Abstract
RNA silencing can function as a virus defense mechanism in a diverse range of eukaryotes, and many viruses are capable of suppressing the silencing machinery targeting them. However, the extent to which this occurs between fungal RNA silencing and mycoviruses is unclear. Here, three Aspergillus dsRNA mycoviruses were partially characterized, and their relationship to RNA silencing was investigated. Aspergillus virus 1816 is related to Agaricus bisporus white button mushroom virus 1 and suppresses RNA silencing through a mechanism that alters the level of small interfering RNA. Aspergillus virus 178 is related to RNA virus L1 of Gremmeniella abietina and does not appear to affect RNA silencing. The third virus investigated, Aspergillus virus 341, is distantly related to Sphaeropsis sapinea RNA virus 2. Detection of mycovirus-derived siRNA from this mycovirus demonstrates that it is targeted for degradation by the Aspergillus RNA silencing machinery. Thus, our results indicate that Aspergillus mycoviruses are both targets and suppressors of RNA silencing. In addition, they suggest that the morphological and physiological changes associated with some mycoviruses could be a result of their antagonistic relationship with RNA silencing.
Mycoviruses are nearly ubiquitous in fungi (2). Although most cause infections that appear innocuous, there are some well-known examples that dramatically alter the phenotype of their host (18). For example, a mycovirus of the tree pathogen Cryphonectria parasitica causes hypovirulence (23), a mycovirus of the edible mushroom Agaricus bisporus causes slow and aberrant growth (20), and a mycovirus of the endophyte Curvularia protuberata allows for the survival of the endophyte and the endophyte's host in geothermal soils (15).
The molecular mechanisms of these and other mycovirus-induced phenotypes are poorly understood. It is possible that at least some are due to host RNA silencing. (For a review, see reference 4.) During RNA silencing a protein called Dicer processes long double-stranded RNA (dsRNA) into small RNA, sometimes referred to as small interfering RNA (siRNA). siRNA is incorporated into an Argonaute-containing effector complex, which can target complementary mRNA for cleavage or translational inhibition. Mycoviruses could thus induce phenotypic change via RNA silencing in a number of ways. For example, if host RNA silencing is involved in gene regulation, mycovirus-based suppression of RNA silencing could interfere with this process. In addition, normal gene expression could be disrupted if there is sufficient complementarity between a mycovirus-derived siRNA and a host gene, resulting in the RNA silencing machinery targeting both host and viral transcripts (31).
A role for RNA silencing in antiviral defense has been well documented for plant viruses and some animal viruses (1, 13, 14, 32), but the only available data concerning mycoviruses and RNA silencing come from studies with C. parasitica. In these studies a C. parasticia Dicer mutant was found to be severely debilitated compared to the wild type when infected with Cryphonectria hypovirus 1 (CHV1) (22), suggesting that RNA silencing normally attenuates CHV1 infections. In addition, an earlier report linked a CHV1 protein to RNA silencing suppression (21). These results suggest that C. parasitica RNA silencing is involved in virus defense.
The main components of RNA silencing—Dicers, Argonautes, and RNA-dependent RNA polymerases (RdRPs)—were previously characterized in the filamentous fungus Aspergillus nidulans (10a, 11). Essentially, A. nidulans was found to have the smallest number of RNA silencing genes among filamentous ascomycetes, none of which were required for growth or developmental processes (10a, 11). Here, an Aspergillus mycovirus is shown to suppress A. nidulans RNA silencing, and another is shown to be processed into siRNA by the A. nidulans RNA silencing machinery. The latter finding is a first for mycovirus research. Thus, two lines of evidence are provided to support mycovirus defense as a role for RNA silencing in A. nidulans.
MATERIALS AND METHODS
Strains and culture conditions.
The A. nidulans strains used in the present study are listed in Table 1. All strains were grown on appropriately supplemented glucose minimal medium (GMM) (24) at 29°C. The mycoviruses were originally identified in A. niger and were transferred into A. nidulans strain 701 via protoplast fusion (30). In the present study, mycovirus-carrying 701 derivatives were used as the initial mycovirus donor strains (Table 1). The mycoviruses were given abbreviated names according to the host strain from which they were originally isolated: virus 178 was from A. niger Ind1.7.8, virus 1816 was from A. niger Ind1.8.16, and virus 341 was from A. niger 341 (30).
TABLE 1.
