Abstract
Salmonella enterica serovar Typhimurium strains of phage types DT104 and U302 are often resistant to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (the ACSSuT resistance type) and are major zoonotic pathogens. Increased consumption of goose meat may enhance the risk of transferring S. enterica serovar Typhimurium and other enteric pathogens from geese to human due to the consumption of meats from infected geese or improper preparation of meats. Therefore, we characterized S. enterica serovar Typhimurium strains isolated from four goose farms (farms A, B, C, and D) and one hatchery farm (farm E) to determine the epidemic and genetic differences among them. Antibiotic susceptibility tests and multiplex PCR confirmed that 77.6% (52/67) of strains were ACSSuT strains isolated from farms A, C, and E. Antibiotic-susceptible strains were isolated mostly from farm B, and no strain was observed in farm D. All ACSSuT strains harbored a 94.7-kb virulence plasmid and contained one 1.1-kb conserved segment identical to that of Salmonella genomic island 1. Four genotypes were determined among these S. enterica serovar Typhimurium isolates by pulsed-field gel electrophoresis analysis of XbaI-digested DNA fragments. Most isolates (85.29%; 29/34) of major genotype Ib were ACSSuT strains isolated mainly from goslings of farm C and egg membranes of farm E, a hatchery farm, suggesting that S. enterica serovar Typhimurium strains in isolates from goslings might originate from its hatchery, from the egg membranes to the gosling fluff after hatching. Multiple phage types, types 8, 12, U283, DT104, and U302, were identified. In conclusion, geese were a reservoir of diverse multidrug-resistant (type ACSSuT) S. enterica serovar Typhimurium strains, and each farm was colonized with genetically closely related S. enterica serovar Typhimurium strains.
Outbreaks of salmonellosis caused by Salmonella enterica serovar Gallinarum result in great economic losses due to the high mortality of young chickens; however, S. enterica serovar Typhimurium appears to have a higher invasion capability than S. enterica serovar Gallinarum (11). As one of the most predominant Salmonella serovars isolated from poultry, S. enterica serovar Typhimurium can be transmitted vertically through eggs and horizontally from the contaminated eggs and meat to humans (1). In ducklings, 93% of Salmonella isolates are S. enterica serovar Typhimurium (30). The infected duckling may be dehydrated and emaciated, have difficulty breathing, and even die in opisthotonus. In the United Kingdom, S. enterica serovar Typhimurium and S. enterica serovar Enteritidis have been reported to cause septicemia in young ducklings with mixed infection with other pathogens (18). In market-ready geese, S. enterica serovar Typhimurium is the major serovar isolated from 60% of fluids from carcass rinsings and 18.4% of cloacal swabs (25). In addition, S. enterica serovar Typhimurium also caused a few cases of chronic salpigitis in geese and ducks (6).
S. enterica serovar Typhimurium usually harbors a 94.7-kb virulence plasmid (pSTV) encoding the 8-kb spv operon (16) that is involved in survival in the macrophage and then assists in systemic infection (21). The ccdAB addition system between the spv operon and RepFIB in pSTV (GenBank accession number AE006471) maintains plasmid stability (34). Since multidrug-resistant (type ACSSuT) phage type DT104 was discovered in 1984 (35), a 43-kb multidrug resistance DNA region responsible for the ACSSuT type has been found and named Salmonella genomic island 1 (SGI1), which is inserted between the thdF and yidY genes and contains two conserved-segment (CS) regions (7). Due to the process of evolution by recombination, insertion, and deletion events, SGI1 variants have been found in several serovars and may enhance the virulence and dissemination of the host strain (27). Recently, we reported multidrug-resistant (type ACSSuT) S. enterica serovar Typhimurium phage types DT104, DT120, and U302 isolated from human in Taiwan (13). To elucidate whether these multidrug-resistant S. enterica serovar Typhimurium phage types can also be isolated from geese, the prevalence of multidrug-resistant S. enterica serovar Typhimurium and the pSTV profile, genotyping, and phage typing of these goose isolates were analyzed. Despite the existence of phage types DT104 and U302 in human and goose isolates, other S. enterica serovar Typhimurium phage types, types 8, 12, and U283, were isolated from geese, which were not of human origin.
MATERIALS AND METHODS
Collection and enrichment of samples.
