Abstract
Signaling by stress-activated mitogen-activated protein kinase (MAPK) pathways influences translation efficiency in mammalian cells and budding yeast. We have investigated the stress-activated MAPK from fission yeast, Sty1, and its downstream protein kinase, Mkp1/Srk1, for physically associated proteins using tandem affinity purification tagging. We find Sty1, but not Mkp1, to bind to the translation elongation factor eukaryotic elongation factor 2 (eEF2) and the translation initiation factor eukaryotic initiation factor 3a (eIF3a). The Sty1-eIF3a interaction is weakened under oxidative or hyperosmotic stress, whereas the Sty1-eEF2 interaction is stable. Nitrogen deprivation causes a transient strengthening of both the Sty1-eEF2 and the Sty1-Mkp1 interactions, overlapping with the time of maximal Sty1 activation. Analysis of polysome profiles from cells under oxidative stress, or after hyperosmotic shock or nitrogen deprivation, shows that translation in sty1 mutant cells recovers considerably less efficiently than that in the wild type. Cells lacking the Sty1-regulated transcription factor Atf1 are deficient in maintaining and recovering translational activity after hyperosmotic shock but not during oxidative stress or nitrogen starvation. In cells lacking Sty1, eIF3a levels are decreased, and phosphorylation of eIF3a is reduced. Taken together, our data point to a central role in translational adaptation for the stress-activated MAPK pathway in fission yeast similar to that in other investigated eukaryotes, with the exception that fission yeast MAPK-activated protein kinases seem not to be directly involved in this process.
Cells need to regulate translation in response to stress conditions for two main reasons. First, protein synthesis is a highly energy-demanding process, consuming one-third of cellular ATP under conditions of active growth. The cell's overall translational activity is rapidly down-regulated under stress conditions as a result of tight control at several levels, including synthesis and modifications of rRNA, transcription of genes encoding ribosomal proteins, and phosphorylation of certain translation initiation factors (43). Second, it is useful to selectively enhance translation of mRNAs encoding proteins required for survival during stress conditions. Such a mechanism is inherently faster than transcriptional initiation, since it acts on a preexisting mRNA population (30). Certain mRNAs have been shown to increase their translational efficiency under conditions of stress and nutrient deprivation, such as Saccharomyces cerevisiae GCN4 (18) or mammalian ATF4 (16). Such mRNAs thus run counter to the overall trend of translational down-regulation under conditions of cell stress.
In mammalian cells, signaling through the stress-activated mitogen-activated protein kinase (MAPK) p38 or Jun N-terminal protein kinase influences both translation rates and mRNA stability. In some instances, this posttranscriptional control has been shown to occur through protein kinases downstream of MAPKs (MAPK-activated protein kinases [MAPKAPKs]). For example, MAPKAPK-2 is required for stabilization of mRNA encoding tumor necrosis factor alpha (28). Hog1 is the sole stress-activated MAPK in budding yeast, and Sty1 is the sole stress-activated MAPK in fission yeast. In budding yeast, two paralogous genes encode the MAPKAPKs Rck1 and Rck2 (8), and in fission yeast the corresponding proteins are Mkp1/Srk1 and Mkp2/Cmk2 (1, 3, 36). Similar to the situation in mammalian cells, Hog1 forms a relatively stable complex with Rck2 in budding yeast (5) and Sty1 with Mkp1/Srk1 in fission yeast (3, 36), and in both organisms, the MAPK phosphorylates the cognate downstream kinase when activated by stress (3-5, 36, 39).
