Abstract
The genetic characteristics and molecular epidemiology of extended-spectrum β-lactamases (ESBLs) among Escherichia coli isolates were investigated at a general hospital and its associated health care facilities in Stockholm, Sweden, during the period from 2001 to 2006. Of 87 consecutive nonduplicate ESBL-positive isolates, 80 isolates encoded CTX-M-type ESBLs, 64 of which were group 1 enzymes. TEM-type and OXA-type β-lactamases were encoded in 63 and 59% of the ESBL isolates, respectively. Pulsed-field gel electrophoresis (PFGE) analysis revealed 40 different pulsotypes, consisting of 11 clones accounting for 66% of all isolates, and 29 unique patterns. Moreover, of the 11 clones, clones 1 and 4 comprised half of the clonally related isolates (28 of 57). Clone 1 was a persistent endemic clone in the area throughout the years, and clone 4 emerged in 2003. However, in recent years, clone 1 isolates were no longer predominant and were gradually replaced by new emerging strains. Concerning β-lactamase gene profiles in relation to PFGE pulsotypes, clone-related bla profiles were observed in certain clones, while in most cases different bla profiles could be observed in the same clone, and the same bla profile could be present in different clones. The molecular epidemiology of ESBL-positive E. coli in the area shows shifts in predominant strains and increased clonal diversity over time. The study also indicated that both clonal spread of epidemic strains and transfer of transposable genetic elements might contribute to the proliferation of ESBLs.
Extended-spectrum β-lactamases (ESBLs) are the major cause of resistance to oxyimino-cephalosporins in Enterobacteriaceae (4). ESBLs are mostly plasmid-mediated bacterial enzymes that are able to hydrolyze a wide variety of penicillins and cephalosporins. Most ESBLs have evolved by genetic mutation from native β-lactamases, particularly TEM-1, TEM-2, and SHV-1. These parent enzymes are commonly found in gram-negative bacteria, particularly in Enterobacteriaceae (4). Until the 2000s, most of the ESBLs were structurally related to the narrow-spectrum TEM- and SHV-type β-lactamases, with one to several amino acid substitutions surrounding their active site (4). The genetic mutations that give rise to ESBLs broaden the parental resistance pattern to a phenotype that includes resistance to broad-spectrum cephalosporins (e.g., cefotaxime [CTX] and ceftazidime [CAZ]) and monobactams (e.g., aztreonam) (9). Furthermore, in the late 1990s, a novel type of ESBLs, the CTX-M enzymes, emerged worldwide, mostly from Escherichia coli (2, 4). The more than 50 CTX-M enzymes thus far reported may be grouped into five main subgroups according to amino acid sequence similarity (CTX-M-1, CTM-M-2, CTX-M-8, CTX-M-9, and CTX-M-25) (2). Most of the CTX-Ms hydrolyze CTX better than CAZ. However, several CTX-Ms, including CTX-M-15, which is now the most widespread CTX-M enzyme worldwide (5, 10, 13, 21, 23), also hydrolyze CAZ efficiently (17). The OXA-type enzymes are another growing family of ESBLs and are unique among the ESBLs because they are most often found in Pseudomonas aeruginosa rather than in members of the Enterobacteriaceae (4, 16). Because ESBL-producing strains are resistant to a wide variety of clinically used antimicrobials, their proliferation poses a serious global health concern that has complicated treatment strategies for a growing number of hospitalized patients (16).
Because ESBL-producing strains often arise in focal outbreaks, their prevalence can vary greatly from one site to another and even over time for a given site (16). As a result, regional and local surveillance are crucial for clinical decision-making and infection control. In the present study, consecutive, nonduplicate clinical isolates of ESBL-positive E. coli were collected over a 6-year period from 2001 to 2006 at the South General Hospital, Stockholm, Sweden, and its associated health care facilities. Our study examined the prevalence and the type of β-lactamase genes among the isolates and investigated clonal relatedness of the strains.
MATERIALS AND METHODS
Bacterial isolates and phenotypic screening for ESBL.
Eighty-seven consecutive, nonduplicate clinical isolates of ESBL-positive E. coli, collected over a 6-year period from 2001 to 2006 at the South General Hospital and its associated health care facilities (Rosenlund Hospital and the Bergsund nursing home), were included in the study. Isolates were identified by conventional biochemical tests. CTX, CAZ, and cefpodoxime (CPD) were used for screening for reduced susceptibility to oxyimino-cephalosporins. The presence of ESBLs was confirmed by the double-disk method as recommended by the Clinical and Laboratory Standards Institute (6) or by the Etest (AB Biodisk, Solna, Sweden), where an eightfold reduction of MIC in the presence of clavulanic acid indicated the presence of ESBL.