The samples were taken from five different goose farms in Chiayi, Taiwan, from 2004 to 2005. The culled sick geese were detected by necropsy and laboratory examination on farm A and farm C according to methods described previously Zander et al. (40). A method employing cloacal swabbing was used to examine S. enterica serovar Typhimurium infection of 4-week-old geese with diarrhea from farms A and C and healthy birds from farms B, C, D, and E (Table 1). All broths and selected agars used for isolating and identifying S. enterica serovar Typhimurium were purchased from Difco (Becton Dickinson Co., Franklin Lakes, NJ). The sampled swabs were enriched in 9 ml of selenite cysteine broth and incubated at 37°C for 24 h. In farm E, 15 goose hatching eggs and three dragging swabs were collected from individual cabinets. The inner membranes of eggshells were separated from the eggshell halves with sterile forceps and then incubated in 5 ml of gram-negative (GN) broth. Before sampling, the dragging swabs were autoclaved within the culture tubes of GN broth and then used to vigorously sample an area of approximately 30 cm2 on the inner surfaces of the hatching cabinets. After sampling, the dragging swabs were put back into the culture tubes, which were incubated for 24 h at 37°C. If the initial subculture was Salmonella negative, delayed secondary enrichment was performed as described previously (37). The negative broth was incubated at room temperature for 5 to 7 days. One milliliter of the broth was then transferred into 9 ml of Rappaport-Vassiliadis broth, and the broth was incubated for 24 h at 37°C.
TABLE 1.
Sampling locations, methods, and clinical conditions of geese used in the study
| Farm [test date (yr/mo)] | Sampling method | Age | Clinical body condition | Test code | No. of isolates |
|---|---|---|---|---|---|
| A (2002/6) | Necropsy | 2 wka | Sick | FAN | 5 |
| B (2003/11) | Cloacal swab | 3 wkb | Normal | CBC | 6 |
| C (2004/8-9) | Necropsy | 2-5 wka | Sick | FCN | 10 |
| Cloacal swab | 1 wkb | Normal | FAC1 | 3 | |
| Cloacal swab | 4 wka | Sick | FAC4 | 27 | |
| D (2004/8-9) | Cloacal swab | 1 wkb | Normal | FBC1 | 0 |
| Cloacal swab | 4 wkb | Normal | FBC4 | 0 | |
| E (2004/8-9) | Cloacal swab | 0 daysb | Normal | HCG | 0 |
| Hatching egg membrane | HHE | 16 | |||
| Hatching cabinet | HHC | 0 |
These geese were sick at the time of clinical examination.
These geese were healthy at the time of clinical examination.
Salmonella isolation.
Selectively enriched samples from selenite cysteine broth and GN broth were streaked onto xylose lysine deoxycholate plates. These plates were incubated at 37°C for 24 h to select typical Salmonella colonies, which were further identified by biochemical tests including triple sugar iron agar and lysine iron agar slants. At least two positive isolates from each plate were maintained on brain heart infusion agar plates.
Serogrouping and serotyping of Salmonella isolates.
S. enterica serovar Typhimurium strains were identified by using commercial Salmonella O and H antisera provided by Difco (Becton Dickinson Co., Franklin Lakes, NJ). All isolates were first serogrouped by the slide agglutination test with the use of O antiserum. With a positive reaction to O antigens 1, 4, 5, and 12, serogroup B Salmonella strains were selected and further examined for their serotypes by the tube agglutination test using H antisera (H antigens i, 1, and 2). While the second-phase antigen was not present in the first agglutination test, strains with second-phase H antigen were selected on the other end of a paper bridge containing first-phase H antiserum across the culture medium.
Antibiotic susceptibility test.
Antimicrobial susceptibility was tested by a standard disk diffusion method, and Escherichia coli (ATCC 25922) was used for validating antibiotic test results (28). The antimicrobial agents used were ampicillin (10 μg), chloramphenicol (30 μg), florfenicol (30 μg), streptomycin (10 μg), trimethoprim-sulfamethoxazole (1.25 μg/23.75 μg), sulfisoxazole (250 μg), and tetracycline (30 μg). Susceptible and resistant isolates were defined according to the criteria suggested by the CLSI (formerly NCCLS) (28). Multiplex PCR was used to amplify genes including spvC for pSTV and the following resistance genes: pse for ampicillin resistance (A), floR for chloramphenicol resistance (C), aadA for streptomycin resistance (S), sulI for sulfonamide resistance (Su), and tetR or tetG for tetracycline resistance (T) (13). In order to differentiate the integron type, PCR was performed to amplify the CS region containing a resistance gene cassette according to a method described previously (29). After PCR amplification, the PCR product of the CS region was sequenced.
