Abstract
Influenza A virus has the ability to overcome immunity from previous infections through the acquisition of genetic changes. Thus, understanding the evolution of the viruses in humans is important for the surveillance and the selection of vaccine strains. A total of 30 influenza A/H3N2 viruses and 35 influenza A/H1N1 viruses that were collected in Vietnam from 2001 to 2006 were used to analyze the evolution of the hemagglutinin (HA), neuraminidase (NA), and matrix protein (M) genes. Phylogenetic analysis of individual gene segments revealed that the HA and the NA genes of the influenza A viruses evolved in a sequential way. However, the evolutionary pattern of the M gene proved to be nonlinear and was not linked with that of the HA and NA genes. Genetic drift in HA1 segments, especially in the antigenic sites of A/H3N2 viruses, occurred more frequently in A/H3N2 viruses than it did in A/H1N1 viruses. Two reassortants, one influenza A/H3N2 strain and one A/H1N1 strain, were found on the basis of the phylogenetic analysis of the three genes. While both genetic mutation and reassortment contributed to their evolution, the frequency of genetic changes and reassortment events differs between the two subtypes. As influenza viruses circulate throughout the year, we emphasize the importance of surveillance in tropical and subtropical zones, where the emergence of new strains may be detected earlier than it is in temperate zones.
The influenza virus is a major viral respiratory pathogen that causes yearly epidemics in tropical and subtropical countries, with epidemic influenza remaining a major cause of morbidity and mortality (27). Recurrent epidemics of influenza are due to the frequent emergence of antigenic variants. With the cocirculation of two influenza A subtypes, genetic reassortment also has an important role in antigenic drift (6, 28). The genome structure of influenza A viruses, consisting of eight segments of negative-sense single-stranded RNA, provides a basis for the remarkable antigenic variability in the human population through mutation and genetic reassortment. The influenza A virus surface glycoproteins, especially hemagglutinin (HA), are under selective pressure for change in order to evade the host's immune system. Thus, the HA and neuraminidase (NA) genes of influenza A viruses mutate at high frequencies (16, 17), resulting in the accumulation of point mutations that may lead to gradual antigenic changes in surface glycoproteins. This is known as antigenic drift. The matrix protein (M) gene, which encodes two viral proteins, M1 and M2, contributes to the control of virulence and growth (14, 36, 43, 44). Mutation in the M2 gene has been correlated with amantadine resistance (23).
The effectiveness of annually applied trivalent influenza vaccines depends on the selection of component strains that offer optimal immunity from the numerous variants in the global influenza virus circulation. Studies based on sequencing analyses of viruses can be utilized as surveillance tools and can contribute to the vaccine selection process when they are combined with classical serological antigenic analysis (10). Continuous monitoring of viral genetic changes throughout the year is necessary for us to develop our ability to precisely define variation in influenza virus.
In the tropical zone, influenza virus circulates throughout the year, as reported in southern China, Indonesia, and Thailand (2, 18, 20). A laboratory-based influenza virus surveillance system has been in place in Hanoi, Vietnam, since 2001, and we have reported on the presence of influenza virus throughout the year, albeit with summer and winter peaks (21). In order to elucidate the evolutionary patterns for influenza viruses in Vietnam, in this study we undertook a genetic analysis of the influenza A viruses circulating from 2001 to 2006, focusing on two external genes (the HA gene and the NA gene) and one internal regulatory gene (the M gene, which codes for the M1 and the M2 channel proteins).
MATERIALS AND METHODS
Virus collection and isolation.
Nasopharyngeal swabs were obtained from outpatients residing in Hanoi and other provinces in Vietnam with symptoms of influenza-like illness from 2001 to 2006. The samples were placed in viral transport medium and transported to the Virology Department at the National Institute of Hygiene and Epidemiology on the day of collection. One hundred-microliter aliquots of the supernatants of the nasopharyngeal swabs were then inoculated onto Madin-Darby canine kidney cells, prepared in 48-well multiple-well plates. The plates were prepared at 37°C with 5% CO2, and virus growth was monitored at 34°C with reference to cytopathic effects. The viruses were passaged three times to obtain sufficient virus titers for virus identification. All isolates were typed and subtyped by the hemagglutination inhibition assay (8). Selected virus isolates with sufficient titers were transferred for further analysis to the Department of Public Health, Niigata University Graduate School of Medical and Dental Science, for genetic analysis.
RNA extraction and PCR.
One hundred-microliter aliquots of the supernatants after the third culture passage were used for viral RNA extraction with an Extragen II kit (Kainos, Tokyo, Japan), according to the manufacturer's instructions. RNA was transcribed to cDNA with the influenza A virus universal primer Uni12, as described elsewhere (24). The HA genes (segment 4), the NA genes (segment 6), and the M genes (segment 7) of H1N1 and H3N2 viruses were amplified with segment-specific primers as described elsewhere (3, 24).
Nucleotide sequencing and phylogenetic analysis.
The PCR products were purified with a MicroSpin S-300 HR column PCR purification kit (Amersham Bioscience, Buckinghamshire, United Kingdom); labeled by use of a BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, Foster, CA), according to the manufacturer's instructions; and then analyzed on an ABI 3100 automatic DNA sequencer. The sequences were assembled by use of the MEGA (version 3.1) program (26), and multiple-sequence alignment was conducted with the Clustal W program for the major coding regions of the three segments: HA1 (906 bp), NA (1,363 bp), and M (with regions of the overlapping reading frames of M1-M2; 923 bp) for H3N2 isolates and HA1 (829 bp), NA (1,363 bp), and M (938 bp) for H1N1 isolates. Phylogenetic trees were constructed by using a neighbor-joining and bootstrap analysis (n = 1,000) program to determine the best fits for the HA, NA, and M genes. Major branches with bootstrap values of >70% were identified as distinct groups. The phylogenetic grouping was not consistent for the three genes. The genomic sequences of the vaccine strains and the other strains used in this study were obtained from the Influenza Sequences Database (http://www.flu.lanl.gov).