Several cases of translational control through stress-activated MAPK pathways have been demonstrated in yeast. In Saccharomyces cerevisiae, the down-regulation and subsequent recovery of global protein synthesis after hyperosmotic shock or oxidative stress are impaired in mutants deficient in the stress-activated MAPK Hog1 (39, 40) or in its binding partner, the downstream protein kinase Rck2 (37, 39). The translation elongation factor eEF2 has been shown to be phosphorylated in an RCK2-dependent way during hyperosmotic shock (39). As for mRNA-specific effects on translation, the MFA2 transcript is less efficiently translated upon glucose addition, and this phenomenon is dependent on an intact stress-activated MAPK pathway (41). Conversely, the TIF51A mRNA is stabilized under the same conditions, and this is likewise HOG1 dependent (42). In Schizosaccharomyces pombe, mutations in the stress-activated MAPK pathway lead to increased phosphorylation of the translation initiation factor eIF2α and reduced translation initiation during oxidative and hyperosmotic stress (12). Further, activation of Sty1 by oxidative stress enhances the stability of atf1+ mRNA (33). This promotes translation of Atf1, a transcription factor important for induction of many stress-induced genes. This control is exerted through the RNA-binding and -stabilizing protein Csx1 (33), which is counteracted by the destabilizing proteins Cip1 and Cip2 (25).
Led by these prior observations, in the present study we investigated physical interactions in fission yeast cells between the stress-activated MAPK Sty1, on one hand, and its downstream kinase Mkp1, the translation elongation factor eEF2, and the translation initiation factor eukaryotic initiation factor 3a (eIF3a), on the other. Sty1 interacts robustly with eEF2 and eIF3a, but Mkp1/Srk1 does not. In sty1Δ mutants, the amount of eIF3a is reduced and the phosphorylation level of eIF3a is decreased. We also examined the impact on total translational activity in wild-type and sty1 cells of oxidative stress, hyperosmotic shock, and nitrogen starvation. All stresses cause translation activity to decrease significantly more in sty1 mutant cells, but only hyperosmotic shock does so in atf1 mutants. After both hyperosmotic shock and nitrogen deprivation, there is a rapid and profound decrease in the amount of polysomes in wild-type cells, followed by a recovery phase. Cells lacking sty1+ lose translation activity more extensively and fail to recover to the degree seen in wild-type cells. At the time when polysomal recovery commences after nitrogen deprivation, coincident with the peak of activation of Sty1, the interactions are maximal between Sty1 and eEF2, as well as between Sty1 and Mkp1.
MATERIALS AND METHODS
Fission yeast techniques.
Standard methods for growth of S. pombe strains, protein preparations, Western analysis, PCR-based genomic epitope tagging, and mating were used as described previously (3). The strains expressing Sty1 or Mkp1 C-terminally marked with a tandem affinity purification (TAP) tag were created as described previously (38). Epitope-tagged proteins in all strains were expressed from the respective endogenous chromosomal loci. The strains used in this study are listed in Table 1.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| DN1 | h−sty1-TAP:kan | This study |
| DN2 | h−mkp1-TAP:kan | This study |
| 972 | h− | P. Russell |
| JM1698 | h−sty1-9myc:ura4+leu1-32 ura4D-18 ade6-M210 | J. Millar |
| JM2766 | h+atf1::kan leu1-32 ura4-D18 ade6-704 | J. Millar |
| CM3 | h−sty1-9myc:ura4+mkp1-3HA:kan leu1-32 ura4D-18 ade6-M210 | 3 |
| EA128 | h+eIF3a-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA129 | h+eEF2-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA136 | h−sty1-9myc:ura4+eEF2-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA137 | h−sty1-9myc:ura4+eIF3a-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA133 | h−mkp1-13myc:hph eIF3a-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA134 | h−mkp1-13myc:hph eEF2-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| DN6 | h+sty1::ura4+eEF2-3HA:kan ura4D-18 leu1-32 | This study |
| DN7 | h+sty1::ura4+eIF3a-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA154 | h−mkp1::ura4+eEF2-3HA:kan ura4-D18 leu1-32 ade6-M216 | This study |
| EA156 | h−mkp1::ura4+ura4-D18 | This study |
| EA98 | h−mkp2::kan | 3 |
| EA157 | h−sty1::ura4+ura4-D18 | This study |
| EA165 | h−mkp1::ura4+ura4-D18 mkp2::kan | This study |
| CM9 | h−mkp1:P3nmt1-3HA:kan | 3 |
| CM10 | h−mkp2:P3nmt1-3HA:kan | 3 |
Single colons denote tagged functional genes; double colons denote disruptions.
