Abstract
A new, efficient molecular method for detection of Bartonella, based on the 16S-23S rRNA intergenic spacer and 16S rRNA amplification by multiplex PCR combined with reverse line blotting, was designed. This assay could simultaneously detect 20 different known species and other Bartonella species not described previously.
Bartonella species are gram-negative facultative intracellular bacteria that can infect erythrocytes and endothelial cells (7). These zoonotic pathogens have complex cycles in the nature, which include different reservoir hosts and arthropod vectors. Bartonella species have been classified as reemerging bacteria with a worldwide distribution, except for B. bacilliformis, which is located in northwestern South America.
More than 20 different Bartonella species have been described so far, and 8 of them have been associated with human disease (14). However, new species have been identified recently as human pathogens, such as B. alsatica (13), B. koehlerae (3), and B. rochalimae (5), as a result of the implementation of more efficient molecular tools for diagnosis. Bartonella spp. can produce a wide variety of clinical manifestations, including severe ones such as encephalitis or endocarditis (14). In fact, the species of this genus are a major cause of culture-negative endocarditis, representing in some studies up to 10% of all the cases (12, 16).
The goal in clinical and environmental studies is not only the identification of positive samples but also the identification of the species involved. Therefore, we describe a new molecular method, based on multiplex PCR combined with reverse line blotting (RLB), for the simultaneous detection of 20 different Bartonella species.
The targets selected for the method were the conservative 16S rRNA, as a generic target for detecting any Bartonella species, and the hypervariable intergenic transcribed spacer 16S-23S rRNA (ITS), which allows distinguishing among the different Bartonella species (4, 9). Also, an internal amplification control (IAC), based on delta-9-tetrahydrocannabinolic acid synthase of Cannabis sativa, was added to evaluate the presence of PCR inhibitors (8).
To design primers and probes (Table 1), available sequences were retrieved from GenBank and were aligned by using ClustalX (6). The ITSs of B. chomelii and B. capreoli were sequenced from strains of the collection of the Institute Pasteur, i.e., B. chomelii A828 (GenBank accession no. EU098133) and B. capreoli IBS 193 (GenBank accession no. EU098130 and EU098131). Interestingly, the latter species had two different ITS sequences, which differed in a 12-bp repetition and were determined after cloning the obtained amplicons from a single colony of B. capreoli. Regions of interest, between 18 and 24 bp long and with melting temperatures above 60°C, were identified by visual analysis. Their feasibility for use as primers and probes was checked with Oligo6 software (Molecular Biology Insights, Inc., West Cascade, CO). The Basic Local Alignment Search Tool (BLASTn) (1) was used for a preliminary assessment of the oligonucleotide specificity. A generic probe for all species was designed based on 16S rRNA, and 17 specific probes were selected from ITSs (Table 1). Given the high homology of the ITS sequences between B. chomelii, B. schoenbuchensis, B. capreoli, and B. birtlesii, a common probe for the four species was designed (S-CHOSCA [Table 1]). The probe and primers for the IAC used in this study have been described previously (8).
TABLE 1.
Probes and primers used in the study
| Target | Organism | Straina | Primer or probe | Sequenceb | Oligonucleotide concn (pmol/μl) |