A. nidulans strains used in this study
| Strain(s)a (source reference) | Codeb | Description or genotype |
|---|---|---|
| 701/8 (30) | - | Nitrate nonutilizing; virus 341 |
| 701/21 (30) | - | Nitrate nonutilizing; virus 1816 |
| 701/23 (30) | - | Nitrate nonutilizing; virus 178 |
| WIM126 (33) | - | pabaA1 yA2 |
| VWIM126A | - | pabaA1 yA2; virus 341 |
| VTMH1 | - | pabaA1 yA2; virus 1816 |
| VTMH3 | - | pabaA1 yA2; virus 178 |
| RTMH13.B3 (11), -VTC | a, b | ΔstcE::argB; veA1 |
| VMDA1-4, -5 | c, d | ΔstcE::argB; veA1; virus 178 |
| VMDA5-1, -2 | e, f | ΔstcE::argB; veA1; virus 1816 |
| VMDA7-1, -2 | g, h | ΔstcE::argB; veA1; virus 341 |
| RTMH13.B1 (11), -VTC | i, j | ΔstcE::argB; aflR(IRT)::trpC veA1 |
| VMDA2-12, -37 | k, l | ΔstcE::argB; aflR(IRT)::trpC veA1; virus 178 |
| VMDA6-1, -2 | m, n | ΔstcE::argB; aflR(IRT)::trpC veA1; virus 1816 |
| RTMH13.B1-G, -H | o, p | ΔstcE::argB; aflR(IRT)::trpC veA1; unstable virus 341 |
| FGSC A773 (16) | - | pyrG89; wA3; pyroA4; veA1 |
| RTMH207.37 (10a) | - | argB2 |
| TTMH190.13 | q | pyrG89; wA3; v341(IRT)::pyroA; veA1 |
| RTMH13.C7 | r | pyrG89; wA3; veA1 |
| VTMH13.C7-E | s | pyrG89; wA3; veA1; virus 341 |
| RDIT9.32 (27) | - | WT |
| VDIT9.32-1 | y | WT; virus 341 |
| RTMH211.14 (10a) | w | WT |
| VTMH211.14-5 | x | WT; virus 341 (lacking short dsRNA band) |
| RTMH218.7 (10a) | aa | ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG |
| VTMH218.7-1 | z | ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG; virus 341 |
| RTMH218.39 (10a) | - | ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG |
| VTMH218.39-1 | u | ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG; virus 341 |
| RTMH242.11 | t | v341(IRT)::pyroA |
| VTMH218.36-9 | v | ΔrsdA::pyrG; virus-341 |
Most strains in this study were assigned a lowercase letter code for reference purposes (Fig. 2 to 5). Strain names that differ by the numbers/letters following their hyphen indicate strains that were single-conidium-derived colonies isolated during the same mycovirus transfer attempt (for example, VMDA2-12 and VMDA2-37). VTC, mycovirus transfer control strain. The aflR(IRT)::trpC transgene used in this study is the previously described aflR(IRT1300)::trpC transgene (11).
-, No code available.
Virus transfers.
Mycovirus transfers to various A. nidulans genotypes were performed by hyphal fusion essentially as described by Coenen et al. (5). Mycovirus donor and acceptor strains were cultured together in media that supported the growth of both strains and then subcultured twice in liquid media that supported acceptor strain growth only, followed by a final culture on solid media that also supported acceptor strain growth only. Pure colonies of the acceptor strain were then obtained by single-spore purification. The length of the culture and subculture steps varied between 5 and 8 days. If genetic recombination between the donor and acceptor strains could influence the genotype of the acceptor strain, the genotype of the mycovirus-infected acceptor strain was confirmed by standard methods. Mycoviruses from donor strains 701/8, 701/21, and 701/23 were first transferred to strain WIM126, and then mycovirus-infected WIM126 derivatives were used as donor strains for subsequent transfers. Virus transfer control strains (strains b and j) were created by putting A. nidulans strains (a and i) through the virus-transfer protocol without coculturing with a mycovirus donor strain.
Mycovirus-free isolates were obtained from mycovirus-infected colonies by single-ascospore purification as follows. First, conidia were harvested from a mycovirus-infected colony in sterile water and qualitatively transferred to solid yeast-glucose-trace element medium (3) and cultured at 29°C in the dark for 10 days. Plates were sealed with parafilm for the first 2 days to promote sexual reproduction. Individual cleistothecia were harvested, along with adjacent conidiophores, by using a sterile needle. A suspension of conidia was obtained (as a control for virus presence) by dipping the conidium-covered cleistothecium in 500 μl of water in a microcentrifuge tube. A suspension of ascospores was obtained by rolling the same cleistothecium on 3% water-agar to remove residual mycelia and conidia and bursting it in a fresh tube of 500 μl of sterile water. Serial dilutions were plated from the spore suspensions to obtain single-conidium-derived colonies and single-ascospore-derived colonies.
Identifying mycovirus-infected strains.
Conidia were qualitatively transferred from a path spanning the diameter of a 5- to 6-day-old colony (previously point inoculated onto solid GMM) to 25.0 ml of liquid GMM and cultured for 2 days at 29°C. For all of the harvesting of fungal tissue described here, fungal mycelium was filtered using standard filter paper. Additional liquid was then removed by squishing the mycelium between dry paper towels, followed by freezing in liquid nitrogen and lyophilization for at least 20 h. Total RNA was extracted from ∼100 mg of ground tissue with TRIzol reagent (Invitrogen). The RNA pellet was dissolved in 100 μl of RNase-free water, and 10 μl was analyzed by gel electrophoresis with a 0.8% agarose-1× TAE gel. The smallest dsRNA fragments reported for virus 341 (one dsRNA) and virus 1816 (two dsRNA) (30) were not detected in our analysis, possibly because they were obscured by A. nidulans rRNA. In addition, the shortest visible dsRNA species of virus 341 (band 4) was occasionally lost during mycovirus transfer (see the supplemental material).
Mycovirus sequencing.
Specific dsRNA bands were purified from 0.8% agarose-1× TAE gels by using a QiaQuik gel extraction kit (Qiagen). Random cDNA clones were obtained and sequenced from the gel-purified dsRNA essentially as described by Marquez et al. (15). In some cases, reverse transcription and PCR were used with specific primers to join random clone-generated contigs. For undetermined reasons, several attempts to link contigs 178A and 178B were unsuccessful. 5′ and 3′ RACE (rapid amplification of cDNA ends) (8) analyses were performed on the largest dsRNA element of virus 341; thus, this sequence is thought to be full length.