Nucleotide sequence accession numbers.
The nucleotide sequence data from this study were deposited in the DDBJ (DNA Data Bank of Japan), with the accession numbers listed in Table 1.
TABLE 1.
Sequence data of influenza isolates used in this study
| Strain group and strain | Collection date (yr.mo.day) | Accession no. of sequences used in this studya
|
||
|---|---|---|---|---|
| Segment 4 HA gene | Segment 6 NA gene | Segment 7 MP gene | ||
| A/H3N2 | ||||
| A/Hanoi/1872/02 | 2002.01.01 | AB281205 | AB281206 | AB281207 |
| A/Hanoi/184/02 | 2002.06.19 | BAE75900 | AB281193 | AB281194 |
| A/Hanoi/190/02 | 2002.06.23 | AB281195 | AB281196 | AB281197 |
| A/Hanoi/197/02 | 2002.06.26 | BAE75901 | AB281198 | AB281199 |
| A/Hanoi/209/02 | 2002.07.01 | AB281200 | AB281201 | AB281202 |
| A/Hanoi/235/02 | 2002.07.10 | BAE75904 | AB281203 | AB281204 |
| A/Hanoi/695/03 | 2003.08.14 | AB221020 | AB281208 | AB281209 |
| A/Tay Nguyen/TN152/03 | 2003.09.18 | AB221034 | NAb | NA |
| A/Tay Nguyen/TN160/03 | 2003.09.25 | AB221035 | NA | NA |
| A/Hanoi/HN3068/04 | 2004.01.18 | AB281210 | AB281211 | AB281212 |
| A/Hanoi/HN3069/04 | 2004.01.18 | BAE75910 | AB281213 | AB281214 |
| A/Hanoi/HN3094/04 | 2004.01.28 | AB281215 | AB281216 | AB281217 |
| A/Hanoi/HN30109/04 | 2004.01.30 | BAE75911 | AB281218 | AB281219 |
| A/Hanoi/HN30138/04 | 2004.02.04 | AB281223 | AB281224 | AB281225 |
| A/Hanoi/HN30135/04 | 2004.02.04 | AB281220 | AB281221 | AB281222 |
| A/Hanoi/ND049/04 | 2004.02.11 | AB281229 | AB281230 | AB281231 |
| A/Hanoi/HN30188/04 | 2004.02.12 | AB281226 | AB281227 | AB281228 |
| A/Hanoi/BG003/04 | 2004.02.13 | AB284161 | AB284162 | AB284163 |
| A/Hanoi/ISBM63/05 | 2005.05.09 | AB281232 | AB281233 | AB281234 |
| A/Hanoi/ISBM69/05 | 2005.05.09 | AB281235 | AB281236 | AB281237 |
| A/Hanoi/TB285/05 | 2005.05.18 | AB281247 | AB281248 | AB281249 |
| A/Hanoi/HN30607/05 | 2005.05.29 | AB281241 | AB281242 | AB281243 |
| A/Hanoi/HN30602/05 | 2005.05.30 | AB281238 | AB281239 | AB281240 |
| A/Hanoi/TN403/05 | 2005.10.11 | AB281256 | AB281257 | AB281258 |
| A/Hanoi/TN405/05 | 2005.10.13 | AB281259 | AB281260 | AB281261 |
| A/Hanoi/TN406/05 | 2005.10.13 | AB281262 | AB281263 | AB281264 |
| A/Hanoi/HN30720/05 | 2005.06.23 | AB281244 | AB281245 | AB281246 |
| A/Hanoi/TN388/05 | 2005.09.26 | AB281250 | AB281251 | AB281252 |
| A/Hanoi/TN410/05 | 2005.10.17 | AB281265 | AB281266 | AB281267 |
| A/Hanoi/TN399/05 | 2005.10.07 | AB281253 | AB281254 | AB281255 |
| A/H1N1 | ||||
| A/Hanoi/1823/01 | 2001.12.21 | AB285934 | NA | AB285935 |
| A/Hanoi/1863/01 | 2001.12.27 | AB285936 | AB285937 | AB285938 |
| A/Hanoi/1873/02 | 2002.01.01 | AB285939 | AB285940 | AB285941 |
| A/Hanoi/1892/02 | 2002.01.06 | AB285942 | NA | AB285943 |
| A/Hanoi/1928/02 | 2002.01.14 | AB285944 | AB285945 | AB285946 |
| A/Hanoi/2006/02 | 2002.01.23 | AB285947 | AB285948 | AB285949 |
| A/Hanoi/188/02 | 2002.06.21 | AB285950 | NA | AB285951 |
| A/Hanoi/191/02 | 2002.06.23 | AB285952 | AB285953 | AB285954 |
| A/Hanoi/337/03 | 2003.01.26 | AB285955 | NA | AB285956 |
| A/Hanoi/784b/03 | 2003.06.25 | AB285967 | AB285968 | AB285969 |
| A/Hanoi/719/03 | 2003.08.26 | AB285957 | AB285958 | AB285959 |
| A/Hanoi/870b/03 | 2003.09.19 | AB285970 | NA | NA |
| A/Hanoi/777/03 | 2003.09.22 | AB285960 | AB285961 | AB285962 |
| A/Hanoi/893b/03 | 2003.10.10 | AB285971 | NA | NA |
| A/Hanoi/898b/03 | 2003.10.12 | AB285972 | NA | NA |
| A/Hanoi/902b/03 | 2003.10.16 | AB285973 | AB285974 | AB285975 |
| A/Hanoi/859/03 | 2003.10.24 | AB285963 | NA | AB285964 |
| A/Hanoi/910b/03 | 2003.10.27 | AB285976 | NA | NA |
| A/Hanoi/867/03 | 2003.10.28 | AB285965 | NA | AB285966 |