|---|---|---|---|---|---|
| ITS | Bartonella spp. | Bart/16-23F | 5′-bio-TTGATAAGCGTGAGGTCGGAGG | 0.4 | |
| Bart/16-23R | 5′-bio-CAAAGCAGGTGCTCTCCCAG | 0.4 | |||
| B. henselae | Bh-Sp1 | S-HENS | 5′-a-ATCGGTTCAATCATATCGCTTT | 3.2 | |
| B. bacilliformis | CIP 77.27 | S-BACI | 5′-a-CCTATGATTGATTTCTAGGC | 0.4 | |
| B. bovis | CIP 106292 | S-BOV2 | 5′-a-CGTTTTGATAGTCTTTTGTGTTGC | 0.4 | |
| B. koehlerae | CIP 107025 | S-KOE | 5′-a-TTAAATTATATCACTTTGGGTCATACG | 0.4 | |
| B. vinsonii subsp. berkhoffii | CIP 104960 | S-VIN-B | 5′-a-TTTCGGACACTATTGATAAA | 3.2 | |
| B. vinsonii subsp. arupensis | CIP 106848 | S-VIN-A1 | 5′-a-ACTTGTTGGAATTGCTTAACC | 3.2 | |
| B. vinsonii subsp. vinsonii | CIP 103738 | S-VIN-A2 | 5′-a-ATGAAAATATTGAGAGATTTG | 3.2 | |
| B. taylori | CIP 107028 | S-TAY | 5′-a-TATCCATTTCGCTTAGGCA | 3.2 | |
| B. doshiae | CIP 107026 | S-DOSH | 5′-a-TTTGAACCTTCTCTCTTTAT | 3.2 | |
| B. grahamii | CIP 107024 | S-GRAH2 | 5′-a-ATTCAAGTTGATGAATTTGGTTAT | 3.2 | |
| B. elizabethae | CIP 103761 | S-ELIZ | 5′-a-TAAGTTCCCTTCAAGAGGATA | 3.2 | |
| B. tribocorum | CIP 105476 | S-TRIB | 5′-a-TTCTATTAAGTTTGTCAAAGGG | 0.4 | |
| B. clarridgeiae | CIP 104772 | S-CLAR | 5′-a-ACGATGCTAAAAGTTGCTAT | 3.2 | |
| B. alsatica | CIP 105477 | S-ALS | 5′-a-GCTGGTGAAACTTGCTTATA | 3.2 | |
| B. quintana | CIP 107027 | S-QUIN | 5′-a-CGCTTATCCATTTGGTTTAA | 3.2 | |
| B. chomelii | CIP 107869 | S-CHOSCA | 5′-a-TTATGATTGCTGATAAGTTTGCTG | 0.4 | |
| B. schoenbuchensis | CIP 107819 | ||||
| B. capreoli | CIP 106691 | ||||
| B. birtlesii | CIP 106294 | ||||
| Bartonella strain from T. europaea | T4c | S-TALP | 5′-a-CAGTCCCTTTAGGTCCATTTAATC | 3.2 | |
| 16S rRNA | Bartonella spp. | 16S-R | 5′-bio-GCCYCCTTGCGGTTAGCACAGCA | 1 | |
| P24Emod | 5′-bio-CCTTCAGTTMGGCTGGATC | 1 | |||
| S-BART16S | 5′-a-CTCGCCCTTAGTTGCCAGCATT | 3.2 |
Strains from the Pasteur Collection used for Fig. 1, except B. henselae Bh-Sp1, which was isolated from blood of a cat and belongs to the Centro Nacional Microbiología collection.
bio, biotin modification; a, amino link modification.
No isolate; sample where this Bartonella strain was detected.
The primers were combined in a multiplex PCR, which was performed in a 50-μl reaction volume with 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (Promega, Madison, WI), 1.5 U of Taq Gold DNA polymerase (Applied Biosystems, Branchburg, NJ), and 0.8 μg/μl of DNase-free bovine serum albumin (Amersham Biosciences, Barcelona, Spain). Primers were used at final concentrations of 0.4 μM for the ITS (Bart/16-23F and Bart/16-23R) and 1 μM for the other two sets of oligonucleotides (16S rRNA and IAC). PCR cycling included an initial denaturing step of 9 min at 94°C, followed by 40 cycles of 30 s at 94°C, 1 min at 64.3°C, and 1 min 30 s at 72°C and a final elongation step of 7 min at 72°C. The amplification was performed in an MJ Research PCT-200 (Ecogen, Barcelona, Spain). The obtained amplicons (438 bp for 16S rRNA, 371 for the IAC, and 176 to 452 bp for the ITS) were further analyzed by RLB. The combination of PCR amplification and hybridization with specific probes allows us to avoid the interference of nonspecific amplifications due to close ubiquitous organisms, such as Mesorhizobium, which was recently described as a cause of difficulty in the detection of Bartonella by PCR (10).
The RLB was performed as previously described (8) with minor modifications as follows: 3.2 or 0.4 pmol/μl of each probe (Table 1) was attached to the membrane, the hybridization was performed at 50°C for 1 h, and the washing steps were done at 44°C. The overall time required for the RLB was 3.5 h.