Among the 87 ESBL isolates of E. coli, 68 were recovered from urine samples, 13 were recovered from wound samples, 4 were recovered from blood samples, and 1 each were recovered from sputum and nephrostomic drainage samples.
PCR amplification for detection of β-lactamase genes.
All isolates were screened for the resistance genes SHV, TEM, CTX-M, and OXA by a multiplex PCR assay using universal primers (Table 1) (3, 8, 12, 15). Bacterial DNA extraction was performed in a MagNA Pure LC System (Roche Diagnostics GmbH, Mannheim, Germany) by using a MagNA Pure LC DNA isolation kit I. PCR amplification reactions were performed in a volume of 25 μl containing 12.5 μl of 2× Qiagen Multiplex PCR Master Mix (Qiagen GmbH, Hilden, Germany), 0.2 μM concentrations of each primer, and 2 μl of DNA template. The cycling parameters were as follows: an initial denaturation at 95°C for 15 min; followed by 30 cycles of 94°C for 30 s, 62°C for 90 s, and 72°C for 60 s; and with a final extension at 72°C for 10 min. The amplified PCR products were subjected to electrophoresis at a 1.5% agarose gel in 1× TAE buffer. Strains with known β-lactamase types were included as references. These were E. coli strains with blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, J62-blaTEM-1, J53-blaTEM-2, J53-blaSHV-1, J53-blaSHV-2, and J53-blaOXA-1 and K. pneumoniae 1204-blaSHV-2 (12), kindly provided by D. Livermore, Health Protection Agency, London, United Kingdom.
TABLE 1.
Primers used for detection of different β-lactamase genes in the multiplex PCR
| Amplicon | Primer sequence (5′ to 3′) | Size (bp) | Reference |
|---|---|---|---|
| blaSHV | CTT TAT CGG CCC TCA CTC AA | 237 | 8 |
| AGG TGC TCA TCA TGG GAA AG | |||
| blaTEM | CGC CGC ATA CAC TAT TCT CAG AAT GA | 445 | 12 |
| ACG CTC ACC GGC TCC AGA TTT AT | |||
| blaCTX-M | ATG TGC AGY ACC AGT AAR GTK ATG GC | 593 | 3 |
| TGG GTR AAR TAR GTS ACC AGA AYC AGC GG | |||
| blaOXA | ACA CAA TAC ATA TCA ACT TCG C | 813 | 15 |
| AGT GTG TTT AGA ATG GTG ATC |
Isolates detected to be CTX-M gene positive in the multiplex PCR were further identified by using in-house designed primers for the CTX-M-1 group (8), the CTX-M-2 group (CM21, 5′-GGA GAA AAG TTC GGG AGG TC-3′; CM22, 5′-GCT TAT CGC TCT CGC TCT GT-3′), and the CTX-M-9 group (CM91, 5′-ACG TGG CTC AAA GGC AAT AC-3′; CM92, 5′-CGG CTG GGT AAA ATA GGT CA-3′), respectively, in PCRs with an annealing temperature at 55°C.
Sequencing of β-lactamase genes.
To confirm and further identify the identities of the β-lactamase genes detected in PCR assays, DNA sequence analysis of the PCR amplicons was performed. TEM, CTX-M group 1, and OXA genes in isolates representing different bla profiles, SHV genes detected in one of the clone 3 isolates (Table 2), and β-lactamase genes (SHV, TEM) detected in a pulsed-field gel electrophoresis (PFGE)-untypeable isolate were subjected to sequence analysis. Respectively amplified PCR products were purified by using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. Bidirectional sequencing was performed by using a BigDye v.1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with an ABI Prism 310 Genetic analyzer (Applied Biosystems).
TABLE 2.
