Abstract
Bronchopneumonia is a population-limiting disease in bighorn sheep in much of western North America. Previous investigators have isolated diverse bacteria from the lungs of affected sheep, but no single bacterial species is consistently present, even within single epizootics. We obtained high-quality diagnostic specimens from nine pneumonic bighorn sheep in three populations and analyzed the bacterial populations present in bronchoalveolar lavage specimens of seven by using a culture-independent method (16S rRNA gene amplification and clone library analyses). Mycoplasma ovipneumoniae was detected as a predominant member of the pneumonic lung flora in lambs with early lesions of bronchopneumonia. Specific PCR tests then revealed the consistent presence of M. ovipneumoniae in the lungs of pneumonic bighorn sheep in this study, and M. ovipneumoniae was isolated from lung specimens of five of the animals. Retrospective application of M. ovipneumoniae PCR to DNA extracted from archived formalin-fixed, paraffin-embedded lung tissues of historical adult bighorn sheep necropsy specimens supported the association of this agent with bronchopneumonia (16/34 pneumonic versus 0/17 nonpneumonic sheep were PCR positive [P < 0.001]). Similarly, a very strong association was observed between the presence of one or more M. ovipneumoniae antibody-positive animals and the occurrence of current or recent historical bronchopneumonia problems (seropositive animals detected in 9/9 versus 0/9 pneumonic and nonpneumonic populations, respectively [P < 0.001]). M. ovipneumoniae is strongly associated with bronchopneumonia in free-ranging bighorn sheep and is a candidate primary etiologic agent for this disease.
Pneumonia epizootics occur frequently among free-ranging bighorn sheep (Ovis canadensis), resulting in mortality due to both all-age pneumonia epizootics and enzootic pneumonia characterized by sporadic or persistent high rates of pneumonia affecting primarily lambs (7, 11, 21, 23). Bacteria most frequently associated with this syndrome include members of the genera Mannheimia and Pasteurella (22). Species within these genera differ in virulence, and the specific types involved with bighorn sheep pneumonia include Mannheimia haemolytica (formerly Pasteurella haemolytica serotype 2) (9), Pasteurella trehalosi serotype 10 (18), and Pasteurella multocida subsp. multocida (26, 27, 31).
A wide variety of bacteria belonging to the family Pasteurellaceae colonize the upper respiratory tracts of bighorn sheep (15, 25, 30, 32). Pathogenic strain types are isolated from the upper respiratory tract flora of apparently healthy bighorn sheep and yet are often absent from the lungs of affected bighorn sheep (2, 6). Frequently, no single pathogenic species is isolated from all or even most affected animals within single epizootics, and instead diverse Pasteurellaceae may be isolated from different animals (27, 31). Therefore, bacteria of the family Pasteurellaceae may be secondary to one or more predisposing infectious agents or other stresses, as is the case with pasteurellosis of domestic ruminants (5).
Several factors may have limited the investigation of the etiology of bighorn sheep bronchopneumonia. The remoteness of the areas where the animals reside and the resulting inevitable delays in discovering the mortality and transporting specimens to the laboratory result in severe tissue autolysis, postmortem bacterial overgrowth, and otherwise poor specimen quality. More generally, conventional bacteriologic culture may offer poor sensitivity for the detection of fastidious agents, often failing to detect the majority of the diverse microbes present in biological samples (12, 13, 24, 28). Culture-independent techniques based on the amplification of small-subunit (16S) rRNA genes may circumvent some of these problems with conventional bacteriology.
In an attempt to clarify the etiology of bronchopneumonia in free-ranging bighorn sheep, we applied culture-independent (16S rRNA gene) and conventional diagnostic analyses to high-quality specimens from lambs collected early in the course of disease during epizootics of bronchopneumonia in the Hells Canyon region of Idaho, Oregon, and Washington.
MATERIALS AND METHODS
Animals.
Epizootic lamb pneumonia was detected in three populations of bighorn sheep in Hells Canyon in 2006 based on observation of respiratory signs (coughing, nasal discharge) and recovery of dead lambs. Seven lambs exhibiting respiratory disease signs were euthanized and submitted to the Washington Animal Disease Diagnostic Laboratory (WADDL) for complete necropsy and ancillary testing. Also in 2006, a radio-collared adult bighorn ram observed with severe respiratory disease signs in the Sheep Mountain population died during sedation and was also submitted to WADDL. In 2007, two lambs were collected prior to the onset of pneumonic signs from a pneumonia-affected population. Finally, one lamb was collected in 2007 from a population in which no pneumonia had ever been detected. Additional specimens from these animals were submitted to the University of Idaho Caine Veterinary Teaching Center for bacteriologic cultures. All animal studies reported here were performed in compliance with all relevant federal guidelines and institutional policies.
Specimens.
Formalin fixed, paraffin-embedded lung tissues from adult bighorn sheep were selected from WADDL records. These included 34 with gross and histologic lesions of bronchopneumonia and 17 without pneumonic lesions. In all selected cases, DNA was extracted from the tissues for subsequent PCR analyses.
Serum specimens were obtained from 18 free-ranging bighorn sheep populations, including 9 populations with a history of respiratory disease and 9 other populations in which respiratory disease had never been observed. In two of the pneumonic populations, sera were available from animals sampled in years both prior to and after the onset of observed pneumonia.
Routine diagnostic procedures. (i) Necropsy.
Lambs were weighed, and postmortem condition and body condition were noted. The percentage of pneumonic lung was estimated for the left and right lung lobes. Tympanic bullae on both sides were examined for exudates. Tissue samples collected included bronchiolar-alveolar lavage fluids, normal and pneumonic lung samples on both the right and left sides, bronchial lymph nodes, tonsils, pharynx, trachea, spleen, liver, kidney, and swabs of tympanic bullae. Tissues fixed in 10% neutral buffered formalin included normal and pneumonic lung, trachea, bronchial lymph nodes, tonsil/pharynx, thymus, thyroid gland, spleen, heart, liver, and kidney. Postmortem blood was collected for serum extraction from large veins or the heart.