| A/Hanoi/979/03 | 2003.12.09 | AB285981 | NA | NA |
| A/Hanoi/981/03 | 2003.12.09 | AB285982 | NA | NA |
| A/Hanoi/949b/03 | 2003.12.10 | AB285977 | NA | NA |
| A/Hanoi/1004/03 | 2003.12.17 | AB285983 | AB285984 | AB285985 |
| A/Hanoi/1007/03 | 2003.12.18 | AB285986 | NA | NA |
| A/Hanoi/959b/03 | 2003.12.19 | AB285978 | AB285979 | AB285980 |
| A/Hanoi/1024/03 | 2003.12.24 | AB285987 | NA | NA |
| A/Hanoi/ISBM15/05 | 2005.03.24 | AB285988 | AB285989 | AB285990 |
| A/Hanoi/ISBM23/05 | 2005.04.01 | AB285991 | AB285992 | AB285993 |
| A/Hanoi/ISBM27/05 | 2005.04.05 | AB285994 | AB285995 | AB285996 |
| A/Hanoi/ISBM31/05 | 2005.04.08 | AB285997 | AB285998 | AB285999 |
| A/Hanoi/Q137/06 | 2006.03.07 | AB286000 | AB286001 | AB286002 |
| A/Hanoi/Q177/06 | 2006.03.15 | AB286003 | AB286004 | AB286005 |
| A/Hanoi/BM344/06 | 2006.05.11 | AB286006 | AB286007 | AB286008 |
| A/Hanoi/BM356/06 | 2006.05.19 | AB286009 | AB286010 | AB286011 |
| A/Hanoi/TX09/06 | 2006.06.08 | AB286012 | AB286013 | AB286014 |
All accession numbers are for the DNA Data Bank of Japan.
NA, not addressed.
RESULTS
Thirty influenza A/H3N2 viruses that were collected from 2002 to 2005 and 35 influenza A/H1N1 viruses from 2001 to 2006 were used in this study. Samples were collected from various regions of Vietnam. No influenza A/H1N2 viruses were found during the study period.
A/H3N2 influenza virus.
For analysis of the sequences of the HA, NA, and M genes of the influenza A/H3N2 viruses, we used the consensus sequence of A/Moscow/10/99 as the phylogenetic root for the HA gene. Other isolates were also used as reference strains for each year's epidemic.
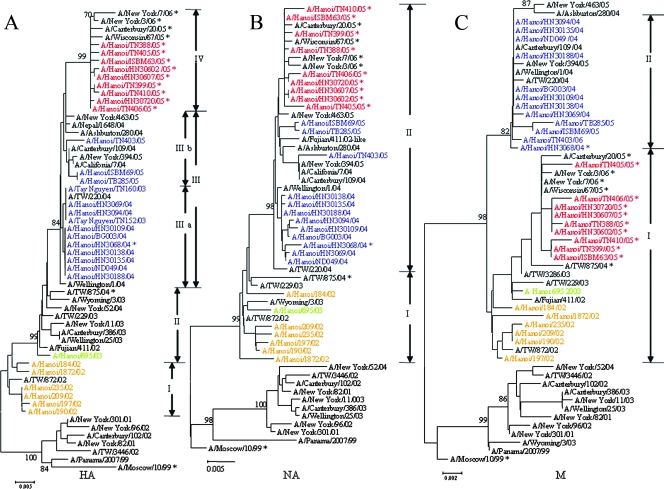
In the phylogenetic tree of the HA genes of the A/H3N2 isolates, the viruses formed a monophyletic group which could be divided into four major subgroups with a bootstrap value of >70% (Fig. 1A). These four groups were concurrent with the influenza seasons: group I in 2002; group II in 2003; group III in 2003, 2004, and 2005; and group IV in 2005. Six isolates circulating during the 2002 season in Hanoi were in positions between the A/Panama/2007/99 and the A/Fujian/411/02 groups of viruses and were clustered in group I. One 2003 isolate and the vaccine strain (A/Fujian/411/02) were clustered in group II. Two isolates circulating in 2003, nine isolates circulating in 2004, and strain A/Wellington/1/04 were classified as group IIIa; and three isolates circulating in 2005 and strain A/California/7/04 were classified as group IIIb. The remaining nine isolates circulating in 2005 and the Northern Hemisphere vaccine strain A/Wisconsin/67/05 clustered in group IV.
FIG. 1.
Phylogenetic analysis of the HA1 domain of HA gene nucleotide sequences (906 bp), NA gene nucleotide sequences (1,363 bp), and M gene (with regions of the overlapping reading frames of M1-M2) nucleotide sequences (923 bp) of influenza A/H3N2 viruses circulating in Vietnam from 2002 to 2005. Reference strains shown in black were obtained from the genetic database. The isolates were assigned to group I (orange), group II (green), group III (purple), or group IV (red) in the phylogenetic tree of the HA gene (A). The grouping of the NA and the M genes was made in accordance with the branching in each tree. The colors of the strains in the phylogenetic trees of the NA gene (B) and the M gene (C) comply with those for the HA gene. Sequence data for the reference strains were obtained from the GenBank database. The asterisks denote the amantadine-resistant strain with the amino acid change S31N in the M2 protein. Bootstrap values of >70% are shown for the main groups.
