivity presented here was determined by utilizing a limited number of experimentally inoculated animals. Further validation of this test will require larger numbers of both infected and noninfected animals, ideally in a natural setting.
Here we describe a TaqMan-based assay designed to detect CaPV in sheep, goats, and cattle and tested on lambs experimentally infected with SPPV (the type species of the genus CaPV). As designed, this assay was capable of detecting LSDV and GTPV in tissue culture samples. However, the clinical effectiveness of this assay for LSDV in cattle and GTPV in goats remains to be determined. Further validation of this assay under field conditions will be necessary to ascertain its viability as a diagnostic tool.
The CaPV real-time PCR assay provides a rapid, sensitive test for CaPV that is able to detect SPPV prior to the onset of clinical disease. For preclinical detection of SPPV, the sample of choice was nasal swabs, which provided better sensitivity than buffy coat samples. For clinical identification of SPPV, scabs or skin lesion biopsies provided high sensitivity and specificity and were easy to collect without the need for euthanizing suspect animals. Postmortem samples included those most likely to be submitted for diagnosis, i.e., lung and regional lymph nodes. These are all likely samples to be submitted for testing during a field investigation.
In conclusion, our results indicate that the CaPV real-time PCR assay is comparable to or exceeds the established method of VI for preclinical detection of SPPV in sheep. Based on our preliminary results, this assay should also be a useful tool for early detection and control of infections by other CaPV viruses, including GTPV and LSDV. This assay can differentiate between CaPV and other viruses causing vesicular disease in ruminants and has a high sensitivity in clinical samples. The CaPV real-time PCR assay represents a significant improvement over other established methods of CaPV detection due to its speed, simplicity, and ability to be carried out in laboratories without the need for tissue culture facilities.
Acknowledgments
We thank E. Hartwig and the PIADC animal care staff for excellent technical assistance, M. Borca and G. Risatti for their technical expertise, and FADDL, APHIS, USDA, for generously providing clinical samples.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1a.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osorio, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76783-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genome of camelpox virus. Virology 2951-9. [DOI] [PubMed] [Google Scholar]
- 1c.Afshar, A., A. Bundza, D. J. Myers, G. C. Dulac, and F. C. Thomas. 1986. Sheep pox: experimental studies with a West African isolate. Can. Vet. J. 27301-306. [PMC free article] [PubMed] [Google Scholar]
- 2.Balinsky, C. A., G. Delhon, C. L. Afonso, G. R. Risatti, M. V. Borca, R. A. French, E. R. Tulman, S. J. Geary, and D. L. Rock. 2007. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J. Virol. 8111392-11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhanuprakash, V., B. K. Indrani, M. Hosamani, and R. K. Singh. 2006. The current status of sheep pox disease. Comp. Immunol. Microbiol. Infect. Dis. 2927-60. [DOI] [PubMed] [Google Scholar]
- 4.Bhanuprakash, V., A. R. Moorthy, G. Krishnappa, R. N. Srinivasa Gowda, and B. K. Indrani. 2005. An epidemiological study of sheep pox infection in Karnataka State, India. Rev. Sci. Tech. 24909-920. [PubMed] [Google Scholar]
- 5.Cao, J. X., P. D. Gershon, and D. N. Black. 1995. Sequence analysis of HindIII Q2 fragment of capripoxvirus reveals a putative gene encoding a G-protein-coupled chemokine receptor homologue. Virology 209207-212. [DOI] [PubMed] [Google Scholar]
- 6.Carn, V. M. 1993. Control of capripoxvirus infections. Vaccine 111275-1279. [DOI] [PubMed] [Google Scholar]
- 7.Carn, V. M., and R. P. Kitching. 1995. The clinical response of cattle experimentally infected with lumpy skin disease (Neethling) virus. Arch. Virol. 140503-513. [DOI] [PubMed] [Google Scholar]
- 8.Davies, F. G. 1976. Characteristics of a virus causing a pox disease in sheep and goats in Kenya, with observation on the epidemiology and control. J. Hyg. (London) 76163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diallo, A., and G. J. Viljoen. 2007. Genus Capripoxvirus, p. 167. In A. A. Mercer, A. Schmidt, and O. Weber (ed.), Poxviruses. Birkhauser, Basel, Switzerland.