Affinity purification and mass spectrometry.
Twenty liters of cell culture grown to an optical density at 595 nm (OD595 nm) of ∼2.0 in 0.5% yeast extract, 3% glucose, and supplements (YES medium) was harvested by centrifugation at 4°C. After one round of washing in cold water, the cells were suspended in 100 ml of HEPES buffer (200 mM KOH-HEPES [pH 7.8], 15 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 15% glycerol, 0.5 mM dithiothreitol [DTT], Complete protease inhibitor cocktail [Roche]) and subjected to 25 cycles of lysis (30 s of beating and 90 s of rest in a Beadbeater [Biospec Inc.]). Cell extracts were centrifuged at 9,000 rpm for 15 min at 4°C. KCl was added to the supernatants to a final concentration of 0.2 M. After mixing for 15 min at 4°C, they were centrifuged at 42,000 rpm for 20 min at 4°C. The supernatants were incubated with immunoglobulin G-Sepharose beads (GE Healthcare) for 1 h at 4°C. Beads were washed in immunoglobulin G buffer (10 mM Tris [pH 8], 150 mM NaCl) on columns and cleaved off with 200 units of TEV protease (Invitrogen) in TEV-cleaving buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 0.5 mM EDTA) at 16°C for 1 h. Eluted proteins were precipitated with trichloroacetic acid at a final concentration of 20%, separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and visualized by Coomassie blue staining. Protein band were excised from gels, treated with trypsin, and subjected to mass spectrometry. Protein bands were analyzed with matrix-assisted laser desorption ionization-time-of-flight mass spectrometry or liquid chromatography-tandem mass spectrometry by the Swegene Proteomics facility at Göteborg University.
Coimmunoprecipitation (co-IP) and detection of phosphoprotein.
Protein extracts were incubated overnight at 4°C with 20 μl of hemagglutinin (HA) probe (Santa Cruz Biotechnology, Inc.), washed extensively in buffer A (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 100 mM KCl, 0.2% Triton X-100, 0.25% NP-40, Complete protease inhibitor cocktail), and resolved by SDS-polyacrylamide gel electrophoresis. For detection of Sty1-myc and Mkp1-myc, anti-myc antibodies (9E10; Santa Cruz Biotechnology, Inc.) were used. For detection of phosphoproteins or total precipitated protein, gels were stained with ProQ Diamond (Invitrogen) or SYPRO Ruby (Invitrogen), respectively, according to the manufacturer's instructions, followed by visualization in a PhosphorImager (Molecular Dynamics). Antibodies used for loading controls were anti-α-tubulin (Sigma) and anti-Cdc2 (100.4; Abcam).
Polysome separation and analysis.
Cells were grown in rich medium or in minimal medium (EMM [27]) without thiamine for studies of overexpression of Mkp1 and Mkp2 from the nmt1 promoter, to an OD595 of 0.6. Cycloheximide was added to 0.1 mg/ml, and the samples were left on ice for 5 minutes. Cells were harvested by centrifugation and lysed in breaking buffer (20 mM Tris-HCl [pH 8], 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 0.1 mg/ml cycloheximide, 0.2 mg/ml heparin) with glass beads in a Fast-Prep (FP120; Bio101 Savant Instruments, Inc., Holbrook, NY) twice for 45 s at speed 4. Polysomes were separated essentially as described previously (37); 10 to 50% sucrose gradients were centrifuged at 4°C for 160 min in SW41Ti rotors at 35,000 rpm. For each profile, the polysome ratio was calculated as the summed area of the peaks corresponding to polysomes 2n and higher divided by the total area representing ribosomal material (40S, 60S, 80S, and all polysomal peaks). In untreated wild-type cells, this ratio was 0.62 (standard deviation = 0.03), and in sty1Δ mutants it was 0.66 (standard deviation = 0.06). The bars presented in Fig. 4 to 6 represent averages of relative values from two to four independent experiments where the ratio in untreated cells has been set to 1.