Because the NA gene for strain Fujian/411/02 was not available from the database, one for a Fujian-like strain was chosen as a reference in the NA gene segment analysis. Analysis of the NA gene in this study showed that the viruses circulating from 2002 to 2005 were divided into two groups (Fig. 1B). Six 2002 isolates and one 2003 isolate clustered in group I (group I and group II for HA). The remaining isolates circulating in 2004 and 2005, an A/Fujian/411/02-like strain and strain A/Wisconsin/67/05, clustered in group II (group III and group IV for HA).
In the phylogenetic tree of the M gene (with regions of the overlapping reading frames of M1-M2), the viruses were divided into two groups. Isolates circulating in 2002, 2003, and 2005 were clustered in group I (group I, group II, and group IV for HA) (Fig. 1C). Nine isolates circulating in 2004 and three isolates circulating in 2005 were clustered in group II (group III for HA).
The amino acid sequences encoded by the HA (HA1 subunit) genes are shown in Table 2. For group I, all six isolates had amino acid changes, at positions S21P, R50G, E83K, N145K, S186G, V202I, W222R, and G225D, in comparison with the sequence of A/Panama/2007/99 strain. Four of the six isolates had additional double changes at positions N144D and G275D, and two of the six had an additional amino acid difference at position A131T. The single isolate circulating in 2003 and some other reference strains circulating in the same year were in group II and had four additional amino acid changes, L25I, H75Q, H155T, and Q156H, compared to the sequence of the isolates in group I. Among the isolates in group IIIa, 11 had additional changes at Y159F, S189N, and S227P and 4 of the 11 had another change at V226I, while among the isolates in group IIIb, three isolates circulating in 2005 further changed at K145N. All the remaining nine isolates in 2005, which were in group IV, had double amino acid changes at S193F and D225N. All isolates in this group were amantadine resistant and had the S31N amino acid change in the M2 gene.
TABLE 2.
Comparison of amino acid sequences for the HA1 subunit of H3N2 viruses collected from 2002 to 2005
| Group | Strain | Substitution at the following amino acid residue in HA1a:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 25 | 50d | 75 | 83 | 131 | 144 | 145 | 155 | 156 | 159 | 186 | 189 | 193 | 202 | 222 | 225 | 226 | 227 | 275 | ||
| A/Panama/2007/99b | S | L | R | H | E | A | N | N | H | Q | Y | S | S | S | V | W | G | V | S | G | |
| I | A/Hanoi/197/02 | P | —c | G | — | K | — | D | K | — | — | — | G | — | — | I | R | D | — | — | D |
| I | A/Hanoi/1872/02 | P | — | G | — | K | T | — | K | — | — | — | G | — | — | I | R | D | — | — | — |
| II | A/Fujian/411/02 | P | I | G | Q | K | T | — | K | T | H | — | G | — | — | I | R | D | — | — | — |
| II | A/Hanoi/695/03 | P | I | G | Q | K | T | — | K | T | H | — | G | — | — | I | R | D | — | — | — |
| IIIa | A/Wellington/7/04 | P | I | G | Q | K | T | — | K | T | H | F | G | N | — | I | R | D | — | P | — |
| IIIa | A/Tay Nguyen/TN152/03 | P | I | G | Q | K | T | — | K | T | H | F | G | N | — | I | R | D | — | P | — |
| IIIa | A/Hanoi/HN3069/04 | P | I | G | Q | K | T | — | K | T | H | F | G | N | — | I | R | D | — | P | — |
| IIIa | A/Hanoi/HN3068/04 | P | I | G | Q | K | T | — | K | T | H | F | G | N | — | I | R | D | I | P | — |
| IIIb | A/Califonia/7/04 | P | I | G | Q | K | T | — | — | T | H | F | G | N | — | I | R | D | I | P | — |
| IIIb | A/Hanoi/ISBM69/05 | P | I | G | Q | K | T | — | — | T | H | F | G | N | — | T | R | D | I | P | — |
| IV | A/Wisconsin/67/05 | P | I | G | Q | K | T | — | — | T | H | F | G | N | F | I | R | N | I | P | — |
| IV | A/Hanoi/TN405/05 | P | I | G | Q | K | T | — | — | T | H | F | G | N | F | I | R | N | I | P | — |
| IV | A/Hanoi/ISBM63/05 | P | I | G | Q | K | T | — | — | T | H | F | G | N | F | I | R | N | I | P | — |
Results are reported as amino acid differences between the sequences of the isolates and the sequence of the A/Panama/2007/99 vaccine strain.
Italic type indicates the reference vaccine strain.
—, no change.
Differences located in the proposed antigenic sites are underlined.
A/H1N1 influenza virus.
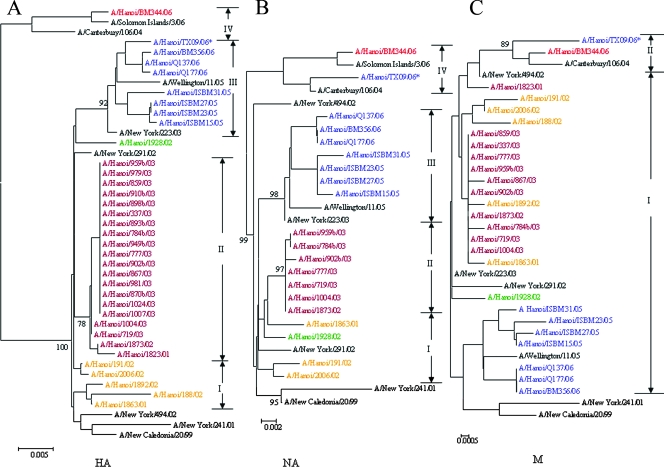
The phylogenetic tree for the HA genes of the A/H1N1 strains, including 35 isolates from 2001 to 2006 and 7 reference strains, showed four groupings (Fig. 2A). The isolates collected in 2004 were not included in our analysis because there was no collection of A/H1N1 viruses at the National Institute of Hygiene and Epidemiology. Among the samples, one isolate collected in 2001 and four isolates collected in 2002 were clustered in group I. The other 2 isolates from 2001 and 2002 and 18 isolates from 2003 clustered in group II. One isolate, A/Hanoi/1928/02, was localized between groups II and III. With the exception of one A/Hanoi/BM344/06 isolate, all samples from 2005 and 2006 clustered in group III. One A/Hanoi/BM344/06 isolate and the A/Solomon Islands/3/06 isolate, which is recommended as an influenza vaccine component in the 2007-2008 season in the Northern Hemisphere, were clustered in group IV, with a high bootstrap value of 100%.