Nucleotide sequences were used to identify related mycoviruses in GenBank (National Center for Biotechnology Information). CLUSTAL W (26) (for viruses 178 and 1816) or MUSCLE (7) (for virus 341) was used to align the translated contigs with sequences of the related mycoviruses (see the supplemental material). The percent identity and similarity were calculated with BioEdit (9) using the BLOSUM62 scoring matrix. Putative protein sequences were also used to search the National Center for Biotechnology Information conserved domain database to identify protein domains.
Radial growth assays and RNA silencing analysis.
A total of 2 μl of a conidial suspension (∼500 conidia per μl in Fig. 2; ∼100 conidia per μl in Fig. 5) were point inoculated onto 25.0 ml of solid GMM and cultured for 6 days at 29°C. A 12-h light/dark cycle was used for veA (wild-type) strains, and a 24-h light cycle was used for veA1 strains. Five or six replicate plates were included for each strain.
FIG. 2.
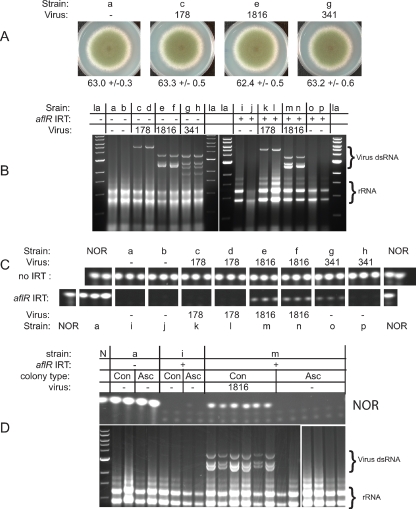
Virus 1816 suppresses RNA silencing. Strains are labeled with a lowercase letter, which can be used with the list in Table 1 to obtain the full genotype of each strain. Pertinent genotype information is provided in each panel. Radial growth and NOR production were determined for non-IRT and aflR (IRT) carrying, mycovirus-infected strains of A. nidulans. (A) None of the three mycoviruses affected radial growth or overall morphology of A. nidulans strains lacking an aflR IRT. Radial growth averages (in mm) with standard deviation values are listed (n = 5 to 6). (B) A single replica plate from the assay for each virus-strain combination was checked for mycovirus by gel electrophoresis. For virus 1816, the two shortest bands sometimes migrate as one band. Virus 341 was lost from strains o and p sometime during or before the experiment. la, DNA ladder. (C) NOR levels were determined in three replica plates for each strain-virus combination. The top row includes data from non-IRT strains, and the bottom row includes data from the corresponding aflR IRT strains. NOR levels were higher in virus 1816-infected strains (m and n) relative to control strains (i and j). The NOR level was also higher in strain o relative to the control strains (i and j), but this phenotype was unstable (see the text). (D) Single-conidium-derived (Con) and single-ascospore-derived (Asc) colonies were obtained from strains a, i, and m and analyzed for mycovirus presence and NOR production. Each lane represents an independent colony. Ascospore passage eliminated virus 1816 from strain m and restored RNA silencing-based suppression of NOR production. N, NOR standard or DNA ladder.
FIG. 5.
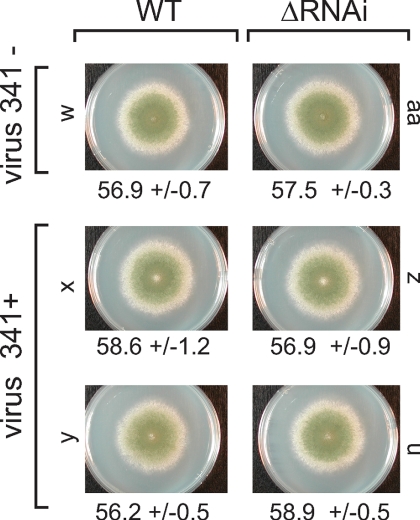
A. nidulans wild-type and RNA silencing mutants infected with virus 341 are morphologically similar. Strains are labeled with a lowercase letter, and the full genotype of each strain is listed in Table 1. Wild-type strains (WT) and strains lacking Dicer, Argonaute, and the two RdRPs (ΔRNAi) were infected with virus 341 and compared in terms of radial growth and overall morphology. Radial growth averages (in mm) with the standard deviation values are listed (n = 5 to 6). No differences were detected between genotypes.
For norsolorinic acid (NOR) analysis, a 1.4-cm-diameter core was taken from the center of a single plate of the radial growth assay, ground in 3 ml of water, and extracted with 3 ml of chloroform. After drying, the residue was redissolved in 100 μl of chloroform. A 5-μl aliquot was then analyzed by thin-layer chromatography as described by Hammond and Keller (11).
Northern blot analysis and v341 IRT construction.
For dclB and rsdA transcript analysis, A. nidulans conidia were inoculated into liquid GMM (2.5 × 107 conidia in 25.0 ml) and cultured under stationary conditions. RNA was extracted from lyophilized tissue with TRIzol reagent. RNA was resolved with a nondenaturing gel for mycovirus detection and with standard formaldehyde denaturing gels for Northern blotting. dclB and rsdA riboprobes were prepared with a MAXIscript kit (Ambion) and 32P-labeled UTP using partial coding sequences as templates (10a). Hybridization was performed in ULTRAhyb buffer (Ambion) at 68°C overnight. Blots were washed twice for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 68°C and twice for 15 min in 0.1× SSC-0.1% SDS at 68°C.