(ii) Histopathologic evaluation.
For routine histopathology, formalin-fixed tissues were trimmed, dehydrated through a series of alcohol and xylene baths, and paraffin embedded. Sections (thickness, 6 μm) were cut, deparaffinized, stained with hematoxylin and eosin, and examined by light microscopy.
(iii) Bacteriologic cultures.
Aerobic bacteriologic cultures were performed using Columbia blood agar, selective Columbia blood agar plates (14), and MacConkey agar plates (media sources were Hardy Diagnostics, Santa Maria, CA, and Becton Dickinson & Co., Sparks, MD) inoculated with bronchoalveolar lavage fluids, swabs of tympanic bullae exudates, and swabs of affected lung tissues and adjacent bronchial lymph nodes. Bacteria were isolated and identified using routine methods at WADDL and at the Caine Veterinary Teaching Center. Mycoplasma cultures were performed on bronchoalveolar lavage (BAL) fluid specimens, swabs of affected lung tissues, and swabs of tympanic bulla exudates using PPLO broth and selective agar plates (R88 and G04; Hardy Diagnostics) and mycoplasma broth and plates (1).
16S rRNA gene analyses.
BAL fluids were obtained aseptically using 10-Fr sterile catheters introduced through tracheal incisions. The catheters were advanced until resistance was encountered, and 20 ml buffered sterile 0.9% NaCl in water was injected and reaspirated immediately afterwards. All chemical buffers were made by using purified water to minimize contamination with external DNA sources.
Bacterial populations of the BAL fluids were evaluated by analysis of 16S rRNA gene sequences, as described elsewhere (17). Briefly, BAL fluids (1.5-ml aliquots) were centrifuged (12,000 rpm for 15 min), DNA was extracted from the pellets (DNeasy tissue kit; Qiagen, Valencia, CA) and an informative ∼917-nucleotide segment of the 16S rRNA gene was amplified, using primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 805R (5′-CCGTCAATTCCTTTRAGTTT-3′), by PCR (50 μl, including 1 μl DNA template, 200 μM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 0.4 μM each primer, 4 μl of bovine serum albumin [10 mg/ml], and 5 U of AmpliTaq Gold [Applied Biosystems, Foster City, CA]). PCR amplification included 35 cycles of denaturing (94°C, 1 min), annealing (55°C, 45 s), and extension (72°C, 1.5 min). Amplification was preceded by an extended denaturing step (94°C, 2 min) and was followed by an extended extension step (72°C, 20 min). PCR products were cloned (TOPO TA cloning kit; Invitrogen Corp., Carlsbad, CA), and 96 clones were picked for each animal in the study. Clone inserts were PCR amplified using vector primers T7 (5′-CCCTATAGTGAGTCGTATTAC-3′) and M13 (5′-CAGGAAACAGCTATGA-3′), and inserts were confirmed by electrophoresis on 1% agarose gels stained with ethidium bromide.
Products were analyzed by restriction fragment length polymorphism (RFLP) analysis following digestion with HinP1I and MspI (New England Biolabs, Ipswich, MA). The digests were examined on ethidium bromide-stained 3% agarose gels, and approximately 30 clones representing the predominant restriction digest patterns from each animal's library were selected for DNA sequencing. DNA from selected clones was isolated from 1.5 ml of LB culture using the PerfectPrep plasmid 96 VAC kit (5 Prime, Inc., Gaithersburg, MD) and was sequenced with vector primers T3 and M13 using BigDye (version 3.1) chemistry on an automated ABI Prism genetic analyzer (Applied Biosystems, Foster City, CA).
DNA sequences were analyzed by BLAST to identify homologous bacterial species, which, with RFLP patterns, were used to determine the predominant bacterial species present in the BAL specimens.
PCR detection of Mycoplasma ovipneumoniae.
Mycoplasma ovipneumoniae-specific 16S rRNA gene sequences were identified by PCR using primers LMF1 (5′-TGAACGGAATATGTTAGCTT-3′) and LMR1 (5′-GACTTCATCCTGCACTCTGT-3′) under amplification conditions described elsewhere (20). Mycoplasma arginini, Mycoplasma bovis, and Acholeplasma laidlawii were used as negative controls.
Indirect hemagglutination serology for M. ovipneumoniae.
Indirect hemagglutination was performed as described elsewhere (4, 8). Antigen was prepared using Mycoplasma ovipneumoniae strain 06OR03, grown in Mycoplasma broth (Hardy Diagnostics; 37°C, 5% CO2, 5 days). Serial twofold dilutions (50 μl; 1:20 through 1:2,540) of test sera were added to paired wells containing 50 μl sensitized and unsensitized erythrocytes, respectively, and were incubated at 25°C for 2 h. Titers were read as the highest dilution demonstrating agglutination of sensitized cells but not unsensitized cells.
Experimental challenge with Mycoplasma ovipneumoniae.
Two bighorn sheep lambs were bottle-reared together in a single isolation room and challenged with M. ovipneumoniae strains 06OR03 and 06WA03 (5 ml containing ∼10e7 CFU of each strain/ml per challenge dose). The lambs were scored for fever and for signs of respiratory tract disease daily; then they were euthanized and necropsied. Lamb 1 received intranasal challenges at the ages of 9, 29, 52, and 59 days and was euthanized and necropsied at 66 days. Lamb 2 received intranasal challenges at the ages of 10, 16, 39, and 46 days and an intratracheal challenge at 69 days and was euthanized and necropsied at 82 days.
Statistical analyses.