FIG. 2.
Phylogenetic analysis of the HA1 domain of HA gene nucleotide sequences (829 bp), NA gene nucleotide sequences (1,363 bp), and M gene (with regions of the overlapping reading frames of M1-M2) nucleotide sequences (938 bp) of influenza A/H1N1 viruses circulating in Vietnam from 2001 to 2006. Reference strains shown in black were obtained from the genetic database. The isolates were assigned to group I (orange), group II (dark red), group III (purple), or group IV (red) in the phylogenetic tree of HA gene (Fig. 1A). The grouping of the NA and M genes was made in accordance with the branching in each tree. The colors of the strains in the phylogenetic trees of the NA gene (B) and the M gene (C) comply with those for the HA gene. The asterisks denote the amantadine-resistant strain with the amino acid change S31N in the M2 protein. Bootstrap values of >70% are shown for the main groups.
Of the total 35 isolates, the NA genes of 20 isolates and the M genes of 26 isolates were examined. In the phylogenetic analysis of the NA genes, the isolates were divided into four groups (Fig. 2B). Intermediate strain A/Hanoi/1928/02 (in the position between groups II and III in the HA phylogenetic tree), two isolates from 2002, and one isolate from 2001 (group I for HA) were clustered in group I. Seven isolates from 2003 were clustered in group II (group II for HA). The isolates from 2005 and 2006 were divided into two groups: group III (group III for HA) and group IV. Isolate A/Hanoi/BM344/06 and isolate A/Hanoi/TX09/06, together with isolate A/Solomon Islands/3/06, which clustered in group III in the HA gene phylogenetic tree, were classified in group IV from the NA analysis.
In the phylogenetic analysis of the M genes (with regions of the overlapping reading frames of M1-M2) of the viruses, all except two isolates from 2001 to 2006 clustered in group I (Fig. 2C); isolates A/Hanoi/BM344/06 and A/Hanoi/TX09/06 clustered in group II with a bootstrap value of 89%.
In comparison to vaccine strain A/New Caledonia/20/99, the HA genes of the A/H1N1 isolates in groups I, II, and III from 2002 to 2006 had mutations at positions 158, 169, 190, 255, and 256. In particular, two amino acid changes at V169A and W255R consistently occurred in the samples (Table 3). At position 158, isolates in group II from 2002 and 2003 demonstrated an N158S change, while an isolate in group III from 2005 had a change at N158K. At position 190, isolates in groups I, II, and III demonstrated N190D and N190V changes. At position 256, isolates in group III from 2005 and 2006 featured Y256F. Isolate A/Hanoi/BM344/06 of group IV, on the other hand, exhibited 12 amino acid changes in comparison with the sequence of A/New Calidonia/20/99; these were E72D, K77R, T86K, Y98H, V132T, K144T, R149K, V169A, V190D, R212K, W255R, and T270N.
TABLE 3.
Comparison of amino acid sequences for the HA1 subunit of H1N1 isolates collected from 2002 to 2006
| Group | Strain | Substitution at the following amino acid residue in HA1 (H3 numbering)a:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 77 | 81d | 90 | 101 | 133 | 144 | 149 | 158 | 169 | 190 | 197 | 212 | 255 | 256 | 270 | ||
| A/New Caledonia/20/99b | E | K | T | Y | V | K | R | N | V | N | T | R | W | Y | T | |
| I | A/Hanoi/1892/02 | —c | — | — | — | — | — | — | — | A | D | — | — | R | — | — |
| I | A/Hanoi/1863/02 | — | — | — | — | — | — | — | — | A | — | — | — | R | — | — |
| II | II A/Hanoi/1873/02 | — | — | — | — | — | — | — | S | A | — | — | — | R | — | — |
| II | A/Hanoi/719/03 | — | — | — | — | — | — | — | S | A | D | — | — | R | — | — |
| II | A/Hanoi/784b/03 | — | — | — | — | — | — | — | S | A | V | — | — | R | — | — |
| III | III A/Hanoi/ISBM15/05 | — | — | — | — | — | — | — | — | A | — | — | — | R | F | — |
| III | A/Hanoi/TX09/06 | — | — | — | — | — | — | — | — | A | D | — | — | R | F | — |
| III | A/Hanoi/Q177/06 | — | — | — | — | — | — | — | K | A | D | — | — | R | F | — |
| IV | IV A/Hanoi/BM344/06 | D | R | K | H | T | E | K | — | A | D | — | K | R | — | N |
| IV | A/Solomon Islands/3/06 | — | R | K | H | T | E | K | — | A | D | K | K | R | — | N |
Results are reported as amino acid differences between the sequences of the isolates and that of the A/New Caledonia/20/99 vaccine strain.
Italic type indicates the reference vaccine strain.
—, no change.
Differences located in the proposed antigenic sites are underlined.