β-Lactamase gene profiles presented in different clones clustered by PFGE and occurrence of the clones over the years
| PFGE type | β-Lactamase gene profilea
|
No. of isolates | Time (yr) of isolation | |||
|---|---|---|---|---|---|---|
| SHV | TEM | CTX-M | OXA | |||
| Clone 1 | − | + | Group 1 | + | 11 | 2001, 2002, 2004, 2005 |
| − | − | Group 1 | + | 5 | 2002-2004 | |
| − | − | Group 1 | − | 2 | 2004 | |
| − | + | Group 1 | − | 1 | 2006 | |
| Clone 2 | − | − | Group 1 | + | 5 | 2002 |
| Clone 3 | + | + | — | − | 1 | 2003 |
| − | + | Group 1 | − | 1 | 2005 | |
| Clone 4 | − | + | Group 1 | − | 6 | 2003-2006 |
| − | − | Group 1 | − | 1 | 2004 | |
| − | + | Group 1 | + | 1 | 2005 | |
| − | + | Group 9 | − | 1 | 2006 | |
| Clone 5 | − | + | Group 1 | + | 2 | 2003, 2006 |
| − | + | Group 1 | − | 1 | 2005 | |
| Clone 6 | − | − | Group 9 | + | 4 | 2004-2006 |
| Clone 7 | − | + | Group 1 | + | 3 | 2005, 2006 |
| Clone 8 | − | + | Group 1 | + | 1 | 2005 |
| − | − | Group 1 | + | 2 | 2006 | |
| Clone 9 | − | − | Group 1 | + | 4 | 2005, 2006 |
| − | − | Group 1 | − | 1 | 2006 | |
| Clone 10 | − | − | Group 1 | + | 2 | 2005 |
| Clone 11 | − | + | Group 9 | − | 2 | 2005 |
−, absence; +, presence.
PFGE.
All isolates were genotyped by PFGE (Bio-Rad GenePath System; Bio-Rad Laboratories, Hercules, CA) after macrorestriction with XbaI. The PFGE banding patterns were analyzed with the Dice coefficient and UPGMA (the unweighted pair-group method with arithmetic averages) by using GelCompar II (Applied Maths, Belgium).
RESULTS
Detection and spread of β-lactamase genes.
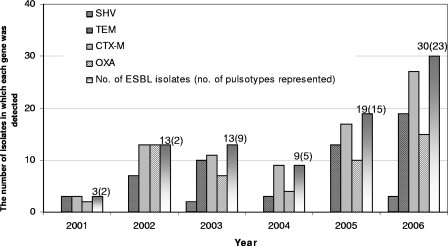
The dissemination of the isolates each year is illustrated in Fig. 1. Among the 87 ESBL isolates recovered from 2001 to 2006, 49 (56%) were isolated during period from 2005 to 2006 (Fig. 1).
FIG. 1.
Distribution of different β-lactamase genes among ESBL-positive E. coli isolates detected in a Swedish hospital and its associated health care facilities from 2001 to 2006.
The multiplex PCR was discriminatory to genes encoding SHV, TEM, CTX-M, and OXA enzymes (Fig. 2). The CTX-M gene was detected in 92% (80 of 87) of the isolates, followed by TEM (55 of 87 [63%]), OXA (51 of 87 [59%]), and SHV (5 of 87 [6%]) (Fig. 1).
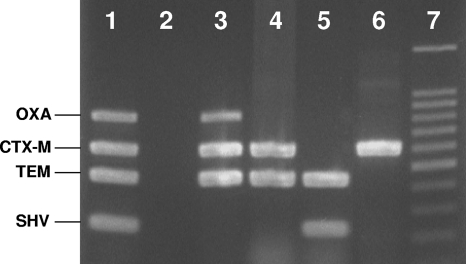
FIG. 2.
Multiplex PCR assay for blaOXA, blaCTX-M, blaTEM, and blaSHV. Lane 1, positive control; lane 2, negative control; lanes 3 to 6, clinical isolates of ESBL-positive E. coli; lane 7, 100-bp DNA ladder (Promega, Madison, WI).
Of the 80 CTX-M alleles, the CTX-M-1 group, CTX-M-2 group, and CTX-M-9 group accounted for 80, 4, and 16%, respectively. Isolates harboring blaCTX-M-1 group were found through the whole investigated period, while the blaCTX-M-2 group-containing isolates were sporadic, and the blaCTX-M-9 group-containing isolates have emerged since 2004.
Clonal relatedness of the isolates.