Identification of virulence plasmid by DNA-DNA hybridization.
The virulence plasmid (pSTV) of S. enterica serovar Typhimurium isolates was checked by Southern blotting hybridization with a probe of the spvC DNA fragment amplified by PCR as previously described (12, 33). Plasmid DNA was subjected to gel electrophoresis as previously described (23). The plasmid DNA was then transferred onto a Zeta-Probe membrane (Bio-Rad, CA) as recommended by the manufacturer. PCR products of spvC were purified by use of a Wizard SV gel and the PCR Clean-Up system (Promega), labeled with digoxigenin (DIG)-11-dUTP (Roche), and used as the DNA probe. After hybridization of the DIG-labeled probe and the addition of anti-DIG antibody conjugated with peroxidase, the membrane was reacted with CSPD (Roche), a chemiluminescent substrate, and then exposed to X-ray film. If both PCR and DNA hybridization were positive for the strain tested, it was concluded that the strain harbored the virulence plasmid.
Genotyping by PFGE and phage typing.
Genotypes of all S. enterica serovar Typhimurium strains were determined by pulsed-field gel electrophoresis (PFGE) analysis using the restriction endonuclease XbaI to digest total genomic DNA. The procedure for the PFGE analysis was described previously (13). The digested DNA was separated by use of CT Mapper XA (Bio-Rad) in 0.5× Tris-borate-EDTA at 14°C for 22 h. Due to the conserved characteristic of the Salmonella genome, strains with a banding pattern difference of more than three bands were designated different genotypes, and strains with at least one band difference were designated different subgenotypes. Representative strains of each S. enterica serovar Typhimurium genotype were phage typed at the French National Center for Salmonella (Institut Pasteur, Paris, France) according to methods described previously by Anderson et al. (2). Phage types and genotypes were analyzed.
RESULTS
S. enterica serovar Typhimurium infection in geese.
The prevalence of S. enterica serovar Typhimurium was 7% (36 of 513 samples) for cloacal swab samples and 10.7% (16 of 150 samples) for hatching eggshell membranes. Although differing in prevalence among sampling sources, the highest isolation rate (22.9%; 27/118) was obtained in diseased sample FAC4 (C-FAC4) isolates and not in normal FAC1 (C-FAC1) isolates of farm C (Table 1). Also, no S. enterica serovar Typhimurium isolate was found in normal farm D and E-HCG as well as E-HHC samples (Table 1).
Antibiotic susceptibility and SGI analysis.
After antibiotic susceptibility analysis by the disk diffusion method and multiplex PCR analysis, 77.6% (52/67 isolates) of S. enterica serovar Typhimurium were of the ACSSuT type (Table 2 and Fig. 1). Most ACSSuT strains were isolated from C-FAC4 (25/52 isolates) and E-HHE (15/52 isolates); however, these resistant strains were not found in B-CBC. Except for one strain that appeared in the 1.3- and 1.1-kb SGI1-specific CS region, all ACSSuT strains contained the 1.1-kb SGI1-specific CS region with identical sequences, which was not amplified from non-ACSSuT strains (Table 2).
TABLE 2.
Antibiotic susceptibilities and distributions of the virulence plasmid of 67 S. enterica serovar Typhimurium strains from different farms
| Type of drug resistance | Total no. of isolates | CS region
|
pSTV (94.7 kb)
|
Sample designation(s) for isolation source(s) | ||
|---|---|---|---|---|---|---|
| Presence | No. of isolates | Presence | No. of isolates | |||
| ACSSuT | 52 | + | 52 | + | 52 | FAC1, FAC4, FAN, FCN, HHE |
| ACSSu | 1 | − | 1 | + | 1 | FAC4 |
| CSSu | 1 | − | 1 | − | 1 | HHE |
| SSu | 1 | − | 1 | + | 1 | FAC4 |
| S | 2 | − | 2 | + | 1 | CBC |
| C | 1 | − | 1 | − | 1 | HHE |
| Nonea | 9 | + | 0 | + | 8 | CBC, FAC1 |
| Total | 67 | 67 | 66 | |||
These strains were isolated from CBC of farm B.
FIG. 1.