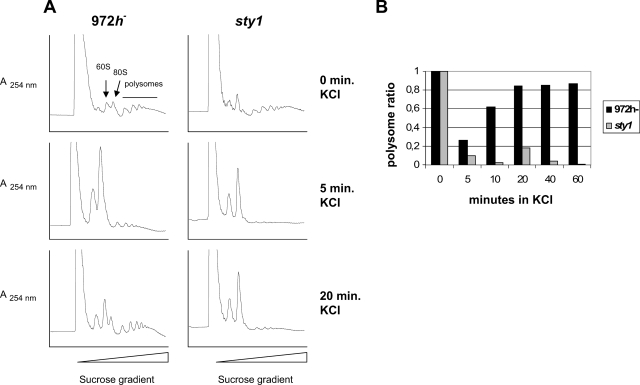
FIG. 4.
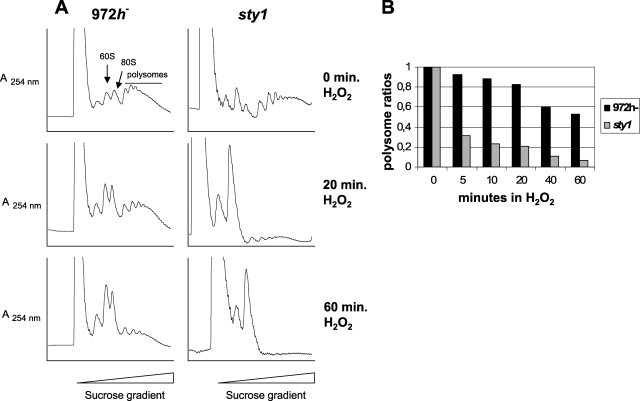
Polysomal contents of wild-type and sty1 cells after hyperosmotic stress. 972h− and sty1 cells were grown to an OD595 of ∼0.5 and subjected to 0.6 M KCl. Extracts were prepared at various times after shock, separated on sucrose gradients, and analyzed online as described in Materials and Methods. (A) Polysome profiles obtained at three different times after hyperosmotic shock. The positions of the 40S, 60S, 80S, and polysomal peaks are indicated in one of the panels. (B) Relative polysome ratios calculated as polysome/(monosome + polysome) areas at five different times after hyperosmotic shock for wild-type and sty1 cells (averages from two to four independent experiments). The ratio before stress (0 min) for each strain was set to 1, and values for subsequent time points are indicated relative to that value.
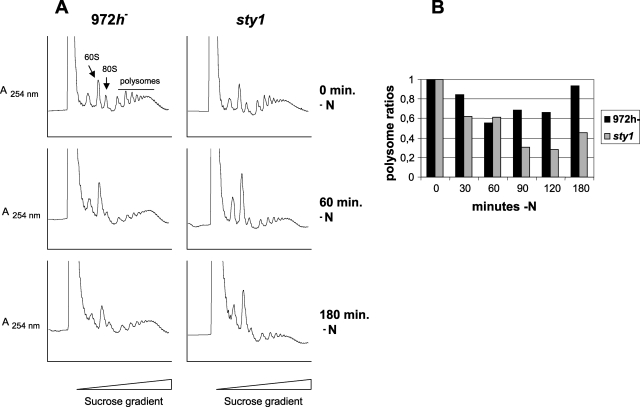
FIG. 6.
Polysomal contents of wild-type and sty1 cells after nitrogen deprivation. 972h− and sty1 cells were grown to an OD595 of ∼0.25 and after a brief centrifugation were resuspended in nitrogen-free EMM medium. (A) Polysome profiles obtained at three different times after nitrogen deprivation as for Fig. 4A. (B) Relative polysome ratios at five times after nitrogen deprivation as for Fig. 4B.