Chi-square analysis was used to compare the frequency of detection of M. ovipneumoniae 16S rRNA gene sequences by PCR in DNA extracted from archived formalin-fixed, paraffin-embedded lung tissues from Hells Canyon bighorn sheep with and without bronchopneumonia. Distributions of antibody titers were compared between populations with and without a history of bronchopneumonia using the nonparametric Mann-Whitney rank sum test, since the F-test results indicated significant departures from normality. All statistical analyses were performed using a commercial software program (Sigma-Stat 2.03; SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
Partial (1,457-nucleotide) 16S rRNA gene sequences from two Mycoplasma ovipneumoniae isolates obtained from bighorn sheep lambs, 06OR03 and 06WA03, are available in GenBank (accession numbers EU265779 and EU265780). All clone library 16S rRNA gene partial sequences obtained in this study are also available in GenBank (accession numbers EU289919 to EU290137).
RESULTS
Nine lambs from three populations of bighorn sheep (Sheep Mountain [n = 4], Imnaha [n = 2], and Black Butte [n = 3]) were collected for necropsy and microbiologic examinations immediately prior to or during pneumonia outbreaks in June and July 2006 and May 2007, and an additional lamb was collected in July 2007 from a population (Asotin) with no history of pneumonia. In October 2006, an adult bighorn ram in the Sheep Mountain population observed with respiratory distress died after sedation for capture and was necropsied and sampled for microbiological testing. The origin, signalment, selected pathological findings, and serologic titers of these animals are presented in Table 1.
TABLE 1.
Origin, signalment, body condition, and observation of otitis media in bighorn sheep collected for necropsy and microbiologic workups in this studya
| Animal | Location | Estimated age | Sex | Wt (kg) | Body condition | Otitis media | M. ovipneumoniae titer |
|---|---|---|---|---|---|---|---|
| 06OR02 | Sheep Mountain | 30 days | F | NR | Good | NR | ND |
| 06OR03 | Sheep Mountain | 28 days | M | NR | Fair | NR | <1:20 |
| 06SM90 | Sheep Mountain | 9 yr | M | NR | Fair | No | 1:2,560 |
| 06OR04 | Imnaha | 10 days | M | 8.0 | Good | No | 1:640 |
| 06OR05 | Imnaha | 28 days | F | 9.0 | Good | Yes | <1:20 |
| 06WA03 | Black Butte | 35 days | F | 10.0 | Poor | Yes | <1:20 |
| 06WA06 | Black Butte | 44 days | F | 12.7 | Good | Yes | <1:20 |
| 06WA08 | Black Butte | 68 days | F | 10.9 | Poor | No | 1:80 |
| 07OR06 | Sheep Mountain | 17 days | M | 9.5 | Good | No | <1:20 |
| 07OR07 | Sheep Mountain | 4 days | M | 6.6 | Good | No | 1:2,560 |
| 07WA03 | Asotin | 28 days | F | 7.7 | Good | No | <1:20 |
NR, not recorded; ND, not determined.
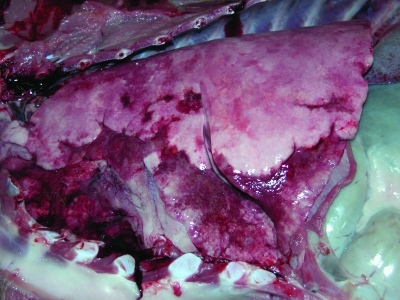
Nine animals demonstrated gross lesions of acute bronchopneumonia, including all animals collected from pneumonic populations, except for animal 07OR07 (which was collected for necropsy at the age of 4 days). The estimated extent of the pneumonic area within the lungs differed among the affected animals, ranging from 15% to 50%. The most extensive lesions were generally in the right anterior lung lobes. Affected areas were dark red to purple and firm, and sections sank in 10% neutral buffered formalin (Fig. 1). In addition, purulent otitis media was detected in three lambs for which the necropsy included examination of the middle ears (Table 1). No other gross pathological lesions were detected. The lamb collected from the nonpneumonic population (07WA03) exhibited no gross lesions.
FIG. 1.
Left lateral thorax of a pneumonia-affected lamb illustrating consolidation of the left ventral lung lobes.
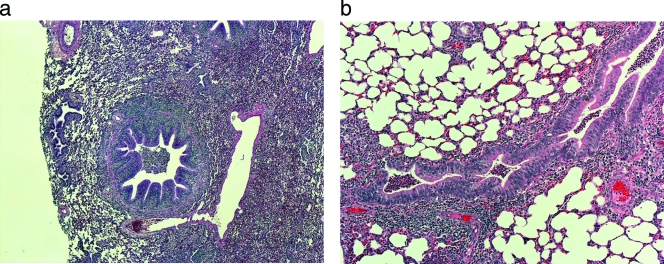
Histopathologic lesions varied with severity and chronicity. Two lambs with grossly normal respiratory tracts were free from detectable histopathologic lesions: 07OR07 (4 days old, from a pneumonic population) and 07WA03 (28 days old, from a nonpneumonic population). Early, mild lesions consisting of hyperplastic bronchiolar epithelia surrounded by lymphocytic infiltrates were observed in more acutely affected animals and in lung tissue adjacent to severely affected areas (Fig. 2a). Bronchial lumina contained neutrophils that also filled surrounding alveoli in more severely affected areas. In more chronic lesions, neutrophil accumulations coalesced into foci of necrosis associated with bacterial colonies (abscess formation). Sections of otitis media had characteristic neutrophil accumulation within the tympanic bullae with extension into the petrous temporal bone (osteomyelitis).
FIG. 2.
Hematoxylin-and-eosin-stained lung tissues illustrating bronchial hyperplasia and lymphocytic cuffing in a naturally affected lamb from Hells Canyon (a) and experimentally challenged lamb 1 (b).