DISCUSSION
Influenza A viruses have segmented genomes consisting of eight RNA segments which encode viral proteins. Because of immunological pressure, genetic variability is mostly confined to regions of the genome responsible for viral surface proteins (33). However, other factors, including interactions of internal and surface proteins, are likely to affect viral fitness in a polygenic manner (25). Our present genetic analysis of A/H3N2 and A/H1N1 viruses isolated in Vietnam from 2001 to 2006 revealed various topologies for the phylogenetic trees of the HA and the NA genes, which encode two external proteins, and the M gene, which encodes an internal protein, indicating that genetic evolution did not occur at the same rate.
During the 5 years, influenza A/H3N2 virus evolved from a strain intermediate between A/Panama/2007/99 and A/Fujian/411/02 to an A/Wiscosin/67/05-like strain. The HA and NA genes evolved independently in a sequential way, whereas the M gene demonstrated a nonlinear pattern not linked to the evolution of the HA and NA genes (29). Furthermore, the HA gene could be divided into four groups with shifts from groups I to IV. The NA and M genes were divided into two groups, but the grouping of the M genes was not in accordance with that of the NA or the HA gene. The results also suggest that the HA gene evolved more rapidly than both the NA and the M genes and that these three genes of influenza A/H3N2 virus change in a nonsynchronous manner.
The genetic drift of the HA genes of the influenza A/H3N2 viruses, located at positions believed to have functional or antigenic significance, was found to have occurred during all epidemic periods. The HA genes of viruses isolated in 2002 were clustered between strains A/Panama/2007/99 and A/Fujian/411/2002 in the phylogeny, with some strains having five amino acid changes within the antigenic sites (38, 39), namely, R50G (site C), E83K (site E), A131T (site A), N144K (site A), and S186G (site D). The same variant began to circulate in other parts of Asia between the end of 2001 and the beginning of 2002 (9). One isolate that was collected in August 2003 showed two key amino acid changes at residues H155T and Q156T (site B). This result indicated invasion of the A/Fujian/411/02 like strain into Vietnam, which was roughly coincident with our antigenic findings from influenza virus surveys in Hanoi (21). Between September 2003 and February 2004, the predominant strain circulating in Asia, the Americas, and Europe was an A/Fujian/411/02-like strain (40). However, two Vietnamese isolates circulating in September 2003 had drifted from the A/Fujian/411/02-like strain and showed additional changes at residues 159 (site B), 189 (site B), and 227 (site D). They had a higher degree of genetic homology to strain A/Wellington/1/04, which was isolated in New Zealand in January 2004 and which also circulated in Vietnam in the winter season of 2003-2004. Moreover, strain A/California/7/04 like, characterized by further changes at residues 145 (site A) and 226 (11, 30, 31), was isolated in Vietnam in May 2005 in this study.
High proportions of amantadine-resistant influenza A/H3N2 viruses with specific amino acid changes at residues 193 and 225 (named clade N) were reported from the 2005-2006 season in Japan and the United States (4, 12, 34, 35). Residue 193 is located within antigenic site B (37), and residue 225 is located within the receptor-binding site. In this study, clade N viruses were found from April to May of 2005 in Vietnam and the surrounding areas (1). However, it was in September and October of 2005 and the subsequent winter in Japan and North America when the strain caused a large community outbreak (5, 12, 34, 35).
The HA genes of the A/H1N1 isolates in groups I, II, and III showed only five amino acid changes from the sequence of the A/New Caledonia/20/99 vaccine strain. The HA1 subunit includes the globular head and contains five major antibody-binding sites, namely, Sa, Sb, Ca1, Ca2, and Cb (7). A substitution at residue 169 (V169A) occurred in the Ca1 antigenic site in all isolates, while substitution at residue 158, located in the Sb site, changed irregularly with the strain. The WHO Influenza Center reported that the majority of H1N1 viruses circulating in 2005 were antigenically closely related to current vaccine strain A/New Caledonia/20/99 (22). One of the strains analyzed at the WHO center, A/Virginia/4/05 (22), is antigenically related to strain A/New Caledonia/20/99 and grouped together with our isolates in group III that were circulating in Vietnam in 2005 and 2006. Therefore, we conclude that the most of the H1N1 viruses circulated in Vietnam from 2001 to 2006 had antigenicity similar to that of strain A/New Caledonia/20/99 and underwent changes more slowly than the influenza A/H3N2 virus subtype, as reported elsewhere (15).
An A/Hanoi/BM344/06 strain collected in May 2006 showed 12 additional amino acid changes in the HA gene in comparison with the sequence of strain A/New Caledonia/20/99. Two of the 12 substitutions, K81R and V169A, were located in antigenic sites Cb and Ca1, respectively. In addition, substitutions at V133T and K144E were located in antibody-binding sites (38). Also, strain A/Solomon Islands/3/06, which was isolated in August 2006 and which was later recommended by WHO as the vaccine strain for the 2007-2008 season (41), clustered with strain A/Hanoi/BM344/06 in the phylogenetic tree of the HA gene. Therefore, it is likely that we obtained an early insight into the arrival of this novel strain of influenza A/H1N1 virus.
In general, reassortment events are reported more often in A/H3N2 viruses than in A/H1N1 viruses (25). Our analysis indicated that reassortment occurred for both H1N1 and H3N2 during this 6-year period. Isolate A/Hanoi/184/02 (H3N2) contained an HA gene intermediate between those of A/Panama/2007/99 and A/Fujian/411/02 and an NA gene similar to that of A/Wyoming/3/03, whereas the M gene proved similar to that of A/Fujian/411/02. For H1N1, isolate A/Hanoi/TX09/06 (H1N1) represents the result of a rarely reported reassortment. The HA gene of the virus was closely related to those in group III, which were antigenically related to the A/New Caledonia/20/99-like strain, but the NA and M genes were closely related to the A/Solomon Islands/3/06 strain. An intersubtype reassortant, A/H1N2, was isolated from Japan as well as from many other areas (13, 19, 32, 42) after 2001, but this was not detected in our present analysis.