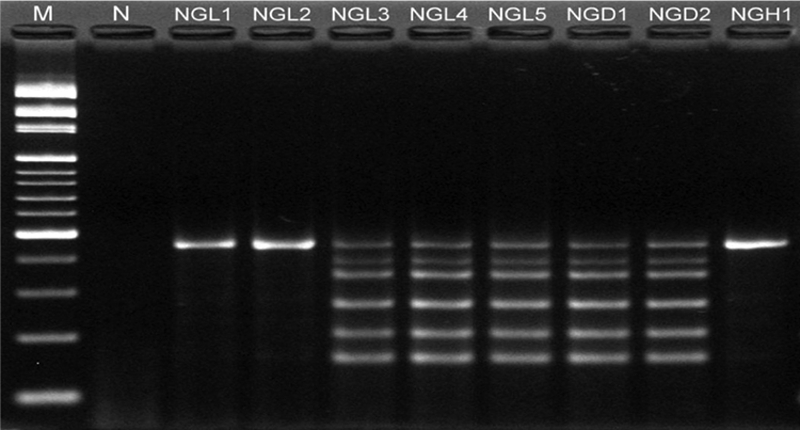
Electrophoretic profiles of the PCR products of drug resistance genes and spvC amplified by a multiplex PCR. M, 100- to 3,000-bp DNA ladder; N, negative. The PCR product sizes are 156 bp for pse, 203 bp for str, 266 bp for floR, 351 bp for sulI, 391 bp for tetG, and 450 bp for spvC.
Virulence plasmid, genotype, and phage types.
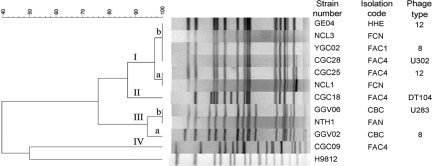
S. enterica serovar Typhimurium usually harbors a 94.7-kb virulence plasmid. Here, all ACSSuT S. enterica serovar Typhimurium isolates (100%; 52/52 isolates) harbored pSTV, which is missing in three strains isolated from E-HHE (Table 2). To understand the genomic variation among these S. enterica serovar Typhimurium isolates, PFGE analysis of XbaI-digested fragments separated 58 strains into four major genotypes (Fig. 2). Genotype distributions varied among isolation sources, and the major genotype, genotype Ib (74%; 43/58 isolates), appeared only in E-HHE as well as C-FAC1 and C-FAC4 (Table 3). Almost all isolates showed identical genotypes, such as genotype III found only in B-CBC samples, or one major genotype at the same farm (Table 3). However, three genotypes (genotypes I, II, and IV) were observed in C-FAC4.
FIG. 2.
Dendrogram and PFGE patterns of XbaI-digested genomic DNA of S. enterica serovar Typhimurium isolates. The dendrogram was constructed based on the unweighted-pair group method using average linkages algorithm and the Dice similarity coefficient by using BioNumerics software with 3% optimization and 1% position tolerance. The genotypes are indicated as I, II, III, and IV. S. enterica serovar Braendrup strain H9812 was the PFGE size marker.
TABLE 3.
Genotype distribution of 58 S. enterica serovar Typhimurium strains from different farms
| Genotype | No. of isolates from:
|
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|
| A-FAN | B-CBC | C-FAC1 | C-FAC4 | D-FCN | E-HHE | ||
| I | |||||||
| Ia | 3 | 1 | 4 | ||||
| Ib | 3 | 21 | 4 | 15 | 43 | ||
| II | 1 | 0 | 1 | ||||
| III | |||||||
| IIIa | 1 | 1 | |||||
| IIIb | 3 | 5 | 8 | ||||
| IV | 1 | 1 | |||||
| Total | 3 | 6 | 3 | 26 | 5 | 15 | 58 |
Phage typing further differentiated these strains and determined the existence of a DT104 strain. Although only seven of the tested strains were analyzed, five phage types, types 8, 12, DT104, U283, and U302, were obtained (Fig. 2). Among these genotypes, genotype Ib was further divided into three phage types, types 8, 12, and U302. In contrast, the same phage type (types 8 and 12) appeared in different genotypes. Moreover, similar to genotype distribution, multiple phage types were observed in C-FAC4 strains.