Routine aerobic cultures revealed diverse bacterial isolates from affected lung tissues (Table 2). The aerobic bacteriologic isolates from affected lung tissues, from BAL fluids, and from swab specimens of purulent middle ear exudates were generally concordant (data not shown). For the Sheep Mountain lambs, Pasteurella multocida was the predominant aerobic bacterial pulmonary pathogen identified in 2006 and P. trehalosi was dominant in 2007, while for Imnaha lambs, bacterial cultures were mixed and no consistent predominant growth was identified. Black Butte bighorn sheep also demonstrated mixed bacterial flora in the lungs, with Pasteurella trehalosi being the most frequently isolated respiratory pathogen. Initial mycoplasma cultures were uniformly negative when attempted on specimens from the study lambs in 2006. However, subsequent repeat culture attempts yielded M. ovipneumoniae from the lungs of three animals collected in 2006 and from both lambs collected from pneumonic populations in 2007.
TABLE 2.
Results of conventional microbiology cultures and 16S rRNA gene library analyses on bighorn sheep BAL specimens
| Animal | Aerobic bacterial culture
|
16S rRNA gene library analysis
|
||
|---|---|---|---|---|
| Species | Level | Species (% identity)a | Proportionb | |
| 06OR02 | Pasteurella multocida | Very many | Uncultured bacterium (89-94) | 0.34 |
| Pasteurella trehalosi | Many | Fusobacterium necrophorum (99) | 0.25 | |
| Moraxella sp. | Many | Uncultured Firmicutes (88-93) | 0.16 | |
| Streptococcus sp. (95-97) | 0.13 | |||
| Cilium-associated respiratory bacterium (98) | 0.09 | |||
| Fusobacterium sp. (96) | 0.03 | |||
| 06OR03 | Pasteurella multocida | Many | Pasteurella multocida (99) | 0.34 |
| Arcanobacterium pyogenes | Few | Mycoplasma ovipneumoniae (99) | 0.31 | |
| Mixed bacterial growth | Moraxella sp. (97) | 0.31 | ||
| Mycoplasma ovipneumoniae (reculture) | Positive | Pasteurella trehalosi (98) | 0.03 | |
| 06SM90 | Pasteurella trehalosi | Many | Uncultured rumen bacterium (93-99) | 0.81 |
| Pasteurella multocida | Few | Uncultured bacterium (89-99) | 0.19 | |
| Moraxella sp. | Very many | |||
| Enterococcus sp. | Very many | |||
| Arcanobacterium pyogenes | Very many | |||
| Mycoplasma ovipneumoniae (reculture) | Positive | |||
| 06OR04 | Pasteurella trehalosi | Moderate | Mycoplasma ovipneumoniae (99) | 1.00 |
| 06OR05 | Mannheimia haemolytica | Moderate | Helcococcus sp. (99) | 0.50 |
| Possible Aerococcus sp. | Many | Fusobacterium necrophorum (99) | 0.33 | |
| Streptococcus suis | Few | Cilium-associated respiratory bacterium (98) | 0.17 | |
| Pasteurella trehalosi | Few | |||
| Coliforms | Few | |||
| 06WA03 | Pasteurella trehalosi | Many | Fusobacterium necrophorum (96-99) | 0.28 |
| Streptococcus sp., beta-hemolytic | Many | Pasteurella trehalosi (96-99) | 0.25 | |
| Mycoplasma ovipneumoniae | Positive | Mycoplasma ovipneumoniae (99-100) | 0.22 | |
| (reculture) | Cilium-associated respiratory bacterium (97-98) | 0.09 | ||
| Unidentified spp. | 0.09 | |||
| Streptococcus suis (99) | 0.06 | |||
| 06WA06 | Pasteurella trehalosi | Very many | Bacteroides pyogenes (98-100) | 0.28 |
| Pasteurella multocida | Few | Uncultured rumen bacterium (92-99) | 0.25 | |
| Mixed bacterial growth | Fusobacterium necrophorum (99) | 0.17 | ||
| Unidentified spp. (86) | 0.17 | |||
| Miscellaneous spp. (98-99) | 0.14 | |||
| 06WA08 | Pasteurella trehalosi | Very many | Uncultured rumen bacterium (89-99) | 0.43 |
| Pseudomonas aeruginosa | Very many | Fusobacterium necrophorum (99) | 0.23 | |
| Streptococcus sp., nonhemolytic | Very many | Unidentified. spp. (83-91) | 0.20 | |
| Arcanobacterium pyogenes | Many | Prevotella sp. (96%) | 0.11 | |
| Bacillus sp. | Low | Miscellaneous spp. (98-99) | 0.03 | |
| Mycoplasma ovipneumoniae (reculture) | Positive | |||
| 07OR06 | Pasteurella trehalosi | Many | Not determined | |
| Mycoplasma ovipneumoniae | Positive | |||
| 07OR07 | Pasteurella trehalosi | Few | Not determined | |
| Alpha-hemolytic Streptococcus sp. | Few | |||
| Mycoplasma ovipneumoniae | Positive | |||
| 07WA03 | Coliform | Few | Not determined | |
| Staphylococcus sp., coagulase negative | Few | |||
Best BLAST matches with identities of >98% are reported to species level, and those with 95 to 98% identity are reported to genus level. Types representing <4% of the sequenced clones are reported as miscellaneous species.
Proportion of clones identified as the corresponding species.
Aseptically collected bronchoalveolar wash fluids from seven bighorn lambs and one adult were analyzed for bacterial populations by amplification, cloning, RFLP analysis, and sequencing of 16S rRNA gene variable regions using primers based on conserved flanking eubacterial or universal sequences. In several cases, these analyses detected the predominant bacterial species identified by aerobic cultures, but for most animals additional species were detected that were not identified by aerobic cultures, and for several animals these noncultured species predominated (Table 2). Of particular interest was the detection of Mycoplasma ovipneumoniae 16S rRNA gene sequences, which were greatly predominant in the lamb with the least advanced pneumonia and prominent in two additional lambs with moderately advanced pneumonia. In the lungs of the other animals affected with severe bronchopneumonia, diverse obligately anaerobic bacterial species predominated.