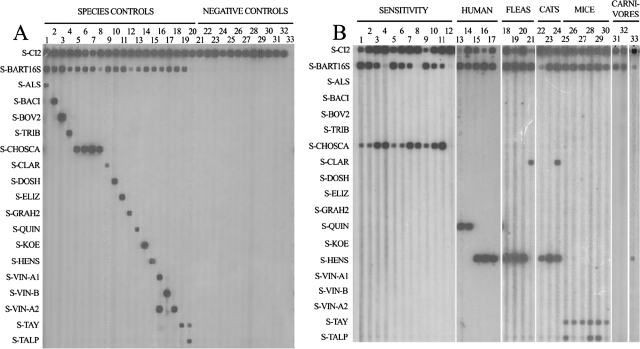
The species specificity of the probes was tested with genomic DNAs from different Bartonella species (Fig. 1), obtained after purification with the QIAamp DNA minikit (IZASA S.A., Barcelona, Spain). One hundred genomic equivalents (GE) of each species and 102 copies of the cloned IAC were amplified by PCR, and amplicons were analyzed by RLB. Positive hybridization signals were obtained with the IAC and the generic probe for all the samples, as well as for each specific probe (Fig. 1A, lanes 1 to 19). B. vinsonii subspecies (B. vinsonii subsp. arupensis, B. vinsonii subsp. berkhoffii, and B. vinsonii subsp. vinsonii) were differentiated by the pattern of hybridization against three probes (P-VIN-A1, P-VIN-A2, and P-VIN-B) (Fig. 1A, lanes 16 to 18).
FIG. 1.
RLB results for different samples. (A) Positive and negative controls. Lanes: 1, B. alsatica; 2, B. bacilliformis; 3, B. bovis; 4, B. tribocorum; 5, B. capreoli; 6, B. chomelii; 7, B. schoenbuchensis; 8, B. birtlesii; 9, B. clarridgeiae; 10, B. doshiae; 11, B. elizabethae; 12, B. grahamii; 13, B. quintana; 14, B. koehlerae; 15, B. henselae; 16, B. vinsonii subsp. arupensis; 17, B. vinsonii subsp. berkhoffii; 18, B. vinsonii subsp. vinsonii; 19, B taylorii; 20, Bartonella sp. from Talpa europaea (cloned DNA); 21, Brucella melitensis biovar melitensis; 22, B. melitensis biovar suis; 23, B. melitensis biovar canis; 24, B. melitensis biovar abortus; 25, B. melitensis biovar neotomae; 26, Borrelia burgdorferi; 27, Chlamydia psitacii; 28, Anaplasma phagocytophilum; 29, Coxiella burnetii; 30, Rickettsia conorii; 31, Francisella tularensis; 32, negative control; 33, water. (B) Sensitivity assay with clinical and environmental samples. Lanes: 1 to 4, B. schoenbuchensis at 103, 102, 10, and 1 GE, respectively; 5 to 8, 103, 102, 10, and 1 GE, respectively, of B. schoenbuchensis plus human DNA; 9 to 12, 103, 102, 10, and 1 GE, respectively, of B. schoenbuchensis plus Ixodes ricinus DNA; 13 to 17, clinical samples, i.e., blood (lanes 13 and 14) and a valve biopsy (lane 15) from patients with endocarditis and lymph node aspirates (lanes 16 and 17) from patients with cat scratch disease; 18 to 21, cat fleas (Ctenocephalides felis); 22 to 24, cat blood; 25 to 30, small mammal blood, i.e., from Mus domesticus (lane 25), Apodemus sylvaticus (lanes 26, 27, and 30), and T. europaea (lanes 28 to 29); 31 to 33, wild carnivore spleen and liver pool samples, i.e., from Vulpes vulpes (lane 31), Meles meles (lane 32), and Felis silvestris (lane 33).
The specificity of the method was tested with 102 copies of IAC plus 102 GE of different related pathogens or an estimated similar amount of DNA from intracellular pathogens. Neither species of Brucella, a genus with rRNA sequences phylogenetically close to those of Bartonella, nor other related pathogens such as Anaplasma, Borrelia, Chlamydia, Coxiella, Francisella, Legionella, Orientia, Rickettsia, and Treponema hybridized with the probes designed for Bartonella (Fig. 1A, lanes 21 to 31). In addition, nonspecific amplification was not observed when negative human samples were assayed (data not shown).