DISCUSSION
S. enterica serovar Typhimurium is widely distributed among diverse ranges of animals, and hence, the prevalence of the pathogen in poultry is not unexpected. In poultry, S. enterica serovar Typhimurium appeared to have a superior invasion capability compared to S. enterica serovar Gallinarum (11). Therefore, S. enterica serovar Typhimurium has been frequently found in young chickens, imported frozen chickens, ducklings, and geese in different countries (6, 18, 20, 25, 26, 28, 30). Although S. enterica serovar Typhimurium may not be a primary cause of disease, it could possibly cause septicemia in young ducklings with complications of other diseases (18). The association of S. enterica serovar Typhimurium with septicemia or diarrhea in young geese (farms A and C) and subclinical infection with S. enterica serovar Typhimurium (farm B) imply that systemic infection with S. enterica serovar Typhimurium with complications of other pathogens may cause the death of geese. Previous reports indicated that the 94.7-kb plasmid pSTV is involved in systemic infection (21). In the present study, most of the S. enterica serovar Typhimurium isolates collected from human and geese harbored pSTV (13) (Table 2), which carries the ccdAB addiction system (34), and the spv operon (21), both responsible for the existence of pSTV in S. enterica serovar Typhimurium and the enhancement of the systemic infection of S. enterica serovar Typhimurium in humans and animals, respectively.
Salmonella can cause food-borne illness in humans due to contaminated meats and/or eggs, where Salmonella localizes on the inner membranes of eggshells in isthmal secretions (8), and the inner membrane also provides a physical barrier to prevent the entry of Salmonella. These studies revealed that S. enterica serovar Typhimurium existed on the inner membrane of goose eggs, as previously reported for duck egg membranes (31), but not from cloacal swabs of 0-day-old goslings and the hatching cabinet (Table 1), which are the most frequently found sites of S. enterica serovar Enteritidis and S. enterica serovar Typhimurium contamination (5, 9, 24) and a useful target to evaluate S. enterica serovar Typhimurium contamination (3, 4). In addition, egg contamination by salmonellae can occur through contact with Salmonella-contaminated equipment, personnel, and the environment and infected wild birds and rodents (14, 15), followed by the entry of Salmonella into hatching eggs in a hatchery. The transmission of S. enterica serovar Typhimurium from the hatchery to goslings was found in farm A since the goslings originated from a hatchery with high levels of contamination of S. enterica serovar Typhimurium on eggshell membranes (Tables 1 and 3). The XbaI-digested PFGE profile indicated that prolonged exposure to the external environment such as a longer feeding period (genotype Ib for C-FAC1 versus genotypes Ia, Ib, II, and IV for C-FAC4) can increase the prevalence of diverse multidrug-resistant Salmonella strains in geese (Fig. 2 and Tables 2 and 3). In addition, identical genotypes in each farm, such as genotype IIIb in A-FAN and genotype Ib in C-FAC1 and E-HHE, and genotype differences among farms (Table 3) suggest that each farm was colonized with genetically closely related S. enterica serovar Typhimurium strains.
Generally, adequate sanitation can reduce the prevalence of Salmonella. For example, proper egg washing may minimize the problem of Salmonella contamination on outer eggshells (5, 22) and transmission from incubator to hatchers (9, 10). However, improper washing or rinsing of eggs, such as processing at lower temperatures, may increase the entrance of S. enterica serovar Typhimurium and S. enterica serovar Enteritidis into egg contents (22), and the washing process cannot remove Salmonella contamination in the inner membrane and egg contents. Therefore, a vaccine is needed to prevent S. enterica serovar Typhimurium and S. enterica serovar Enteritidis infection.
Since the discovery of a multidrug-resistant ACSSuT S. enterica serovar Typhimurium DT104 strain in United Kingdom (35), this strain has spread around the world, and it is prominently distributed in Europe and Northern America (17, 20), is less dispersed in Asia, such as in Korea (39) and Japan (32), and causes gastroenteritis and occasional outbreaks in human and wild or domestic animals. Type ACSSuT S. enterica serovar Typhimurium DT104 isolates not only have been frequently found in human (13) but have been commonly found in geese in the present study (Table 2). In Asia, several phage types of S. enterica serovar Typhimurium have been isolated from poultry and animals, such as phage types 13, 120, and DT104 in chickens (19) and phage type 90 in humans and animals (36). In this study, diverse phage types were found mainly in type ACSSuT S. enterica serovar Typhimurium isolates belonging to genotypes I and II, and some phage types, types 8, 12, and U283, were found only in geese (Fig. 2), suggesting that geese are a reservoir of diverse multidrug-resistant S. enterica serovar Typhimurium isolates.