Continuous monitoring of viral genetic changes throughout the year is warranted to monitor the variations of influenza viruses. As influenza viruses circulate throughout the year in the tropical and subtropical zones (18, 20), the results for viruses from those areas are very suitable for monitoring purposes. This applies especially in Asia, where the emergence of new strains may be detected earlier than the emergence of new strains in temperate zones, where there is only a single peak of activity in each year.
Acknowledgments
We thank all the staff in the Respiratory Virus Section of the Virology Department, National Institute of Hygiene and Epidemiology, Hanoi, Vietnam; Akemi Watanabe and the staff in the Department of Public Health, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan; Akinori Miyashita and Ryozo Kuwano, Department of Molecular Genetics, Brain Research Institute, Niigata University; and the staff of the Virology Section in Niigata Prefectural Institute of Public Health and Environmental Sciences, Niigata, Japan. We also express our appreciation to the Vietnamese medical staff who supported this investigation at health care facilities. We thank Clyde Dapat for editing of the manuscript.
We declare that none of the authors have any conflict of interest.
This work was supported by Grants-in-Aid for Scientific Research from Monbu Kagakusho (Ministry of Education, Culture, Sports, Science and Technology, Japan) and by the Acute Respiratory Infections Panels, United States-Japan Cooperative Medical Science Program (which is supported by the U.S. Department of Health and Human Services and the U.S. Department of State in the United States and the Ministry of Foreign Affairs; the Ministry of Health, Labor, and Welfare; and the Ministry of Education, Culture, Sports, Science and Technology in Japan).
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Barr, I. G., A. C. Hurt, P. Iannello, C. Tomasov, N. Deed, and N. Komadina. 2007. Increased amantadine resistance in influenza A (H3) viruses in Australia and neighboring countries in 2005. Antivir. Res. 73112-117. [DOI] [PubMed] [Google Scholar]
- 2.Beckett, C. G., H. Kosasih, C. Ma'roef, E. Listiyaningsih, I. R. Q. Elyazar, S. Wuryadi, D. Yuwono, J. L. McArdle, A. L. Corwin, and K. R. Porter. 2004. Influenza surveillance in Indonesia: 1999-2003. Clin. Infect. Dis. 39443-449. [DOI] [PubMed] [Google Scholar]
- 3.Besselaar, T. G., L. Botha, J. M. McAnerney, and B. D. Schoub. 2004. Antigenic and molecular analysis of influenza A (H3N2) virus strain isolated from a localized influenza outbreak in South Africa in 2003. J. Med. Virol. 7371-78. [DOI] [PubMed] [Google Scholar]
- 4.Bright, R. A., M. J. Medina, X. Xu, G. P. Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 3661175-1181. [DOI] [PubMed] [Google Scholar]
- 5.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295891-894. [DOI] [PubMed] [Google Scholar]
- 6.Buonagurio, D. A., S. Nakada, J. D. Parvin, M. Krystal, P. Palese, and W. M. Fitch. 1986. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science 232980-982. [DOI] [PubMed] [Google Scholar]
- 7.Caton, A., G. G. Brownlee, J. W. Yewell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31417-427. [DOI] [PubMed] [Google Scholar]
- 8.Center for Disease Control. 1982. Concepts and procedures for laboratory based influenza surveillance. Center for Disease Control, Atlanta, GA.
- 9.Chi, X. S., T. V. Bolar, P. Zhao, J. S. Tam, R. Rappaport, and S. M. Cheng. 2005. Molecular evolution of human influenza A/H3N2 virus in Asia and Europe from 2001 to 2003. J. Clin. Microbiol. 436130-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, N. J., Z. S. Bai, and A. P. Kendal. 1983. Laboratory-based surveillance of influenza A (H1N1) and A (H3N2) viruses in 1980-81: antigenic and genomic analyses. Bull. W. H. O. 61143-152. [PMC free article] [PubMed] [Google Scholar]
- 11.Daum, L. T., M. W. A. Shaw, I. Klimov, L. C. Canas, E. A. Macias, D. Niemeyer, J. P. Chambers, R. Renthal, S. K. Shrestha, R. P. Acharya, S. P. Huzdar, N. Rimal, K. S. Myint, and P. Gould. 2005. Influenza A (H3N2) outbreak, Nepal. Emerg. Infect. Dis. 111186-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deyde, V. M., X. Xu, R. A. Bright, M. Shaw, C. B. Smith, Y. Zhang, Y. Shu, L. V. Gubareva, N. J. Cox, and A. I. Klimov. 2007. Surveillance of resistance to adamantanes among influenza A (H3N2) and A (H1N1) viruses isolated worldwide. J. Infect. Dis. 196249-257. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, J. S., A. Alvarez-Aguero, V. Gregory, Y. P. Lin, A. Hay, and M. C. Zambon. 2003. Influenza A (H1N2) viruses and their impact during the 2001-02 influenza season in the United Kingdom. Emerg. Infect. Dis. 9304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami, K., Y. Qiao, R. Fukuda, and M. Enami. 1993. An influenza virus temperature-sensitive mutant defective in the nuclear-cytoplasmic transport of the negative-sense viral RNAs. Virology 194822-827. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, N. M., A. P. Galvani, and R. M. Bush. 2003. Ecological and immunological determinants of influenza evolution. Nature 422428-433. [DOI] [PubMed] [Google Scholar]