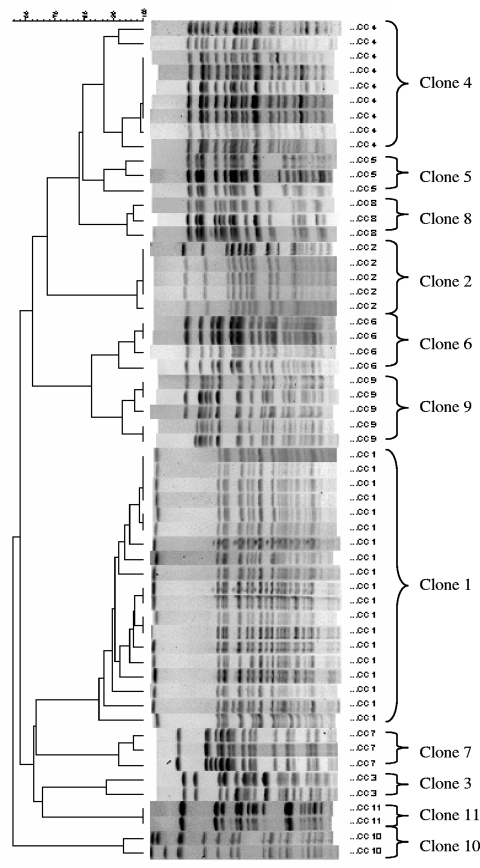
Clonal relatedness was determined by PFGE. PFGE banding patterns were obtained for 86 of the 87 ESBL isolates. One isolate was consistently untypeable by the applied method. A pulsotype was defined as PFGE banding pattern(s) that were ≥85% similar (18, 20). A clone was designated when a pulsotype was presented by at least two isolates. Thus, isolates within a clonal group were ≥85% similar. Pulsotypes presented by single isolates were called unique patterns. A total of 40 pulsotypes were obtained. Genotyping by PFGE revealed that 66% (57 of 86) of the typeable isolates were nonunique and could be separated into 11 clones (Fig. 3 and Table 2). Twenty-nine isolates presented unique banding patterns that differed more than 15% from every other isolate.
FIG. 3.
The 11 clones detected by XbaI-digested PFGE analysis. Strains were clustered with the Dice coefficient and UPGMA. The scale indicates the percentage of similarity.
Among the 11 clones discovered during the period from 2001 to 2006, clones 1 and 4 accounted for 33% (19 of 57) and 16% (9 of 57), respectively. The occurrence of different clones over the years is shown in Table 2.
β-Lactamase genes in relation to PFGE patterns.
All of the clone 1 isolates harbored genes encoding CTX-M-1 group ESBLs, and blaCTX-M-15 was identified by sequencing analysis among the representative isolates, one isolate from each bla profile of the clonal group. In addition, blaTEM and/or blaOXA were present in 89% (17 of 19) of the isolates belonging to clone 1 (Table 2).
Clone 2 was composed of five isolates, harboring blaCTX-M-15 and blaOXA-1/-30.
Two isolates, detected in 2003 and 2005, respectively, belonged to clone 3. One clone 3 isolate harbored blaSHV-5/-12 ESBL and blaTEM-1, while genes coding for TEM and CTX-M-1 group β-lactamases were detected in the other clone 3 isolate (Table 2).
Of the nine clone 4 isolates, a profile of TEM plus CTX-M-1 group β-lactamases was present in six isolates. Addition of OXA-type β-lactamase, deletion of TEM, or the presence of CTX-M-9 group ESBL was observed among the remaining three isolates (Table 2).
Isolates belonging to clone 5, clone 7, clone 8, clone 9, or clone 10 universally encoded CTX-M-1 group ESBLs. Also, genes encoding TEM-and/or OXA-type β-lactamases were present in most isolates (Table 2).
Clones 6 and 11 were composed of isolates encoding CTX-M-9 group ESBLs, while clone 6 isolates also harbored genes coding for OXA β-lactamases and clone 11 isolates also encoded TEM β-lactamases (Table 2).
The PFGE-untypeable E. coli isolate exhibited blaSHV-5/-12 ESBL and blaTEM-1.
Three isolates harboring genes encoding CTX-M group 2 enzymes were clonally unrelated, while the same bla profile of TEM plus CTX-M-2 group was observed among them. Sequencing analysis on one of the TEM alleles revealed ESBL TEM-52.
Geographic dissemination of the isolates.
Of the 87 isolates investigated in the study, 74 were isolated from the South General Hospital, 7 were isolated from Rosenlund Hospital, and 6 were isolated from the Bergsund nursing home. The latter two health care facilities were located in the same part of town as the South General Hospital.
Among the seven isolates from Rosenlund Hospital, three were involved in the outbreak in Stockholm 2002 (8), belonging to clone 1 or 2, and four were detected in 2006, belonging to clone 1, clone 7, or unrelated PFGE pulsotypes. The six isolates from the Bergsund nursing home all belonged to clone 1.