Finally, the sensitivity of the technique was determined with 102 copies of IAC plus 103, 102, 10, and 1 GE of B. schoenbuchensis (Fig. 1B, lane 1 to 4). The test was repeated in the presence of foreign DNA free of pathogens (300 ng of human DNA [Fig. 1B, lanes 5 to 8] or 300 ng of DNA from an Ixodes ricinus specimen [Fig. 1B, lanes 9 to 12]). As expected, in all the samples the IAC was amplified, and the sensitivity for B. schoenbuchensis was 10 GE with the 16S rRNA probe and 1 GE with the ITS probe. In the presence of human DNA there was no loss of sensitivity, while with arthropod DNA we lost a logarithmic unit with the ITS probe (Fig. 1B, lane 9 to 12) but still detected until 10 GE with the 16S rRNA probe.
It has been suggested that the simultaneous presence of different Bartonella species in the same sample could be underestimated using the current diagnostic methods (11). Cases of mixed infections with B. vinsonii subsp. berkoffii and B. henselae in patients have been described. Using our method, we have observed a preferential amplification of B. vinsonii subsp. berkoffii, although 10 GE of B. henselae or B. quintana could be amplified and detected in the presence of 100 or 1,000 GE of B. vinsonii subsp. berkoffii (data not shown).
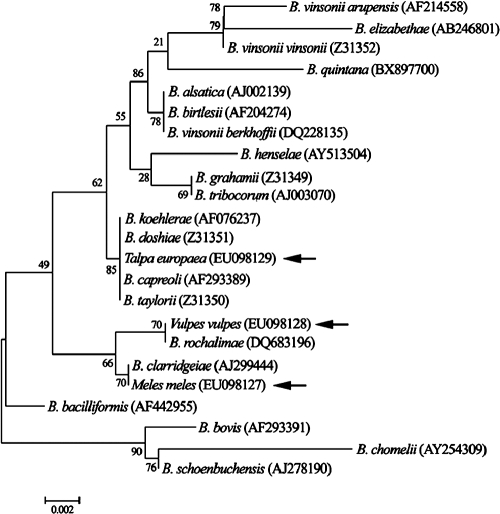
The method was later validated with clinical and environmental samples (Fig. 1). In clinical samples, we detected B. quintana and B. henselae from patients with endocarditis and lymphadenopathies (Fig. 1B, lanes 13 to 17). In environmental samples, we detected B. henselae and B. clarridgeiae in cat fleas (Ctenocephalides felis), cats (Felis felis), and a wild cat (F. silvestris), including a mixed infection of both species in a cat (Fig. 1B, lanes 18 to 24 and 33). In small mammals, B. taylorii was detected (Fig. 1B, lanes 25 to 30). However, when the ITS amplicons were sequenced, an undescribed 452-bp ITS fragment (GenBank accession no. EU098135) different from the sequence of the ITS of B. taylorii was detected in two moles (Talpa europaea) and one house mouse (Mus domesticus). In the BLAST server from the NCBI, this fragment showed an 81% homology with an uncultured Bartonella (accession no. AJ269794) which had been detected in a wood mouse (4). The 16S rRNA amplicons from these samples were also sequenced (GenBank accession no. EU098129) and grouped in the dendrogram in the same clade as B. capreoli, B. taylorii, and B. doshiae (Fig. 2). As it was detected in a mole, it could correspond to B. talpae, although there are currently no available strains or sequences of this species. Consequently, we designed a new probe for this agent, and we cloned the corresponding ITS amplicon to be used as positive control (Fig. 1A, lane 20). We were then able to distinguish this species from B. taylorii in the RLB (Fig. 1B, lanes 25, 27, and 28).
FIG. 2.
Phylogenetic tree built with partial Bartonella 16S rRNA sequences. Tree inference for the phylogeny reconstruction was done with the neighbor-joining method with the interior branch test by using Mega version 4 software (15).
Finally, in samples from wild carnivores we found hybridization signals only with the generic probe and not with any other specific probe, suggesting the presence of other Bartonella species. In fact, the 422-bp ITS sequence (GenBank accession no. EU098134) obtained from a fox (Vulpes vulpes) (Fig. 1B, lane 31) had 99% homology with B. rochalimae, a new species which has recently been detected in a tourist visiting Peru (5). Also, the ITS sequence from a badger (Meles meles) (Fig. 1B, lane 32), showed a previously undescribed 427-bp sequence (GenBank accession no. EU98132) with 88% homology with B. clarridgeiae. In the dendrogram (Fig. 2), both 16S rRNA sequences (GenBank accession no. EU98127 and EU098128) from these samples grouped in the same clade as B. clarridgeiae and B. rochalimae.