DNAs extracted from BAL fluids or affected lung tissue specimens from the 11 animals analyzed in this study were tested for the presence of Mycoplasma ovipneumoniae rRNA gene sequences by PCR. All nine lambs and the adult ram from populations with pneumonia were positive for M. ovipneumoniae sequences by this test, while the single lamb (07WA03) from the nonpneumonic Asotin population was negative.
PCR to detect M. ovipneumoniae-specific 16S rRNA gene sequences was then applied to DNA extracted from archived formalin-fixed, paraffin-embedded lung tissues of 51 adult bighorn sheep originating from Hells Canyon populations with a history of pneumonia occurrence. Of 34 animals tested that had gross and histologic lesions of bronchopneumonia, 16 were PCR positive, while the 17 animals without bronchopneumonia were uniformly PCR negative (χ2 = 9.18, 1 df; P = 0.02).
With the detection of M. ovipneumoniae-specific DNA sequences in all pneumonic lambs collected in this study and in a significant proportion of adult bighorn sheep with bronchopneumonia, serology was then used to detect exposure to this agent in additional bighorn sheep populations. Banked serum samples were obtained from animals from 18 populations, including 8 Hells Canyon populations and 10 other bighorn sheep populations located in three Western states and two Canadian provinces (3 in California, 2 in Washington, 2 in Oregon, 2 in Alberta, 1 in British Columbia) outside the Hells Canyon region. The 18 populations included 9 populations that had experienced moderate to severe pneumonia losses and 9 populations in which pneumonia had never been observed. More than 60% of apparently healthy animals from pneumonic populations were seropositive for M. ovipneumoniae with indirect hemagglutination titers of 1:80 to 1:2,560, whereas no animals tested from nonpneumonic populations had titers of 1:80 or greater. Geometric mean titers from pneumonic and nonpneumonic populations differed significantly (Table 3) (P < 0.001 by the Mann-Whitney rank sum test).
TABLE 3.
Indirect hemagglutination serology for detection of antibodies to Mycoplasma ovipneumoniae in pneumonic and healthy bighorn sheep populations in western North America
| Populationa | Sample date(s) | Statusb | No. seropositive (total)c | GMTd (SD) |
|---|---|---|---|---|
| Asotin, WA (HC) | 2001, 2003, 2006 | Healthy | 0 (23) | 1.45 (0.96) |
| Cadomin, AB | 2000 | Healthy | 0 (15) | 1.00 (0) |
| Clemans Mt., WA | 2006 | Healthy | 0 (22) | 1.30 (0.79) |
| Coglan Butte, OR | 2002 | Healthy | 0 (19) | 1.64 (1.32) |
| Hall Mt., WA | 1987, 1989, 1996 | Healthy | 0 (12) | 1.00 (0) |
| Imnaha, OR (HC) | 2000 | Healthy | 0 (20) | 1.18 (0.56) |
| John Day, OR | 2002 | Healthy | 0 (20) | 1.30 (0.73) |
| Lostine, OR (HC) | 1982, 1983, 1986 | Healthy | 0 (31) | 1.32 (0.88) |
| Ram Mt., AB | 1999 | Healthy | 0 (20) | 1.00 (0) |
| Sierra Nevada, CA | 2005 | Healthy | 0 (17) | 1.16 (0.65) |
| Spence's Bridge, BC | 1997 | Healthy | 0 (24) | 1.00 (0) |
| Total, healthy populations | 0 (223) | 1.24 (0.79) | ||
| Black Butte, WA (HC) | 1995, 1997, 2000, 2003, 2006 | Pneumonia | 26 (39) | 5.78 (2.03) |
| Imnaha, OR (HC) | 2003, 2006, 2007 | Pneumonia | 6 (21) | 3.44 (1.67) |
| Lostine, OR (HC) | 1987, 2003, 2005, 2006 | Pneumonia | 26 (35) | 5.52 (2.44) |
| Muir Creek OR (HC) | 2003 | Pneumonia | 1 (1) | 7.85 |
| Peninsular, CA | 2005 | Pneumonia | 5 (6) | 6.46 (1.58) |
| Redbird, ID (HC) | 2003, 2006 | Pneumonia | 8 (19) | 3.50 (2.33) |
| Sheep Mountain, OR (HC) | 2006 | Pneumonia | 11 (12) | 6.15 (1.91) |
| Wenaha, OR (HC) | 2003, 2006 | Pneumonia | 8 (14) | 4.50 (2.66) |
| White Mountains, CA | 2005, 2006 | Pneumonia | 10 (11) | 6.27 (1.58) |
| Total, pneumonic populations | 101 (158) | 5.21 (2.35) |
AB, Alberta; BC, British Columbia; HC, population located in the Hells Canyon region.
Healthy, populations without documented or observed bronchopneumonia; pneumonia, populations with documented bronchopneumonia.
No. seropositive, number of sera with indirect hemagglutination titers of >1:40; total, number of sera tested.
GMT, geometric (log base 2) mean titers.
In two pneumonic populations (Imnaha and Lostine), serum samples were available from bighorn sheep sampled both before and after the first observed occurrence of pneumonia. All sera collected from Imnaha or Lostine animals in the years prior to the onset of pneumonic disease were negative for M. ovipneumoniae antibodies.
Experimental challenge with Mycoplasma ovipneumoniae.
Two bighorn sheep lambs were removed from their M. ovipneumoniae-negative captive dams between the ages of 12 and 24 h and were bottle-reared together in a single isolation room. The lambs were challenged with intranasal M. ovipneumoniae (two-strain mixture; ∼10e7 CFU per challenge) approximately every 2 weeks beginning at the age of 5 to 7 days. Lamb 2 in addition received a single intratracheal challenge at the age of 69 days. Blood samples for serology were obtained prechallenge and at approximately 2-week intervals thereafter. Oropharyngeal swabs from both lambs became consistently culture positive for M. ovipneumoniae beginning at the age of 21 days (lamb 1) or 14 days (lamb 2). While both lambs were observed to occasionally exhibit coughs, nasal discharges, and fevers of >103.5°F, no sustained clinical illness was observed. Lamb 1 developed an M. ovipneumoniae titer of 1:320 at the age of 48 days, which increased to 1:2,560 at the age of 62 days. Lamb 2 remained consistently seronegative.