Two-dimensional (2D) gel electrophoresis analysis of eEF2.
Ten micrograms of total protein extract from sty1+ or sty1Δ cells expressing eEF2-(HA)3 from its endogenous promoter were separated in the first dimension on 14-cm pH 3 to 11 strips and in the second dimension on 8% SDS-polyacrylamide gels at the Swegene Proteomics facility at Göteborg University. The separated proteins were blotted to nitrocellulose membranes and detected with anti-HA antibodies (lab stock) as described above for Western analysis.
Stress treatment.
Cells were grown to mid-log phase (OD595 = 0.5) in rich medium and subjected either to oxidative stress by the addition of 1 mM H2O2 or to hyperosmotic stress by the addition of an equal volume of prewarmed YES medium containing 1.2 M or 2 M KCl. For nitrogen starvation, the cells were grown to early log phase (OD595 = 0.25) in YES medium, centrifuged for 2 min at 2,000 rpm, and resuspended in EMM minimal medium without NH4Cl.
RESULTS
Sty1 interacts with proteins from the translational machinery.
In order to find novel proteins that interact with Sty1 in the stress signaling pathway, we performed affinity purification with TAP-tagged Sty1 and Mkp1 (38). To verify that the tagged proteins were functional, we assessed growth of the strains carrying TAP tag fusions after exposure to UV irradiation or to hyperosmotic stress (0.6 M KCl). In both cases, growth was indistinguishable from that of wild-type cells (not shown).
We then proceeded to purify the TAP-tagged proteins and their binding partners. As expected, the predominant Sty1-interacting protein was found to be the MAPKAPK Mkp1, which was already known to exist and function in part in a complex with Sty1 (3, 36). Likewise, Sty1 was the main interacting protein found in the Mkp1 TAP purification. Additional proteins were found to interact with Sty1 and Mkp1 in substoichiometric amounts. A prominent group among these were proteins involved in translation initiation: eIF3a, interacting with both Sty1-TAP and Mkp1-TAP, and translation elongation factors eEF2 and eEF3, interacting with Sty1-TAP) (Table 2).
TABLE 2.
Copurified proteins using TAP tagging
| S. pombe protein | Open reading frame(s) | Mol mass (kDa) |
|---|---|---|
| Sty1-TAP | ||
| Mkp1/Srk1 | SPCC1322.08 | 66 |
| eIF3a | SPBC17D11.05 | 107 |
| eEF2 | SPAC513.01c, SPCP31B10.07 | 94 |
| eEF3 | SPCC417.08 | 117 |
| Mkp1-TAP | ||
| Sty1 | SPAC24B11.06c | 40 |
| eIF3a | SPBC17D11.05 | 107 |
eEF3 is a fungus-specific elongation factor, in contrast to most of the highly conserved proteins involved in protein synthesis in eukaryotes (2). We wanted to focus on proteins widely conserved in evolution, and for this reason we investigated only the interactions with eEF2 and eIF3a further. eIF3 functions as a scaffold for several eIFs in the initiation of protein synthesis. It is the most complex of the eIFs, consisting of five core subunits (the largest of them being eIF3a) and different subsets of the four noncore subunits (17). The elongation factor eEF2 catalyzes the translocation of the nascent protein chain in the ribosome. eEF2 in fission yeast is encoded by two different genes, eft201+ (SPAC513.01c) and eft202+ (SPCP31B10.07), with identical predicted amino acid sequences (26).