DISCUSSION
The results of this study provide insights into the genetic characteristics and molecular epidemiology of ESBLs among E. coli isolates at a general hospital and its associated health care facilities in Stockholm, Sweden.
The ESBL-positive E. coli isolates investigated here encoded mainly CTX-Ms (92%), followed by TEM-type (63%), OXA-type (59%), and SHV-type (6%) β-lactamases. The blaCTX-M genes were widespread among the ESBL-positive E. coli (92%), which was similar to the level reported in Switzerland (91%) (10), Norway (90%) (21), and Austria (85%) (7). The majority of the blaCTX-M belonged to CTX-M group 1 (80%), mostly CTX-M-15, as also reported in Switzerland (10), France (11), Austria (7), Norway (21), and Amsterdam (1). The remainders encoded CTX-M group 9 enzymes (16%) and group 2 enzymes (4%). TEM-1 and OXA-1/-30 were the most common β-lactamases found in blaTEM/blaOXA-positive strains. SHV-5/-12 ESBLs were only observed in sporadic isolates. Generally, the findings in the present study mirror the prevalence of ESBLs reported in other European countries (1, 7, 10, 11, 14, 21). Moreover, it was disclosed in the present study that more than half of the ESBL producers in Stockholm carried genes for OXA-type β-lactamases in addition to CTX-M enzymes.
PFGE analysis revealed totally 40 different pulsotypes, consisting of 11 clones and 29 unique patterns. Of the 11 clones, clones 1 and 4 were predominant. Clone 1 was a persistent endemic clone in the area throughout the years, all of the isolates of which encoded CTX-M group 1 ESBLs. Furthermore, the most common β-lactamase gene profile of clone 1 isolates was CTX-M gene combined with genes encoding for TEM and OXA enzymes. All isolates from the Bergsund nursing home, however, lacked blaTEM, and certain isolates detected after 2004 lacked blaOXA. Thus, this endemic strain might have undergone genetic alterations over time and site. Clone 1 was also one of the clones involved in the first ESBL outbreak reported in Sweden 2002 (8). Clone 2 was another clone involved in the outbreak 2002; however, no more such strains were recovered after the outbreak during this study period. The disappearance of clone 2 isolates in the area may be attributed to the aggressive infection control measures after the first outbreak. In recent years, although existing, clone 1 isolates no longer dominated and were gradually replaced by new emerging strains. Clone 4 emerged in 2003 and persisted in the following years. The molecular epidemiology of ESBL-positive E. coli in the area shows shifts in predominant strains over time. Furthermore, increased clonal diversity was observed during the years, with 15 and 23 different pulsotypes observed in 2005 and 2006, respectively, in comparison to a total of 14 pulsotypes during 2001-2004. Therefore, the spread of ESBL-positive E. coli isolates in the area was not related to the spread of a single clone.
Concerning β-lactamase gene profiles in relation to PFGE pulsotypes, clone-related bla profiles were found in isolates belonging to clones 2, 6, 7, or 11 (Table 2), while in most cases different bla profiles could be observed in a same clone, and the same bla profile could be present in different clones. This fact supports the suggestion that the ESBL-encoding genes have been disseminated either by proliferation of epidemic strains or by transposable genetic elements carrying the resistance traits (24). Thus, the same active infection control efforts should be taken in combating ESBL cases of either clonal spread or spread by transposable genetic elements. Clinical laboratories in Sweden have been required to submit notification of cases involving ESBL-producing Enterobacteriaceae strains to the Swedish Institute of Infectious Disease Control since February 2007 (19). The ability to produce ESBL confers resistance against broad-spectrum cephalosporins, which are widely used for the treatment of serious bacterial infections. Still, the carbapenems offer an alternative for treatment, but resistance to cabapenems has been reported, and such multiresistance poses even more serious problems for treatment (22).
In conclusion, CTX-M group 1 enzymes dominated in ESBL-positive E.coli isolates in a Swedish general hospital and its associated health care facilities. TEM-type and OXA-type β-lactamase genes were also present in more than half of the ESBL isolates, and SHV-type in a few strains. One endemic ESBL clone persisted in the area throughout the years, while clonal shifts and increased clonal diversity were observed. This study also indicated that both clonal spread of epidemic strains and transfer of transposable genetic elements might contribute to the proliferation of ESBLs.