The molecular method described, which is being patented (2; P. Anda, 29 June 2007, Oficina Española de Patentes y Marcas, pending patent P200701830), has shown an excellent specificity and sensitivity and is a powerful tool for the study of environmental samples and of the etiology of human bartonellosis. Moreover, the technique allows the identification of new Bartonella species and could be updated progressively with the design of probes for new species, as has been done in the case of the Bartonella detected in small mammals. New probes are being tested for new Bartonella species detected in wild carnivores as well as for other species, such B. rattimassiliensis or B. phoceensis.
Nucleotide sequence accession numbers.
The sequences obtained in this study has been submitted to GenBank under the following accession numbers: EU098127 and EU98132 (16S rRNA and ITS of a Bartonella sp. detected in M. meles), EU098128 and EU098134 (16S rRNA and ITS of a Bartonella sp. detected in V. vulpes), EU098129 and EU98135 (16S rRNA and ITS of a Bartonella sp. detected in T, europaea), EU098130 and EU098131 (ITSs from B. capreoli), and EU098133 (ITS from B. chomelii).
Acknowledgments
This study has been supported by the Fondo de Investigación Sanitaria (P1050901) and RTIC EBATRAG (G03/057) from the Instituto de Salud Carlos III.
We thank Santos Jiménez and Azucena Pérez from Consejería de Salud del Gobierno de La Rioja for supplying samples from cats and fleas.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anda, P., R. Escudero, M. Rodríguez-Moreno, I. Jado, and M. I. Jimenez-Alonso. December 2006. Method and kit for the detection of bacterial species by means of DNA. Patent WO/2006/136639.
- 3.Avidor, B., M. Graidy, G. Efrat, C. Leibowitz, G. Shapira, A. Schattner, O. Zimhony, and M. Giladi. 2004. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J. Clin. Microbiol. 423462-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtles, R. J., S. Hazel, K. Bown, D. Raoult, M. Begon, and M. Bennett. 2000. Subtyping of uncultured bartonellae using sequence comparison of 16 S/23 S rRNA intergenic spacer regions amplified directly from infected blood. Mol. Cell. Probes 1479-87. [DOI] [PubMed] [Google Scholar]
- 5.Eremeeva, M. E., H. L. Gerns, S. L. Lydy, J. S. Goo, E. T. Ryan, S. S. Mathew, M. J. Ferraro, J. M. Holden, W. L. Nicholson, G. A. Dasch, and J. E. Koehler. 2007. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 3562381-2387. [DOI] [PubMed] [Google Scholar]
- 6.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73237-244. [DOI] [PubMed] [Google Scholar]
- 7.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 98-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jado, I., R. Escudero, H. Gil, M. I. Jiménez-Alonso, R. Sousa, A. L. García-Pérez, M. Rodríguez-Vargas, B. Lobo, and P. Anda. 2006. Molecular method for identification of Rickettsia species in clinical and environmental samples. J. Clin. Microbiol. 444572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, W. A., M. Z. Fall, J. Rooney, D. L. Kordick, and E. B. Breitschwerdt. 2000. Rapid identification and differentiation of Bartonella species using a single-step PCR assay. J. Clin. Microbiol. 381717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maggi, R. G., and E. B. Breitschwerdt. 2005. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J. Clin. Microbiol. 431171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maggi, R. G., A. W. Duncan, and E. B. Breitschwerdt. 2005. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J. Clin. Microbiol. 432651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raoult, D., P. E. Fournier, F. Vandenesch, J. L. Mainardi, S. J. Eykyn, J. Nash, E. James, C. Benoit-Lemercier, and T. J. Marrie. 2003. Outcome and treatment of Bartonella endocarditis. Arch. Intern. Med. 163226-230. [DOI] [PubMed] [Google Scholar]
- 13.Raoult, D., F. Roblot, J. M. Rolain, J. M. Besnier, J. Loulergue, F. Bastides, and P. Choutet. 2006. First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J. Clin. Microbiol. 44278-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolain, J. M., P. Brouqui, J. E. Koehler, C. Maguina, M. J. Dolan, and D. Raoult. 2004. Recommendations for treatment of human infections caused by Bartonella species. Antimicrob. Agents Chemother. 481921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 16.Znazen, A., J. M. Rolain, N. Hammami, S. Kammoun, A. Hammami, and D. Raoult. 2005. High prevalence of Bartonella quintana endocarditis in Sfax, Tunisia. Am. J. Trop. Med. Hyg. 72503-507. [PubMed] [Google Scholar]