Lamb 1 was euthanized for necropsy at the age of 62 days. Gross lesions were limited to the right anterior lung lobe, which had small (<2-cm2) areas of consolidation. Histologically these areas (Fig. 2b) demonstrated bronchiolar hyperplasia and peribronchiolar lymphoid aggregates resembling those of pneumonic lambs from Hells Canyon. Lamb 2 had a few scattered bronchioles in the anterior lung lobes with small peribronchiolar lymphoid aggregates but no evidence of bronchiolitis or consolidation. M. ovipneumoniae isolation from middle ears, lung tissue, bronchial lymph node tissue, and BAL fluids from the experimentally challenged lambs was attempted; all these sites were culture negative for lamb 1 but culture positive for lamb 2.
DISCUSSION
The observations reported here strongly suggest an etiologic role for the bacterium M. ovipneumoniae in bronchopneumonia in bighorn sheep both in Hells Canyon and elsewhere in western North America. This agent was initially detected by 16S rRNA gene analysis of the bacterial population, and the association was further supported by gross necropsy findings and histopathologic studies, bacteriologic cultures, PCR analyses, and serologic studies. In Hells Canyon, the association includes respiratory disease at the individual animal level in both juvenile and adult bighorn sheep. In bighorn sheep populations outside of Hells Canyon, the association is at the population level based on serologic data obtained from adult bighorn sheep only. A previous study provided strong evidence of a role for M. ovipneumoniae in a pneumonia outbreak in a captive population of a closely related sheep species (Dall's sheep [Ovis dalli]), and the results of that study provided a valuable guide for this investigation (4).
Further work will be required to determine the specific role of Mycoplasma ovipneumoniae in bighorn sheep respiratory disease. Mycoplasma infections are frequently associated with middle ear infections (19), and M. ovipneumoniae may therefore also contribute to the etiology of the severe otitis media documented here in some of the study lambs and most (7 of 9) other lambs with pneumonia found dead or close to death during the study (data not shown). Our results also suggest that this agent may serve as a sole pneumonia pathogen in some bighorn lambs, as suggested by its predominance by 16S rRNA gene analysis in lamb 06OR04 (Table 2). However, in most animals, polymicrobial infections were present, and M. ovipneumoniae may more commonly act as a primary agent that increases the susceptibility of infected bighorn sheep to secondary bronchopneumonia.
Exposure to M. ovipneumoniae as detected serologically differed markedly among different bighorn sheep populations and roughly correlated with disease occurrence. For example, the recent seroprevalence in the Imnaha population was quite low, and this population lost less than half the lamb crop to pneumonia during 2006 and 2007 (data not shown). In contrast, seroprevalence was much higher in the Black Butte and Sheep Mountain populations, both of which lost nearly their entire lamb crops during the same period. These data from Imnaha suggest that M. ovipneumoniae transmission within populations is not necessarily fast or complete.
The experimental lamb challenge studies, though limited to two animals, clearly demonstrated that M. ovipneumoniae infection alone is insufficient to consistently cause fatal bronchopneumonia within the time course of the natural disease. For one lamb, intranasal challenges resulted in oropharyngeal colonization, lung infection, and seroconversion, and the lung lesion was histologically very similar to that seen in the natural disease in Hells Canyon. However, the extent of the bronchopneumonia was limited, and it is likely that the lamb would have recovered if not euthanized. For a second lamb, given both a series of intranasal challenges and a subsequent intratracheal challenge, the oropharynx became colonized but no lower respiratory tract disease or seroconversion was observed. At necropsy this lamb did have purulent otitis media, but no gross or microscopic signs of pneumonia were observed. Other experimental challenge studies of domestic sheep lambs with M. ovipneumoniae have reported difficulty in reproducing clinical disease, attributed to hypothesized strain variation in virulence or virulence attenuation in laboratory culture (10, 16).
Mannheimia haemolytica, demonstrated experimentally to cause fulminant lethal bronchopneumonia in bighorn sheep and long considered a major cause of pneumonia in wild bighorn sheep, was not detected here by the 16S rRNA gene studies and was isolated by conventional bacteriology from only a single animal in this study (9, 22). Moreover, the rather chronic pneumonia lesions observed in most of the study animals and the demonstration of advanced pneumonic lesions in lambs collected before or at the onset of clinical signs are inconsistent with the rapid course of experimental infection of bighorn sheep with M. haemolytica (9). While lamb mortality in Hells Canyon populations with pneumonia peaks at the age of 42 to 70 days (7), significant pneumonic lesions were already present in all nine lambs collected between the ages of 10 and 44 days, suggesting that the typical course of this disease is at least several weeks. Taken together, the molecular analyses, bacteriologic cultures, and epidemiological evidence gathered in this study suggest that Mannheimia haemolytica does not play a major role in the etiology of bighorn lamb pneumonia in these Hells Canyon populations.
The results of the 16S rRNA gene studies were largely discordant with the results of aerobic bacterial cultures. The predominant aerobic bacterial growth was detected in the 16S rRNA gene library sequences of only two animals (06OR03 and 06WA03). In 06OR04, the lamb with the least severe pneumonia, the sole initial aerobic bacterial isolate was P. trehalosi, whereas the sole 16S rRNA gene sequence identified was that of M. ovipneumoniae. For the remaining animals, the 16S rRNA gene libraries detected only bacterial sequences absent from the aerobic culture results. The failure to isolate and identify M. ovipneumoniae in initial cultures was apparently due to the unfamiliarity of the laboratories with this species, its very low rate of growth on the media used, and its unusual colony morphology, which was not initially recognized as Mycoplasma.