An inverted-repeat transgene (IRT) for virus 341 was designed by cloning two similar ∼750-bp fragments of virus 341 in an inverted orientation into pTMH78.1 (see the supplemental material), an IRT construction plasmid for targeting IRTs to the A. nidulans pyroA locus (10a). The IRT was transformed to A. nidulans strain FGSC A773 and a single correct transformant, TTMH190.13, was identified by Southern blotting (data not shown). TTMH190.13 was crossed to A. nidulans strain RTMH207.37 to obtain the v341 IRT-carrying prototroph RTMH242.11.
For siRNA detection A. nidulans conidia were inoculated into liquid GMM (106 conidia in 25.0 ml) and cultured under stationary conditions for 3 days. Fungal tissue was harvested as described above but was not lyophilized. Approximately 3.5 g of the semidry fungal tissue was ground under liquid nitrogen with a mortar and pestle. Total RNA was isolated with TRIzol reagent. From this point, the protocol for siRNA isolation and detection as described by Hamilton and Baulcombe (10) was followed. Low-molecular-weight RNA was dissolved in formamide after the ethanol-based precipitation step, and the total volume was divided between two polyacrylamide gels for fractionation. After electrophoresis and transfer to nylon membranes, the RNA was hybridized to single-stranded riboprobes in ULTRAhyb hybridization buffer (Ambion) at 37°C overnight. Blots were washed twice for 30 min in 2× SSC-0.2% SDS at 50°C.
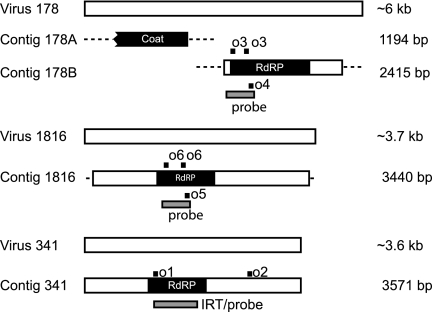
Mycovirus and aflR specific riboprobes were prepared with a MAXIscript kit and 32P-labeled UTP, followed by hydrolysis to an average length of 50 nucleotides (10). The templates for the mycovirus riboprobes are indicated in Fig. 1. The aflR template was described previously (11). For some gels, oligonucleotides were loaded as positive controls for riboprobe specificity, migration distance, and nucleic acid transfer efficiency (see Fig. 1 and the supplemental material).
FIG. 1.
Aspergillus virus 178, virus 1816, and virus 341. The sizes of the longest visible dsRNA elements associated with virus 178 (∼6 kb), virus 1816 (∼3.7 kb), and virus 341 (∼3.6 kb) were estimated by their migration distance relative to a DNA ladder during gel electrophoresis. Sequencing of cDNA from these dsRNA elements resulted in contigs that span part (virus 187), most (virus 1816), or all (virus 341) of their lengths. Two separate contigs were obtained for virus 178, and their predicted positions are indicated. Putative domains are shown (coat protein [pfam05518] and RdRP [pfam02123]). The Expect value for the virus 178 coat protein domain is 2e-84 and for the virus 178, 1816, and 341 RdRP domains are 6e-97, 2e-43, and 3e-11, respectively. The positions of oligonucleotides o1 to o6, which were used as controls for siRNA screening, are indicated. Sense oligonucleotides are depicted above the contig, and antisense oligonucleotides are depicted below the contig. Probe templates and the region used for v341 IRT construction are also indicated.
GenBank accession numbers.
The following sequences have been deposited in GenBank: for Aspergillus virus 178, contig-178a, EU289894, and contig-178b, EU289895; for Aspergillus virus 1816, contig-1816, EU289896; and for Aspergillus virus 341, contig-341, EU289897.
RESULTS
Three Aspergillus mycoviruses have distinct ancestry.
A single, unique 6-kb band was observed in total RNA isolated from A. nidulans strains infected with virus 178 (Fig. 1 and 2B) (30). Sequencing of cDNA derived from this band resulted in contigs of 1,194 and 2,415 bp, encoding a putative coat protein domain and an RNA directed-RNA polymerase (RdRP) domain, respectively (Fig. 1 and see S1 in the supplemental material). RNA virus L1 of Gremmeniella abietina (28), the causative agent of Scleroderris canker on coniferous trees, is virus 178's closest known relative. Amino acid identity and similarity levels between the partial virus 178 sequence and G. abietina RNA virus L1 were 63.7 and 75.7%, respectively (see Fig. S2 in the supplemental material). G. abietina RNA virus L1 is similar to members of the family Totiviridae (28).
Four unique bands between 2 and 3.7 kb were observed in total RNA from virus 1816-infected strains (Fig. 1 and 2B) (30). Sequencing of cDNA derived from the longest band produced a contig of 3,440 bp with an RdRP domain (Fig. 1 and see Fig. S3 in the supplemental material). The closest known relative of virus 1816 is white button mushroom virus 1 of Agaricus bisporus (29). The amino acid identity and similarity levels between the partial virus 1816 sequence and the white button mushroom virus 1 RdRP were 29.0 and 45.8%, respectively (see Fig. S4 in the supplemental material). To our knowledge, A. bisporus white button mushroom virus 1 has not been assigned to a specific virus family.