Acknowledgments
We are grateful to Shahnaz Askarian Nameghi and Evrim Inekci for laboratory assistance.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.al Naiemi, N., A. Bart, M. D. de Jong, C. M. Vandenbroucke-Grauls, P. J. Rietra, Y. J. Debets-Ossenkopp, P. C. Wever, L. Spanjaard, A. J. Bos, and B. Duim. 2006. Widely distributed and predominant CTX-M extended-spectrum beta-lactamases in Amsterdam, The Netherlands. J. Clin. Microbiol. 443012-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 483758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canton, R., and T. M. Coque. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9466-475. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2005. Methods for dilution antimicrobial susceptibility tests for bacteria that grow anaerobically, 6th ed. Approved standard M7-A6. CLSI, Wayne, PA.
- 7.Eisner, A., E. J. Fagan, G. Feierl, H. H. Kessler, E. Marth, D. M. Livermore, and N. Woodford. 2006. Emergence of Enterobacteriaceae isolates producing CTX-M extended-spectrum beta-lactamase in Austria. Antimicrob. Agents Chemother. 50785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang, H., C. Lundberg, B. Olsson-Liljequist, G. Hedin, E. Lindback, A. Rosenberg, and J. Struwe. 2004. Molecular epidemiological analysis of Escherichia coli isolates producing extended-spectrum beta-lactamases for identification of nosocomial outbreaks in Stockholm, Sweden. J. Clin. Microbiol. 425917-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., and I. Carreras. 1990. Activities of beta-lactam antibiotics against Escherichia coli strains producing extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 34858-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lartigue, M. F., C. Zinsius, A. Wenger, J. Bille, L. Poirel, and P. Nordmann. 2007. Extended-spectrum beta-lactamases of the CTX-M type now in Switzerland. Antimicrob. Agents Chemother. 512855-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavigne, J. P., H. Marchandin, J. Delmas, N. Bouziges, E. Lecaillon, L. Cavalie, H. Jean-Pierre, R. Bonnet, and A. Sotto. 2006. qnrA in CTX-M-producing Escherichia coli isolates from France. Antimicrob. Agents Chemother. 504224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monstein, H. J., Å. Östholm-Balkhed, M. V. Nilsson, M. Nilsson, K. Dornbusch, and L. E. Nilsson. 2007. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM, and blaCTX-M genes in Enterobacteriaceae. APMIS 1151400-1408. [DOI] [PubMed] [Google Scholar]
- 13.Naseer, U., O. B. Natas, B. C. Haldorsen, B. Bue, H. Grundt, T. R. Walsh, and A. Sundsfjord. 2007. Nosocomial outbreak of CTX-M-15-producing Escherichia coli in Norway. APMIS 115120-126. [DOI] [PubMed] [Google Scholar]
- 14.Nyberg, S. D., M. Osterblad, A. J. Hakanen, P. Huovinen, J. Jalava, and T. F. Resistance. 2007. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002-2004. Scand. J. Infect. Dis. 39417-424. [DOI] [PubMed] [Google Scholar]
- 15.Ouellette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl. Acad. Sci. USA 847378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., and J. Segreti. 2006. Overview of the epidemiological profile and laboratory detection of extended-spectrum beta-lactamases. Clin. Infect. Dis. 42(Suppl. 4)S153-S163. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 453355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh, A., R. V. Goering, S. Simjee, S. L. Foley, and M. J. Zervos. 2006. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 19512-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderblom, T., and J. Struwe. 2007. Extended spectrum beta-lactamase-producing Enterobacteriaceae: the first six months of notifications according to the Swedish communicable disease act. Euro. Surveill. 12E071018.1. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tofteland, S., B. Haldorsen, K. H. Dahl, G. S. Simonsen, M. Steinbakk, T. R. Walsh, and A. Sundsfjord. 2007. Effects of phenotype and genotype on methods for detection of extended-spectrum-beta-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Clin. Microbiol. 45199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodford, N., J. W. Dallow, R. L. Hill, M. F. Palepou, R. Pike, M. E. Ward, M. Warner, and D. M. Livermore. 2007. Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. Int. J. Antimicrob. Agents 29456-459. [DOI] [PubMed] [Google Scholar]
- 23.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54735-743. [DOI] [PubMed] [Google Scholar]
- 24.Yu, W. L., Y. C. Chuang, and J. Walther-Rasmussen. 2006. Extended-spectrum beta-lactamases in Taiwan: epidemiology, detection, treatment, and infection control. J. Microbiol. Immunol. Infect. 39264-277. [PubMed] [Google Scholar]