The remaining discrepancies between 16S rRNA gene studies and conventional culture are largely explained by two factors. First, since the goal of the 16S rRNA gene studies was to identify the predominant bacterial species present in the lesions rather than to explore the full range of microbial flora present, we analyzed only approximately 30 clones per library. Therefore, we were unlikely to detect any species that composed <5% of the BAL flora. Second, since only aerobic bacteriologic studies were performed, the obligately anaerobic bacterial flora was not detected by culture. The combination of anaerobic flora in high numbers and the limited sensitivity of the 16S rRNA gene method as applied here resulted in the failure of many bacteria isolated in aerobic cultures to be represented among the 16S rRNA gene library clones analyzed. Additional limiting factors of conventional cultures included the presence of aerobically growing bacterial species not typically associated with respiratory disease, which may have been regarded as likely contaminants, and the presence of some genuinely nonculturable bacterial species.
One such bacterium that is nonculturable on conventional bacteriologic media is the cilium-associated respiratory bacillus (CAR bacillus), which was detected by 16S rRNA gene analyses in three lambs. CAR bacilli are nonclassified gliding bacteria that require cell culture or serum-supplemented cell culture medium for growth. They are reported to infect the tracheae and middle ear tissues of a number of mammalian species, including cervidae (3). These infections are asymptomatic in many hosts but produce chronic respiratory disease in laboratory rats, often in coinfections with Mycoplasma pulmonis (29). It is possible that CAR bacilli similarly contribute to the etiology of bighorn sheep respiratory disease, possibly in synergy with M. ovipneumoniae. However, CAR bacilli were not observed in silver-stained histologic sections of the airways of affected lung tissues of the animals in the present study, suggesting that it does not play a predominant role in the bighorn sheep pneumonic disease complex in Hells Canyon.
In summary, the findings reported here provide evidence of a strong association between exposure to M. ovipneumoniae and the occurrence of bronchopneumonia in free-ranging bighorn sheep. However, the limited transmission of this agent observed in some bighorn sheep populations and the results of the pilot experimental challenge study reported here indicate that further investigation is required to clarify the pathogenicity of this agent. Factors including virulence differences among M. ovipneumoniae strains, the presence of other bacterial or viral coinfections, mineral deficiencies that may compromise phagocyte function, environmental stressors including extremely hot or dry conditions, or other unknown factors may be critical to the virulence of M. ovipneumoniae. Nevertheless, the consistent lack of M. ovipneumoniae exposure for the healthy bighorn sheep populations from widely separated regions in western North America strongly suggests that this agent plays a necessary role in the development of this disease.
Acknowledgments
This work was supported by the Idaho Department of Fish and Game, the Oregon Department of Fish and Wildlife, the Washington Department of Fish and Wildlife, the Bureau of Land Management, the University of Idaho Caine Veterinary Teaching Center, Federal Aid in Wildlife Restoration Project W-160-R, and the Foundation for North American Wild Sheep (National, Eastern, Idaho, Oregon, and Washington chapters).
Additional bighorn sheep serum specimens were generously provided by W. J. Foreyt (Washington State University), B. J. Gonzales (California Department of Fish and Game), J. T. Jorgenson and J. Kneteman (Alberta Sustainable Resource Development), J. Bernatowicz and K. G. Mansfield (Washington Department of Fish and Wildlife), and H. Schwantje (British Columbia Ministry of Environment). Field support was provided by V. L. Coggins, P. Fowler, B. Frude, M. Hansen, K. Karash, N. Myatt, B. Ratliff, C. Strobl, R. Torland, M. Vekasy, P. Wik, and P. Zager.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Atlas, R. M. 1993. Handbook of microbiological media, p. 424. CRC Press, Boca Raton, FL.
- 2.Aune, K. A., N. Anderson, D. Worley, L. Stackhouse, J. Henderson, and J. Daniel. 1998. A comparison of population and health histories among seven Montana bighorn sheep populations. Bienn. Symp. North. Wild Sheep Goat Counc. 1146-69. [Google Scholar]
- 3.Bergottini, R., S. Mattiello, L. Crippa, and E. Scanziani. 2005. Cilia-associated respiratory (CAR) bacillus infection in adult red deer, chamois, and roe deer. J. Wildl. Dis. 41459-462. [DOI] [PubMed] [Google Scholar]
- 4.Black, S. R., I. K. Barker, K. G. Mehren, G. J. Crawshaw, S. Rosendal, L. Ruhnke, J. Thorsen, and P. S. Carman. 1988. An epizootic of Mycoplasma ovipneumoniae infection in captive Dall's sheep (Ovis dalli dalli). J. Wildl. Dis. 24627-635. [DOI] [PubMed] [Google Scholar]
- 5.Brogden, K. A., H. D. Lehmkuhl, and R. C. Cutlip. 1998. Pasteurella haemolytica complicated respiratory infections in sheep and goats. Vet. Res. 29233-254. [PubMed] [Google Scholar]
- 6.Cassirer, E. F. 2005. Ecology of disease in bighorn sheep in Hells Canyon, USA. Ph.D. dissertation. University of British Columbia, Vancouver, Canada.
- 7.Cassirer, E. F., and A. R. E. Sinclair. 2007. Dynamics of pneumonia in a bighorn sheep metapopulation. J. Wildl. Manag. 711080-1088. [Google Scholar]
- 8.Cho, H. J., H. L. Ruhnke, and E. V. Langford. 1976. The indirect hemagglutination test for the detection of antibodies in cattle naturally infected with mycoplasmas. Can. J. Comp. Med. 4020-29. [PMC free article] [PubMed] [Google Scholar]
- 9.Foreyt, W. J., K. P. Snipes, and R. W. Kasten. 1994. Fatal pneumonia following inoculation of healthy bighorn sheep with Pasteurella haemolytica from healthy domestic sheep. J. Wildl. Dis. 30137-145. [DOI] [PubMed] [Google Scholar]
- 10.Gilmour, J. S., G. E. Jones, and A. G. Rae. 1979. Experimental studies of chronic pneumonia of sheep. Comp. Immunol. Microbiol. Infect. Dis. 1285-293. [DOI] [PubMed] [Google Scholar]
- 11.Hobbs, N. T., and M. W. Miller. 1992. Interactions between pathogens and hosts: simulation of pasteurellosis epidemics in bighorn sheep populations, p. 997-1007. In D. R. McCullough and R. H. Barrett (ed.), Wildlife 2001: populations. Elsevier Applied Science, Essex, England.