Four unique bands between 1.5 and 3.6 kb were observed in total RNA from virus 341-infected strains (Fig. 1 and 2B) (30). Sequencing of cDNA from the longest dsRNA molecule, in addition to 5′ and 3′ RACE analyses, produced a single contig of 3,571 bp encoding an RdRP domain (Fig. 1 and see Fig. S5 in the supplemental material). The closest known relative of virus 341 is a totivirus from the pine tree pathogen Sphaeropsis sapinea (19). The amino acid identity and similarity levels between virus 341 and the RdRP of S. sapinea RNA virus 2 were 15.2 and 27.8%, respectively (see Fig. S6 in the supplemental material).
Aspergillus virus 1816 correlates with suppression of experimental RNA silencing.
A. nidulans strains with an aflR IRT are inhibited in the production of the secondary metabolite NOR due to IRT-induced RNA silencing of the transcription factor aflR (11). Hence, the level of NOR production can be used as a measure of RNA silencing functionality in aflR IRT strains. Virus 178, virus 1816, and virus 341 did not alter NOR production, radial growth rate, or morphology of A. nidulans non-IRT strains (Fig. 2A and C and see Fig. S7 in the supplemental material), suggesting that these mycoviruses were ideally suited for a NOR-based RNA silencing-suppression screen.
The NOR-deficient phenotype induced by the aflR IRT was stable in virus 178-infected colonies (Fig. 2C), suggesting that virus 178 does not suppress RNA silencing. This phenotype was also similar in five of five single-conidium-derived colonies from a virus 178-infected parent colony, all of which remained infected after single-spore purification (see Fig. S9 in the supplemental material). For undetermined reasons, the presence of virus 178 correlated with a reduction in radial growth rate and an increased production of aerial hyphae in the aflR IRT genetic background (see Fig. S8 in the supplemental material). These phenotypic changes were not observed in virus 178 infections of strains lacking an aflR IRT (Fig. 2A).
Unlike virus 178, virus 1816 did not alter growth or morphological characteristics when in the aflR IRT genetic background (see Fig. S8 in the supplemental material). Interestingly however, the presence of virus 1816 in aflR IRT strains did correlate with an increase in NOR production (Fig. 2C). This phenotype was stable in five of five single-conidium-derived colonies, all of which remained infected after single-spore purification (see Fig. S9 in the supplemental material). To confirm that the increase in NOR production was dependent on virus 1816 presence, the virus was eliminated from the strain by ascospore passage (5). As predicted, ascospore-passage cured the virus infection and restored RNA silencing-based NOR suppression (Fig. 2D).
Virus 341 interacted differently than the other two mycoviruses in aflR IRT strains. For example, although virus 341 infections were asymptomatic in non-IRT strains (Fig. 2A and see Fig. S7 in the supplemental material), infections of aflR IRT strains correlated with a sectoring phenotype and the partial remediation of NOR production (Fig. 2C and see Fig. S8 in the supplemental material, strains o and p). However, subsequent analysis could not identify mycovirus in these colonies (Fig. 2B, strains o and p), despite data from the virus transfer attempt indicating that these strains carried virus 341 (data not shown). Single-conidium-derived colonies from strain o were also found to have undetectable levels of mycovirus (see Fig. S9 in the supplemental material), but these were phenotypically normal in morphology and RNA silencing-based NOR suppression (see Fig. S9 in the supplemental material). It is thus unknown whether the observed transient morphological abnormalities and NOR production phenotype were due to an interaction with virus 341 or if they were an artifact related to the mycovirus transfer attempt. Despite several independent trials, it was not possible to obtain an aflR IRT strain with a stable virus 341 infection (data not shown).
Virus 341-derived siRNA is detected in an A. nidulans Argonaute mutant.
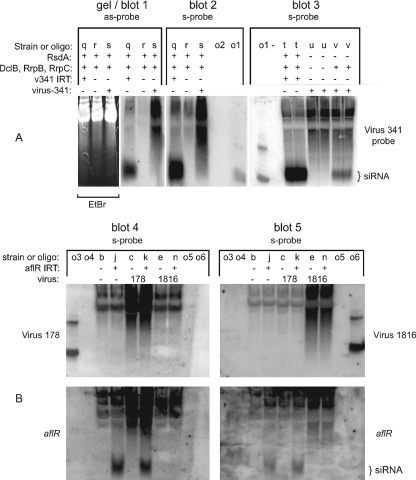
The experiments described above demonstrated that at least one of the three investigated Aspergillus mycoviruses is capable of RNA silencing suppression, suggesting that Aspergillus RNA silencing and some mycoviruses have an antagonistic relationship. If Aspergillus RNA silencing does indeed function in mycovirus defense, then mycovirus-derived siRNA should be present in infected strains. To test this hypothesis, low-molecular-weight RNA extracts were screened for virus 341-, virus 178-, and virus 1816-derived siRNA. As a positive control for siRNA isolation and detection, an IRT containing virus 341 sequences (v341 IRT) was constructed and transformed into A. nidulans. (The IRT should produce hairpin RNA that is processed into siRNA by the RNA silencing machinery.) Although the normal mycovirus dsRNA pattern was visible in high-molecular-weight RNA from infected strains (data not shown), siRNA was typically only detectable in low-molecular-weight RNA from the v341 IRT strain (Fig. 3). This suggested that siRNA levels might be too low for detection by Northern blotting. To examine this possibility, we borrowed a method previously used with Neurospora crassa. Nolan et al. (17) reported that use of an N. crassa Argonaute mutant allowed for the detection of transposon-derived siRNA that could not otherwise be detected by Northern blotting. Therefore, an A. nidulans Argonaute (ΔrsdA) mutant (10a) was infected with virus 341 and its low-molecular-weight RNA was screened for siRNA. In the ΔrsdA genetic background, virus 341-derived siRNA was clearly detected. This result was similar in two independent experiments (Fig. 3, blot 3, and data not shown). These data suggest that mycovirus-derived siRNA is present at relatively low levels in at least some mycovirus-infected strains.