Acknowledgments
This work was funded by grants from the Council of Agriculture, grant 93AS-1.8.1-BQ-BC (C.-Y.Y.), and the National Science Council (NSC95-2314-B-415-002), Executive Yuan, Taiwan (C.C.), and by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract N01-AI-30054, project no. ZC002-03 (Y.-F.C.).
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Aabo, S., J. P. Christensen, M. S. Chadfield, B. Carstensen, J. E. Olsen, and M. Bisgaard. 2002. Quantitative comparison of intestinal invasion of zoonotic serotypes of Salmonella enterica in poultry. Avian Pathol. 3141-47. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, M. J. De Saxe, and J. D. De Sa. 1977. Bacteriophage typing designations of Salmonella typhimurium. J. Hyg. (London) 78297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, J. S., R. J. Buhr, N. A. Cox, and M. E. Berrang. 1996. Effect of hatching sanitation treatments on Salmonella cross-contamination and hatchability of broiler eggs. Poult. Sci. 75191-196. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, J. S., N. A. Cox, and M. E. Berrang. 1994. Hatchery-acquired salmonellae in broiler chicks. Poult. Sci. 731153-1157. [DOI] [PubMed] [Google Scholar]
- 5.Baker, R. C., R. A. Quershi, T. S. Sandhu, and J. F. Timoney. 1985. The frequency of Salmonella on duck eggs. Poult. Sci. 644646-652. [DOI] [PubMed] [Google Scholar]
- 6.Bisgaard, M. 1995. Salpingitis in web-footed birds: prevalence, aetiology and significance. Avian Pathol. 24443-452. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 461714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, J. D., F. V. Immerseel, G. Meulemans, F. Haesebrouck, and R. Ducatelle. 2003. Adhesion of Salmonella enterica serotype Enteritidis isolates to chicken isthmal glandular secretions. Vet. Microbiol. 93223-233. [DOI] [PubMed] [Google Scholar]
- 9.Cason, J. A., J. S. Bailey, and N. A. Cox. 1993. Location of Salmonella typhimurium during incubation and hatching of inoculated eggs. Poult. Sci. 722064-2068. [DOI] [PubMed] [Google Scholar]
- 10.Cason, J. A., N. A. Cox, and J. S. Bailey. 1994. Transmission of Salmonella typhimurium during hatching of broiler chicks. Avian Dis. 38583-588. [PubMed] [Google Scholar]
- 11.Chadfield, M. S., D. J. Brown, S. Aabo, J. P. Christensen, and J. E. Olsen. 2003. Comparison of intestinal invasion and macrophage response of Salmonella Gallinarum and other host-adapted Salmonella enterica serovars in the avian host. Vet. Microbiol. 9249-64. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, C. H., and J. T. Ou. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 342619-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu, C. H., L. H. Su, C. H. Chu, M. H. Wang, C. M. Yeh, F. X. Weill, and C. Chu. 2006. Detection of multidrug-resistant Salmonella enterica serovar Typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J. Clin. Microbiol. 442354-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, N. A., M. E. Berrang, and J. A. Cason. 2000. Salmonella penetration of egg shells and proliferation in broiler hatching eggs—a review. Poult. Sci. 791571-1574. [DOI] [PubMed] [Google Scholar]
- 15.Davies, R. H., R. A. J. Nicholas, I. M. McLaren, J. D. Corkish, D. G. Lanning, and C. Wray. 1997. Bacteriological and serological investigation of persistent Salmonella enteritidis infection in an integrated poultry organization. Vet. Microbiol. 58277-293. [DOI] [PubMed] [Google Scholar]
- 16.Gulig, P. A., H. Danbara, D. G. Guiney, A. Lax, F. Norel, and M. Rhen. 1993. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 7825-830. [DOI] [PubMed] [Google Scholar]
- 17.Helms, M., S. Ethelberg, K. Molbak, and the DT104 Study Group. 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg. Infect. Dis. 11859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry, R. R. 2000. Salmonella infection in ducks, p. 157-167. In C. Wray and A. Wray (ed.), Salmonella in domestic animals. CABI Publishing Press, New York, NY.