- 12.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14190-197. [DOI] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1804765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaworski, M. D., A. C. S. Ward, D. L. Hunter, and I. V. Wesley. 1993. Use of DNA analysis of Pasteurella haemolytica biotype T isolates to monitor transmission in bighorn sheep (Ovis canadensis canadensis). J. Clin. Microbiol. 31831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaworski, M. D., D. L. Hunter, and A. C. S. Ward. 1998. Biovariants of isolates of Pasteurella from domestic and wild ruminants. J. Vet. Diagn. Investig. 1049-55. [DOI] [PubMed] [Google Scholar]
- 16.Jones, G. E., J. S. Gilmour, and A. G. Rae. 1982. The effects of different strains of Mycoplasma ovipneumoniae on specific pathogen-free and conventionally-reared lambs. J. Comp. Pathol. 92267-272. [DOI] [PubMed] [Google Scholar]
- 17.Kelley, S. T., U. Theisen, L. T. Angenent, A. St. Amand, and N. R. Pace. 2004. Molecular analysis of shower curtain biofilm microbes. Appl. Environ. Microbiol. 704187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraabel, B. J., M. W. Miller, J. A. Conlon, and H. J. McNeil. 1998. Evaluation of a multivalent Pasteurella haemolytica vaccine in bighorn sheep: protection from experimental challenge. J. Wildl. Dis. 34325-333. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, T., T. Shibahara, K. Kimura, Y. Wada, K. Sato, Y. Imada, Y. Ishikawa, and K. Kadota. 2003. Mycoplasma bovis-associated suppurative otitis media and pneumonia in bull calves. J. Comp. Pathol. 129100-110. [DOI] [PubMed] [Google Scholar]
- 20.McAuliffe, L., F. M. Hatchell, R. D. Ayling, A. I. King, and R. A. Nicholas. 2003. Detection of Mycoplasma ovipneumoniae in Pasteurella-vaccinated sheep flocks with respiratory disease in England. Vet. Rec. 153687-688. [DOI] [PubMed] [Google Scholar]
- 21.McCarty, C. W., and M. W. Miller. 1998. Modeling the population dynamics of bighorn sheep: a synthesis of literature. Colorado Division of Wildlife special report 73. Colorado Division of Wildlife, Denver, Colorado.
- 22.Miller, M. W. 2001. Pasteurellosis, p. 330-339. In E. S. Williams and I. K. Barker (ed.), Infectious diseases of wild mammals. Iowa State University Press, Ames.
- 23.Monello, R. J., D. L. Murray, and E. F. Cassirer. 2001. Ecological correlates of pneumonia epizootics in bighorn sheep herds. Can. J. Zool. 791423-1432. [Google Scholar]
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276734-740. [DOI] [PubMed] [Google Scholar]
- 25.Queen, C., A. C. S. Ward, and D. L. Hunter. 1994. Bacteria isolated from nasal and tonsillar samples of clinically healthy Rocky Mountain bighorn and domestic sheep. J. Wildl. Dis. 301-7. [DOI] [PubMed] [Google Scholar]
- 26.Rudolph, K. M., D. L. Hunter, W. J. Foreyt, E. F. Cassirer, R. B. Rimler, and A. C. S. Ward. 2003. Sharing of Pasteurella spp. between free-ranging bighorn sheep and feral goats. J. Wildl. Dis. 39897-903. [DOI] [PubMed] [Google Scholar]
- 27.Rudolph, K. M., D. L. Hunter, R. B. Rimler, E. F. Cassirer, W. J. Foreyt, W. J. deLong, G. C. Weiser, and A. C. S. Ward. 2007. Microorganisms associated with a bighorn sheep pneumonia epizootic in Hells Canyon, USA. J. Zoo Wildl. Med. 38548-558. [DOI] [PubMed] [Google Scholar]
- 28.Safaee, S., G. C. Weiser, E. F. Cassirer, R. R. Ramey, and S. T. Kelley. 2006. Molecular survey of host-associated microbial diversity in bighorn sheep. J. Wildl. Dis. 42545-555. [DOI] [PubMed] [Google Scholar]
- 29.Schoeb, T. R., M. K. Davidson, and J. K. Davis. 1997. Pathogenicity of cilia-associated respiratory (CAR) bacillus isolates for F344, LEW, and SD rats. Vet. Pathol. 34263-270. [DOI] [PubMed] [Google Scholar]
- 30.Ward, A. C. S., D. L. Hunter, M. D. Jaworski, P. J. Benolkin, M. P. Dobel, J. B. Jeffress, and G. A. Tanner. 1997. Pasteurella spp. in sympatric bighorn and domestic sheep. J. Wildl. Dis. 33544-557. [DOI] [PubMed] [Google Scholar]
- 31.Weiser, G. C., W. J. DeLong, J. L. Paz, B. Shafii, W. J. Price, and A. C. S. Ward. 2003. Characterization of Pasteurella multocida associated with pneumonia in bighorn sheep. J. Wildl. Dis. 39536-544. [DOI] [PubMed] [Google Scholar]
- 32.Wild, M. A., and M. W. Miller. 1991. Detecting nonhemolytic Pasteurella haemolytica infections in healthy Rocky Mountain bighorn sheep (Ovis canadensis canadensis): influences of sample site and handling. J. Wildl. Dis. 2753-60. [DOI] [PubMed] [Google Scholar]