FIG. 3.
Mycovirus-derived siRNA is present in a virus 341-infected Argonaute mutant. Strains are labeled with a lowercase letter, which can be used with Table 1 to obtain the full genotype of each strain. In addition, pertinent genotype information and virus infection status are indicated above each lane with plus (+) and minus (−) symbols. (A) Low-molecular weight RNA from various A. nidulans strains was hybridized to a probe made of virus 341 sequences. Either antisense (as-probe)- or sense (s-probe)-specific riboprobes were used. For blot 1, the ethidium bromide-stained gel is shown to demonstrate relative RNA levels. siRNA is only detected in the v341 IRT strain (blots 1 to 3, strain q) and a virus 341-infected Argonaute mutant (blot 3, strain v). (B) Low-molecular-weight RNA from various A. nidulans strains was hybridized to a probe consisting of virus 178 (top left) and virus 1816 (top right) sequences, followed by a probe consisting of aflR sequences (bottom). Sense-specific riboprobes were used for each analysis. Mycovirus-derived siRNA and aflR siRNA were not detected for virus 1816, indicating that virus 1816 interferes with IRT-derived siRNA levels. Mycovirus-derived siRNA was also not detected for virus 178, but aflR siRNA was not affected by this mycovirus. For blots 2, 3, 4, and 5, 50 to 100 pmol of oligonucleotides were loaded as controls (see Fig. 1). In blots 3, 4, and 5, two bands can be seen in some oligonucleotide control lanes. It appears that the higher band is due to impurities in the oligonucleotide preparation, since it is present even when only one oligonucleotide is loaded as a control (blot 3). This also suggests that only one of the two oligonucleotides is recognized by the riboprobe in blots 4 and 5. This may be an artifact related to a decreased specific activity of the riboprobe at increasing distances from the transcription start site, thus causing the oligonucleotide closest to the transcription start site to be the only band detected by the riboprobe.
Virus 178 and virus 1816 were not transferred to an ΔrsdA genetic background; thus, it is not known whether siRNA from these mycoviruses can also be detected by such a method. As described above, low-molecular-weight RNA from virus 178 and virus 1816 rsdA+ strains was screened for mycovirus-derived siRNA without success (Fig. 3B). To determine that this was not a technical problem with siRNA detection, an aflR probe was used to locate the aflR siRNA which should be present in the noninfected and mycovirus-infected aflR IRT strains (Fig. 3B, strains j, k, and n). As predicted, aflR siRNA was detected in the control aflR IRT strain and in the virus 178-infected aflR IRT strain. However, the virus 1816-infected aflR IRT strain did not have detectable levels of siRNA (Fig. 3B, strains j, k, and n). These results were identical in two independent experiments and are consistent with the RNA silencing suppression activity observed for virus 1816. Thus, these data suggest that the lack of mycovirus-derived siRNA for virus 178 and 1816 was not due to a technical problem and that the aflR IRT suppression mechanism of virus 1816 involves a loss or reduction in IRT-derived siRNA.
Virus 341 infections of A. nidulans RNA silencing mutants are symptomless.
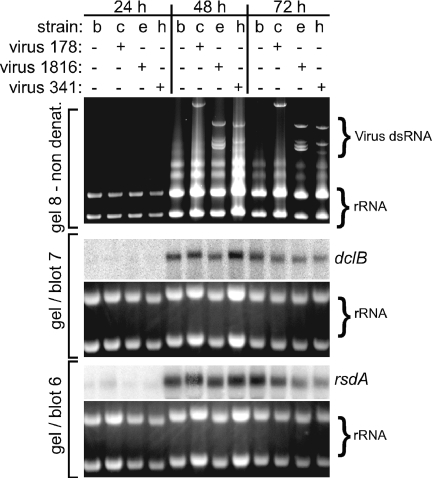
Northern blot analyses indicate that the A. nidulans RNA silencing genes Dicer (dclB) and Argonaute (rsdA) are temporally regulated (Fig. 4). However, none of the three mycoviruses appear to significantly alter transcript levels for these genes in 24-, 48-, and 72-h cultures (Fig. 4). If Aspergillus RNA silencing functions in mycovirus defense, it seems possible that mycoviruses may have evolved strategies to avoid stimulating the RNA silencing machinery. Such an evolutionary strategy may be a clue as to why virus 341 infections of A. nidulans strains lacking the core components of RNA silencing (ΔRNAi = ΔdclB, ΔrsdA, ΔrrpB and ΔrrpC) (10a) have no affect on gross morphology and radial growth (Fig. 5 and see Fig. S10 in the supplemental material). Because virus 178 and virus 1816 have not been transferred to ΔRNAi strains, it is not yet known what affect these mycoviruses may have on A. nidulans strains lacking functional RNA silencing.