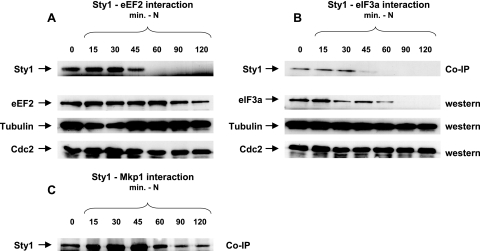
To verify the interactions between Sty1, Mkp1, and these translation factors, we performed co-IP with Sty1 or Mkp1 and eIF3a or eEF2. Strains were constructed with myc-tagged Sty1 or Mkp1 coexpressed with either HA-tagged eIF3a or eEF2 (SPAC513.01c). As seen in Fig. 1 and 2, we could detect a robust interaction between Sty1 and eEF2, as well as eIF3a, under standard growth conditions. Based on the amounts of coprecipitated and input protein, we estimate that 2 to 3% of eIF3a is bound to Sty1 under these conditions (not shown). Under the same conditions of culture and stringency of washing, an interaction between Mkp1 and eEF2, or between Mkp1 and eIF3a, is hardly detectable (Fig. 1 and 2).
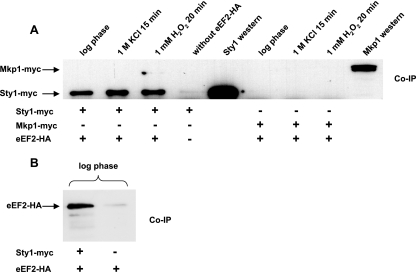
FIG. 1.
Sty1, but not Mkp1, is coimmunoprecipitated with eEF2 in undisturbed log-phase cells and after oxidative and hyperosmotic stress. Strains expressing HA-tagged eEF2, myc-tagged Sty1, or myc-tagged Mkp1, alone or in combinations as indicated, were grown to an OD595 of ∼0.5 in YES medium. Protein preparations were made from cells without stress or after exposure to 1 mM H2O2 for 20 min or 1 M KCl for 15 min. Following bicinchoninic acid protein quantification, equal amounts from each sample were used for IP of eEF2-HA with HA probe. (A) Interacting Sty1-myc and Mkp1-myc were detected with anti-myc antibodies. A strain expressing only Sty1-myc (JM1698) was used as a negative control for co-IP. Western blots of total protein (smaller amounts loaded than for the co-IP lanes) show the positions of myc-tagged Sty1 and Mkp1. (B) eEF2 interacting with Sty1 was detected with anti-HA antibodies. A strain expressing only eEF2-HA (EA129) was used as a negative control.
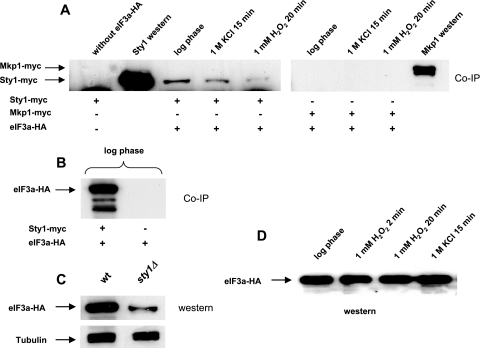
FIG. 2.
Sty1, but not Mkp1, is coimmunoprecipitated with eIF3a in undisturbed log-phase cells, and the interaction weakens after oxidative and hyperosmotic stress. (A) Cells were grown and exposed to stress, and protein samples were prepared and loaded as for Fig. 1 except that strains expressing eIF3a-HA were used in place of eEF2-HA. (B) eIF3a interacting with Sty1 was detected with anti-HA antibodies. A strain expressing only eIF3a-HA (EA128) was used as a negative control. (C) Western blots of cells expressing eIF3a-HA in either a wild-type (EA128) or sty1Δ (DN7) background. (D) Western blot of cells expressing eIF3a-HA under stress conditions, to show that the level is unchanged.
The Sty1-eEF2 interaction is unaffected by oxidative and hyperosmotic stress.