- 10.Garner, M. G., S. D. Sawarkar, E. K. Brett, J. R. Edwards, V. B. Kulkarni, D. B. Boyle, and S. N. Singh. 2000. The extent and impact of sheep pox and goat pox in the state of Maharashtra, India. Trop. Anim. Health Prod. 32205-223. [DOI] [PubMed] [Google Scholar]
- 11.Gulbahar, M. Y., M. Cabalar, Y. Gul, and H. Icen. 2000. Immunohistochemical detection of antigen in lamb tissues naturally infected with sheeppox virus. J. Vet. Med. B 47173-181. [DOI] [PubMed] [Google Scholar]
- 12.Hailat, N., O. al-Rawashdeh, S. Lafi, and Z. al-Bateineh. 1994. An outbreak of sheep pox associated with unusual winter conditions in Jordan. Trop. Anim. Health Prod. 2679-80. [DOI] [PubMed] [Google Scholar]
- 13.Kara, P. D., C. L. Afonso, D. B. Wallace, G. F. Kutish, C. Abolnik, Z. Lu, F. T. Vreede, L. C. Taljaard, A. Zsak, G. J. Viljoen, and D. L. Rock. 2003. Comparative sequence analysis of the South African vaccine strain and two virulent field isolates of lumpy skin disease virus. Arch. Virol. 1481335-1356. [DOI] [PubMed] [Google Scholar]
- 14.Kitching, R. P. 1999. Capripoxviruses, p. 1376-1381. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, vol. 3. Academic Press, San Diego, CA. [Google Scholar]
- 15.Kitching, R. P., J. M. Hammond, and D. N. Black. 1986. Studies on the major common precipitating antigen of capripoxvirus. J. Gen. Virol. 67139-148. [DOI] [PubMed] [Google Scholar]
- 16.Kitching, R. P., and W. P. Taylor. 1985. Clinical and antigenic relationship between isolates of sheep and goat pox viruses. Trop. Anim. Health Prod. 1764-74. [DOI] [PubMed] [Google Scholar]
- 17.Kitching, R. P., and W. P. Taylor. 1985. Transmission of capripoxvirus. Res. Vet. Sci. 39196-199. [PubMed] [Google Scholar]
- 18.Mangana-Vougiouka, O., P. Markoulatos, G. Koptopoulos, K. Nomikou, N. Bakandritsos, and O. Papadopoulos. 1999. Sheep poxvirus identification by PCR in cell cultures. J. Virol. Methods 7775-79. [DOI] [PubMed] [Google Scholar]
- 19.Mellor, P. S., R. P. Kitching, and P. J. Wilkinson. 1987. Mechanical transmission of capripox virus and African swine fever virus by Stomoxys calcitrans. Res. Vet. Sci. 43109-112. [PubMed] [Google Scholar]
- 20.Munz, E., and K. Dumbell. 1994. Sheeppox and goatpox, p. 613-615. In J. A. Coetzer, G. R. Thomson, and R. C. Tustin (ed.), Infectious diseases of livestock, vol. 1. Oxford University Press, Cape Town, South Africa. [Google Scholar]
- 21.Murray, M., W. B. Marin, and A. Koylu. 1973. Experimental sheep pox: a histological and ultrastructural study. Res. Vet. Sci. 15208-214. [PubMed] [Google Scholar]
- 22.OIE. 2004. Sheep pox and goat pox, p. 211. In Manual of diagnostic tests and vaccines for terrestrial animals, 5th ed. World Organisation for Animal Health, Paris, France.
- 23.OIE. 2002. Sheep pox and goat pox. Animal diseases data bulletin A100. World Organisation for Animal Health, Paris, France.
- 24.OIE. 2005. Sheep pox and goat pox, p. 215. In Terrestrial animal health code. World Organisation for Animal Health, Paris, France.
- 25.Plowright, W., W. G. Macleod, and R. D. Ferris. 1959. The pathogenesis of sheep pox in the skin of sheep. J. Comp. Pathol. 69400-413. [DOI] [PubMed] [Google Scholar]
- 26.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2001. Genome of lumpy skin disease virus. J. Virol. 757122-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, J. H. Sur, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genomes of sheeppox and goatpox viruses. J. Virol. 766054-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuppurainen, E. S., E. H. Venter, and J. A. Coetzer. 2005. The detection of lumpy skin disease virus in samples of experimentally infected cattle using different diagnostic techniques. Onderstepoort J. Vet. Res. 72153-164. [DOI] [PubMed] [Google Scholar]
- 29.Wesley, R. D., J. C. Quintero, and C. A. Mebus. 1984. Extraction of viral DNA from erythrocytes of swine with acute African swine fever. Am. J. Vet. Res. 451127-1131. [PubMed] [Google Scholar]