FIG. 4.
Aspergillus Dicer and Argonaute transcript patterns during mycovirus infection. Strains are labeled with a lowercase letter, and the full genotype of each strain is listed in Table 1. RNA from 24- to 72-h stationary cultures was analyzed for mycovirus and dclB and rsdA transcript levels. Ethidium-stained rRNA is shown to indicate the relative RNA levels.
DISCUSSION
Several years ago, van Diepeningen et al. demonstrated that stable mycovirus infections of A. nidulans could be obtained by protoplast fusion with naturally infected Aspergillus niger isolates (30). Three of the mycoviruses successfully transferred to A. nidulans in that study were partially sequenced here. Their predicted RdRP sequences suggest that they have diverse ancestral backgrounds, with each identifying a different best match in GenBank. More significantly, we demonstrate here that at least some Aspergillus mycoviruses are capable of RNA silencing suppression and that at least some others are targeted by the A. nidulans RNA silencing machinery. These data support the hypothesis that one function of A. nidulans RNA silencing is mycovirus defense.
Virus 178 and virus 341 either correlated with an abnormal morphology and/or were unstable in aflR IRT strains but not in isogenic control strains. Although the cause of these phenomena is unknown, it is interesting that they both involve an IRT-expressing strain of A. nidulans. It seems that the presence of an IRT somehow modified the virus-host interaction. For example, RNA silencing could be less efficient in the presence of an IRT, if RNA silencing proteins are engaged in IRT processing and unable to fulfill their role in mycovirus defense. Supporting circumstantial evidence that the IRT may be a factor in these phenomena comes from the observation that virus 1816, which was not affected by IRT presence, is the only one demonstrated to suppress IRT-RNA silencing.
Virus 1816 clearly correlates with RNA silencing suppression. Although the exact suppression mechanism is not known, it seems that dsRNA is protected, Dicer is inhibited, or siRNA is more quickly turned over (because aflR siRNA is not detected in a virus 1816-infected aflR IRT strain). Future work will investigate each of these possibilities. There are thought to be six major dsRNA elements associated with virus 1816 (30), four of which were detected by our methods. It is possible that one of these elements encodes an RNA silencing suppressor, as has been found for the C. parasitica hypovirus CHV1 (21) and several viruses of plants and animals (for reviews, see references 14 and 32).
A hallmark of RNA silencing-based virus control is the presence of virus-derived siRNA. Since virus 1816 suppresses IRT-derived siRNA levels, it is not surprising that siRNA was not detected from this mycovirus. However, virus 178- and virus 341-derived siRNA was also not detected in RNA silencing-capable strains of A. nidulans. Thus, if these mycoviruses are normally targeted and degraded by RNA silencing, their siRNA must be below the threshold of detection by Northern blotting. This hypothesis is supported by the fact that virus 341 siRNA was detected in an Argonaute mutant, which is thought to allow for the accumulation of siRNA above normal levels. It is not yet known whether a similar experiment with virus 178 and virus 1816 would allow for the detection of siRNA from these mycoviruses.
Virus 341 siRNA may normally be present at a low level because virus 341 is likely successful at avoiding RNA silencing. All known true dsRNA viruses do not expose their genome to the host cell, but rather keep it sequestered in the virion. Such a strategy may enhance RNA silencing avoidance and contribute to the low siRNA level of virus 341. Avoidance of silencing is also supported by the finding that virus 341, as well as viruses 178 and 1816, did not stimulate Dicer or Argonaute gene expression.
Virus 341 did not affect the growth rate or overall morphology of strains lacking RNA silencing capability. These data suggest that A. nidulans RNA silencing is not crucial for keeping virus 341 replication levels below a threshold above which fitness (as determined by overall morphology and radial growth) is affected. It is unknown whether virus 341 RNA levels are increased in the RNA silencing mutants, but qualitative analysis suggests such an increase may be small at best (see Fig. S10 in the supplemental material).
A significant finding of this study is that mycovirus-derived siRNAs exist. Some mycovirus-derived siRNAs may have a major effect on host phenotype. This may occur if the siRNA targets the RNA silencing machinery toward a host mRNA. Such a scenario would require that approximately 25 bp of the mycoviral genome—the approximate length of siRNA in A. nidulans (11)—have a high level of identity with that of a host gene. Given the high mutation rate characteristic of RNA viruses (6), it is possible that the occurrence of mycovirus-derived siRNA with host gene complementarity is common in nature. In cases where a newly evolved mycovirus-derived siRNA decreases host gene expression in a manner that was beneficial to the host and the mycovirus, a mutualistic symbiosis could develop. It is also possible that some mycovirus-induced hypovirulence phenotypes of phytopathogenic fungi are due to mycovirus-derived siRNA. In this scenario, natural selection may promote siRNA that manipulates fungal virulence in a manner that enhances the survival of all partners in the interaction.