In S. cerevisiae, the Sty1 homolog Hog1 and the Mkp1 homolog Rck2 are required for the translational shutdown in response to hyperosmotic shock and for recovery of translation after adaptation (39, 40). During hyperosmotic shock, an RCK2-dependent phosphorylation of eEF2 has been reported (39). To test for any change in interaction between Sty1 and eEF2 after the activation of Sty1, we compared co-IPs from cells exposed to hyperosmotic or oxidative stress with those from undisturbed cells. The fraction of eEF2-bound Sty1 was unchanged after treatment with 1 M KCl for 15 min or 1 mM H2O2 for 20 min (Fig. 1). The total protein levels of eEF2 did not seem to change after oxidative or osmotic stress (data not shown).
The Sty1-eIF3a interaction decreases after oxidative or hyperosmotic stress.
The corresponding investigation of the interaction with Sty1 was also undertaken for eIF3a. In Fig. 2A it is seen that this interaction diminished after hyperosmotic stress (1 M KCl for 15 min) and to an even greater extent after oxidative stress (1 mM H2O2 for 20 min). To test whether any differences in eIF3a protein levels could be the reason for the reduced interaction between Sty1 and eIF3a after stress, we performed Western blotting on total proteins. However, the S. pombe eIF3a protein levels did not change after oxidative or osmotic stress (Fig. 2D).
The interactions of Sty1 with eEF2 and eIF3a are regulated during nitrogen starvation.
The Sty1 pathway is also known to be involved in nutrient sensing and the meiotic program. Sty1 is phosphorylated and maximally activated at 30 to 60 min after nitrogen withdrawal, while the total Sty1 protein level is constant (35). To find out if this has any effect on the Sty1-eIF3a and Sty1-eEF2 interactions, we subjected cells growing in early log phase to nutrient stress by resuspending them in minimal medium without nitrogen and collected samples at various time points thereafter. For both eEF2 and eIF3a, the interaction with Sty1 changes during nutrient stress. The Sty1-eIF3a interaction was maintained for up to 30 min after nitrogen deprivation and then disappeared (Fig. 3B). Western analysis of eIF3a in the same time interval revealed a decrease after 15 min; eIF3a was present up to 60 min after nitrogen deprivation and then vanished (Fig. 3B). The eEF2 protein level was unchanged after nutrient limitation except for a small reduction at the last time point, 120 min. This does not reflect general protein degradation after nitrogen withdrawal, since both α-tubulin and Cdc2 levels, which have previously been shown to be constant under these conditions (15), did not change (Fig. 3A and B, lower panels). The Sty1-eEF2 interaction initially grew stronger and peaked at 30 min after nitrogen withdrawal (Fig. 3A). At this time interval, the eEF2 level remained unchanged, indicating that this reflects a true strengthening of the interaction.
FIG. 3.
The intensities of the Sty1-eEF2, Sty1-eIF3a, and Sty1-Mkp1 interactions change after nitrogen deprivation. Strains expressing eEF2-HA and Sty1-myc (A), eIF3a-HA and Sty1-myc (B), or Mkp1-HA and Sty1-myc (C) were grown to an OD595 of ∼0.25 in YES medium. After a brief centrifugation, the cells were resuspended in EMM minimal medium without NH4Cl. Protein samples were prepared and loaded as for Fig. 1 at the times indicated after transfer to nitrogen-free medium. (A and B) Top rows, Sty1-myc coprecipitated with eEF2-HA (A) or eIF3a-HA (B). Second rows, Western blots from the same protein preparations as above, to show the total amount of eEF2-HA (A) or eIF3a-HA (B). Third and fourth rows, Western blots from the same protein preparations probed for α-tubulin and Cdc2 as loading controls. (C) Sty1-myc coprecipitated with Mkp1-HA.
The Sty1 interaction with Mkp1 peaks at the time of maximal Sty1 activation after nitrogen deprivation.
We have previously identified Mkp1 as a downstream target of Sty1 that is involved in nutrient sensing in S. pombe. Mkp1 is dephosphorylated after nitrogen withdrawal (3), and to see if this influenced the interaction with Sty1, we performed co-IP of Mkp1-HA and detected Sty1-myc after transferring the cells to nitrogen-free minimal medium. The interaction transiently increased to reach a maximum at 30 to 45 min, the time of Sty1 activation (35), and fell back to normal levels after 60 min (Fig. 3C).