- 16.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long term trends in the evolution of H (3) HA1 human influenza type A. Proc. Natl. Acad. Sci. USA 947712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitch, W. M., R. M. Bush, C. A. Bender, K. Subbarao, and N. J. Cox. 2000. Predicting the evolution of human influenza A. J. Hered. 91183-185. [DOI] [PubMed] [Google Scholar]
- 18.Gachara, G., J. Ngeranwa, J. M. Magana, J. M. Simwa, P. W. Wango, S. M. Lifumo, and E. O. Ochieng. 2006. Influenza virus strains in Nairobi, Kenya. J. Clin. Virol. 35117-118. [DOI] [PubMed] [Google Scholar]
- 19.Gregory, V., M. Bennett, M. H. Orkhan, A. I. Hajjar, N. Varsano, E. Mendelson, M. Zambon, J. Ellis, A. Hay, and Y. P. Lin. 2002. Emergence of influenza A H1N2 reassortant viruses in the human population during 2001. Virology 3001-7. [DOI] [PubMed] [Google Scholar]
- 20.Hampson, A. W. 1999. Epidemiological data on influenza in Asian countries. Vaccine 17(Suppl. 1)S19-S23. [DOI] [PubMed] [Google Scholar]
- 21.Hang, L. K. N., R. Saito, K. N. Ha, M. Nishikawa, Y. Shobugawa, C. N. Doan, T. H. Long, P. H. Lien, and H. Suzuki. 2007. Epidemiology of influenza in Hanoi, Vietnam, from 2001 to 2003. J. Infect. 5558-63. [DOI] [PubMed] [Google Scholar]
- 22.Hay, A. J., and Y. Lin. 2005. Characteristics of human influenza A H1N1, A H3N2 and B viruses isolated February to July 2005. WHO Influenza Centre, London, United Kingdom.
- 23.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 43021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, E. C., E. Ghedin, N. Miller, J. Taylor, Y. Bao, K. S. George, B. T. Grenfell, S. L. Salzberg, C. M. Fraster, D. J. Lipman, and J. K. Taubenberger. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 31579-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 27.Li, C. K., B. C. Choi, and T. W. Wong. 2006. Influenza related deaths and hospitalizations in Hong Kong: a subtropical area. Public Health 120517-524. [DOI] [PubMed] [Google Scholar]
- 28.Lin, Y. P., V. Gregory, M. Bennett, and A. Hay. 2004. Recent changes among human influenza viruses. Virus Res. 10347-52. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom, S. E., Y. Hiromoto, R. Nerome, K. Omoe, S. Sugita, Y. Yamazaki, T. Takahashi, and K. Nerome. 1998. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J. Virol. 728021-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza IV. Elsevier Science, Amsterdam, The Netherlands.
- 31.Mar, A., R. Rahmanian, V. Lei, D. Lawrence, M. Krajden, R. C. Brunham, D. Skowronski, Y. Li, T. Booth, S. H. Goh, and M. Petric. 2006. Longitudinal analysis of genotype distribution of influenza A virus from 2003 to 2005. J. Clin. Microbiol. 443583-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishikawa, F., and T. Sugiyama. 1983. Direct isolation of H1N2 recombinant virus from a throat swab of a patient simultaneously infected with H1N1 and H3N2 influenza A viruses. J. Clin. Microbiol. 18425-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 Spanish influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 976785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito, R., D. Li, C. Shimomura, H. Masaki, Q. E. Mai, L. K. N. Hang, T. N. Hien, T. N. Tien, V. P. Tu, T. K. N. Tien, M. Sato, Y. Suzuki, and Y. Suzuki. 2006. An off-seasonal amantadine resistant H3N2 influenza outbreak in Japan. Tohoku J. Exp. Med. 21021-27. [DOI] [PubMed] [Google Scholar]
- 35.Saito, R., D. Li, and H. Suzuki. 2007. Amantadine-resistant influenza A (H3N2) virus in Japan, 2005-2006. N. Engl. J. Med. 356312-313. [DOI] [PubMed] [Google Scholar]
- 36.Smeenk, C. A., and E. G. Brown. 1994. The influenza virus variant A/FM/1/47-MA possesses single amino acid replacements in the hemagglutinin, controlling virulence, and in the matrix protein, controlling virulence as well as growth. J. Virol. 68530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, D. J., A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305371-376. [DOI] [PubMed] [Google Scholar]
- 38.Wiley, D. C., I. A. Wilson, and J. J. Skehel. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza hemagglutinin and their involvement in antigenic variation. Nature 289373-378. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, I. A., and N. J. Cox. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8737-771. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization. 2004. Recommended composition of influenza virus vaccines for use in the 2004-2005 influenza season. Wkly. Epidemiol. Rec. 7988-92. [PubMed] [Google Scholar]
- 41.World Health Organization. 2007. Recommended composition of influenza virus vaccines for use in the 2007-2008 influenza season. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/influenza/vaccinerecommendations/en/index.html.
- 42.Xu, X., C. B. Smith, B. A. Mungall, S. E. Lindstrom, H. E. Hall, K. Subbarao, N. J. Cox, and A. Klimov. 2002. Intercontinental circulation of human influenza A (H1N2) reassortant viruses during the 2001-2002 influenza season. J. Infect. Dis. 186490-493. [DOI] [PubMed] [Google Scholar]
- 43.Yasuda, J., D. J. Bucher, and A. Ishihama. 1994. Growth control of influenza A virus by M1 protein: analysis of transfectant viruses carrying the chimeric M gene. J. Virol. 688141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuda, J., T. Toyoda, M. Nakayama, and A. Ishihama. 1993. Regulatory effects of matrix protein variations on influenza virus growth. Arch. Virol. 133283-294. [DOI] [PubMed] [Google Scholar]