Natural mycovirus infections of A. nidulans appear to be rare in nature (5). In fact, A. niger mycoviruses were used in the present study because, to our knowledge and that of other researchers (5), there are no known Aspergillus mycoviruses of A. nidulans origin. One possibility is that the sexual cycle of A. nidulans, along with the associated formation of new somatic incompatibility groups, contributes to the low rate of natural A. nidulans mycovirus infections (5). Our results are consistent with this hypothesis since ascospore-derived colonies of virus 1816- and virus 178-infected strains were free of the mycovirus observed in their parent colonies (Fig. 2D and data not shown). It is conceivable that RNA silencing suppression by some mycoviruses may block the sexual cycle. For example, N. crassa requires some RNA silencing proteins for its sexual cycle (12, 25). Thus, in some instances, mycovirus suppression of RNA silencing may serve the interest of the mycovirus by limiting a life cycle stage that is inhibitory to mycoviral spread through the host population. This scenario may select for the separation of RNA silencing processes from sexual reproduction, as appears to be the case for A. nidulans (10a). In addition, any coupling of RNA silencing to housekeeping functions may be detrimental to an organism that frequently encounters RNA silencing-suppressing viruses. In a companion paper we show that A. nidulans strains lacking the core RNA silencing components are completely normal in growth and development, suggesting that this organism has separated RNA silencing from general cellular processes (10a). The identification of a mycovirus suppressor of fungal RNA silencing in the present study (and another study [21]) provides a motivating factor for such separation.
Supplementary Material
Acknowledgments
We gratefully acknowledge Alfons Debets (Wageningen University, The Netherlands) for the mycovirus-infected A. nidulans isolates.
Funding for this project was provided by a College of Agricultural and Life Sciences Wisconsin Distinguished Graduate Fellowship (T.M.H.), the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (2005-35201-15350) (N.P.K.), the National Science Foundation (MCB-0236393) (N.P.K.), and The Samuel Roberts Noble Foundation (M.J.R.).
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Baulcombe, D. 2004. RNA silencing in plants. Nature 431356-363. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1986. Fungal virology. CRC Press, Inc., Boca Raton, FL.
- 3.Calvo, A. M., J. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 704733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerutti, H., and J. A. Casas-Mollano. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 5081-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coenen, A., F. Kevei, and R. F. Hoekstra. 1997. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet. Res. 691-10. [DOI] [PubMed] [Google Scholar]
- 6.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10859-864. [DOI] [PubMed] [Google Scholar]
- 7.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 858998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 10.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286950-952. [DOI] [PubMed] [Google Scholar]
- 10a.Hammond, T. M., J. W. Bok, M. D. Andrewski, Y. Reyes-Dominguez, C. Scazzocchio, and N. P. Keller. 2008. RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 7339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond, T. M., and N. P. Keller. 2005. RNA Silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, D. W., R. J. Pratt, M. McLaughlin, and R. Aramayo. 2003. An Argonaute-like protein is required for meiotic silencing. Genetics 164821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, H. W., and S. W. Ding. 2005. Antiviral silencing in animals. FEBS Lett. 5795965-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquez, L. M., R. S. Redman, R. J. Rodriguez, and M. J. Roossinck. 2007. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315513-515. [DOI] [PubMed] [Google Scholar]
- 16.McCluskey, K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52245-262. [DOI] [PubMed] [Google Scholar]
- 17.Nolan, T., L. Braccini, G. Azzalin, A. De Toni, G. Macino, and C. Cogoni. 2005. The posttranscriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 331564-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3632-642. [DOI] [PubMed] [Google Scholar]
- 19.Preisig, O., B. D. Wingfield, and M. J. Wingfield. 1998. Coinfection of a fungal pathogen by two distinct double-stranded RNA viruses. Virology 252399-406. [DOI] [PubMed] [Google Scholar]
- 20.Romaine, C. P., and B. Schlagnhaufer. 1995. PCR analysis of the viral complex associated with La France disease of Agaricus bisporus. Appl. Environ. Microbiol. 612322-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segers, G. C., R. van Wezel, X. Zhang, Y. Hong, and D. L. Nuss. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5896-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segers, G. C., X. Zhang, F. Deng, Q. Sun, and D. L. Nuss. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 10412902-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapira, R., G. H. Choi, and D. L. Nuss. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiu, P. K., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107905-916. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsitsigiannis, D. I., R. Zarnowski, and N. P. Keller. 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 27911344-11353. [DOI] [PubMed] [Google Scholar]
- 28.Tuomivirta, T. T., and J. Hantula. 2003. Two unrelated double-stranded RNA molecule patterns in Gremmeniella abietina type A code for putative viruses of the families Totiviridae and Partitiviridae. Arch. Virol. 1482293-2305. [DOI] [PubMed] [Google Scholar]
- 29.van der Lende, T. R., E. H. Duitman, M. G. Gunnewijk, L. Yu, and J. G. Wessels. 1996. Functional analysis of dsRNAs (L1, L3, L5, and M2) associated with isometric 34-nm virions of Agaricus bisporus (white button mushroom). Virology 21788-96. [DOI] [PubMed] [Google Scholar]
- 30.van Diepeningen, A. D., A. J. Debets, and R. F. Hoekstra. 1998. Intra- and interspecies virus transfer in aspergilli via protoplast fusion. Fungal Genet. Biol. 25171-180. [DOI] [PubMed] [Google Scholar]
- 31.Varga, J., B. Toth, and C. Vagvolgyi. 2003. Recent advances in mycovirus research. Acta Microbiol. Immunol. Hung. 5077-94. [DOI] [PubMed] [Google Scholar]
- 32.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6206-220. [DOI] [PubMed] [Google Scholar]
- 33.Yager, L. N. 1992. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans, p. 19-41. In J. Bennett and M. Klich (ed.), Aspergillus: biology and industrial applications. Butterworth-Heinemann, Boston, MA. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.