- 19.Hernandez, T., C. Rodriguez-Alvarez, M. P. Arevalo, A. Torres, A. Sierra, and A. Arias. 2002. Antimicrobial-resistant Salmonella enterica serovars isolated from chickens in Spain. J. Chemother. 14346-350. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, T., A. Sierra, C. Rodriguez-Alvarez, A. Torres, M. P. Arevalo, M. Calvo, and A. Arias. 2005. Salmonella enterica serotypes isolated from imported frozen chicken meat in the Canary islands. J. Food Prot. 682702-2706. [DOI] [PubMed] [Google Scholar]
- 21.Hoertt, B. E., J. T. Ou, D. J. Kopecko, L. S. Baron, and R. L. Warren. 1989. Novel virulence properties of the Salmonella typhimurium virulence-associated plasmid: immune suppression and stimulation of splenomegaly. Plasmid 148-58. [DOI] [PubMed] [Google Scholar]
- 22.Hutchison, M. L., J. Gittins, A. W. Sparks, T. J. Humphrey, C. Burton, and A. Moore. 2004. An assessment of the microbiological risks involved with egg washing under commercial conditions. J. Food Prot. 674-11. [DOI] [PubMed] [Google Scholar]
- 23.Kado, C., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1741365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limawongpranee, S., H. Hayashidani, A. T. Okatani, K. Ono, C. Hirota, K. Kaneko, and M. Ogawa. 1999. Prevalence and persistence of Salmonella in broiler chicken flocks. J. Vet. Sci. 61255-259. [DOI] [PubMed] [Google Scholar]
- 25.Mann, E., and C. D. McNabb. 1984. Prevalence of Salmonella contamination in market-ready geese in Manitoba. Avian Dis. 284978-983. [PubMed] [Google Scholar]
- 26.Mario, P. N. 1990. Salmonella typhimurium outbreak in broiler chicken flocks in Mexico. Avian Dis. 34221-223. [PubMed] [Google Scholar]
- 27.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 81915-1922. [DOI] [PubMed] [Google Scholar]
- 28.NCCLS. 2003. Performance standards for antimicrobial disk susceptibility tests: approved standard M2-A8, 8th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 29.Ng, L.-K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 433018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price, J. I., E. Dougherty III, and D. J. W. Bruner. 1962. Salmonella infection in white pekin duck: a short summary of the years 1950-60. Avian Dis. 6145-147. [Google Scholar]
- 31.Saitanu, K., C. Koowatananukul, J. Jerngklinchan., and J. Sasipreeyajan. 1994. Detection of salmonellae in hen eggs in Thailand. Southeast Asian J. Trop. Med. Public Health 25324-327. [PubMed] [Google Scholar]
- 32.Sameshima, T., M. Akiba, H. Izumiya, J. Terajima, K. Tamura, H. Watanabe, and M. Nakazawa. 2000. Salmonella typhimurium DT104 from livestock in Japan. Jpn. J. Infect. Dis. 5315-16. [PubMed] [Google Scholar]
- 33.Southern, E. M. 1975. Detection of specific sequence among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98503-517. [DOI] [PubMed] [Google Scholar]
- 34.Tam, J., and B. C. Kline. 1989. The F plasmid ccd autorepressor is a complex of CcdA and CcdB protein. Mol. Gen. Genet. 21926-32. [DOI] [PubMed] [Google Scholar]
- 35.Threlfall, E. J., B. Rowe, and L. R. Ward. 1993. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol. Infect. 111189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vo, A. T., E. van Duijkeren, A. C. Fluit, M. E. Heck, A. Verbruggen, H. M. Maas, and W. Gaastra. 2006. Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet. Microbiol. 113153-158. [DOI] [PubMed] [Google Scholar]
- 37.Waltman, W. D., and E. T. Mallinson. 1995. Isolation of Salmonella of poultry tissue and environmental samples: a nationwide survey. Avian Dis. 3945-54. [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Yang, S. J., K. Y. Park, S. H. Kim, K. M. No, T. E. Besser, H. S. Yoo, S. H. Kim, B. K. Lee, and Y. H. Park. 2002. Antimicrobial resistance in Salmonella enterica serovars Enteritidis and Typhimurium isolated from animals in Korea: comparison of phenotypic and genotypic resistance characterization. Vet. Microbiol. 86295-301. [DOI] [PubMed] [Google Scholar]
- 40.Zander, D. V., A. J. Bermudez, and E. T. Mallinson. 1997. Principles of disease prevention: diagnosis and control, p. 3-45. In B. W. Calnek (ed.), Disease of poultry, 10th ed. Iowa State University Press, Ames, Iowa.