Sty1, but not Mkp1 or Mkp2, contributes to recovery of general translation after osmotic and oxidative stress or nitrogen deprivation.
Oxidative, osmotic, and nutrient stresses cause down-regulation of overall protein translation in yeast (9). In S. pombe, the stress-activated protein kinase pathway is important for supporting translation initiation and also for the translational adaptation during osmotic and oxidative stress (12). To further investigate Sty1 and the two Sty1-activated protein kinases Mkp1 and Mkp2 and their role in regulating translation after stress, we performed polysomal profile analyses of wild-type and sty1Δ, mkp1Δ, mkp2Δ, or mkp1Δ mkp2Δ mutant cells as well as cells overexpressing Mkp1 or Mkp2 from the nmt1 promoter.
Wild-type cells treated with 0.6 M KCl responded very rapidly, inhibiting the translation within 5 min, resulting in heavily reduced polysome levels (26%) and an increase in the free ribosome level (Fig. 4A and B). This transient translational response to osmotic stress was already reversed after 20 min, with almost restored (85%) polysome levels. In sty1 cells, the translational efficiency dropped even more drastically, with polysomes almost totally absent (below 10%) and an even higher accumulation of free ribosomes. In contrast to wild-type cells, sty1Δ cells did not recover from the translational shutdown after osmotic stress, as at 20 min after KCl addition there was still no increase in the polysomal fraction (Fig. 4A and B).
In oxidative stress (1 mM H2O2), polysome profiles of wild-type cells displayed a continuous lowering of translational efficiency with time (Fig. 5). The picture in sty1Δ mutants was similar, but there the translational decrease was even more pronounced.
FIG. 5.
Polysomal contents of wild-type and sty1 cells after oxidative stress. 972h− and sty1 cells were grown to an OD595 of ∼0.5 and exposed to 1 mM H2O2. Polysomal extracts were prepared and analyzed as for Fig. 4. (A) Polysome profiles obtained at three different times after oxidative stress as for Fig. 4A. (B) Relative polysome ratios at five times after oxidative stress as for Fig. 4B.
We also tested the effect on translation after nitrogen starvation in both wild-type and sty1 cells (Fig. 6). Wild-type cells showed a 50% reduction in polysome ratio at 1 h. At this time, however, recovery of translation efficiency started, and at 3 h without external nitrogen, it had increased almost to the same level as before stress was applied. In the sty1Δ mutant, polysome levels kept decreasing for 2 h and dropped to 25% of the starting value. Some recovery was seen only at 3 h, and the polysomal ratio stayed below 50%. This again indicates an important role for Sty1 both in maintaining translation immediately after stress and in long-term adaptation of translation to nitrogen-poor conditions. To rule out that the recovery of polysome content was due to a selective inhibition of translation elongation, which would result in decreased ribosome runoff in the absence of increased translation, we measured protein synthesis by metabolic labeling. The results showed that amino acid uptake and incorporation into protein roughly paralleled the recovery in polysomal levels seen at later time points, including the delay observed for sty1 mutant cells (see Fig. S1 in the supplemental material).
Finally, we wanted to investigate whether the Sty1 binding MAPKAPK Mkp1 or Mkp2 had an influence on translational efficiency. Neither the mkp1Δ mutant, the mkp2Δ mutant, nor the mkp1Δ mkp2Δ double mutant showed any difference in the translation response to osmotic or oxidative stress or to nitrogen deprivation. The same was true for strains overexpressing either of the kinases (see Fig. S2 in the supplemental material). Thus, it seems plausible that the Sty1 translational effect is not mediated by the downstream MAPKAPK Mkp1 or Mkp2.
Atf1 is required for maintenance of translation after hyperosmotic shock but not after oxidative or nutrient stress.
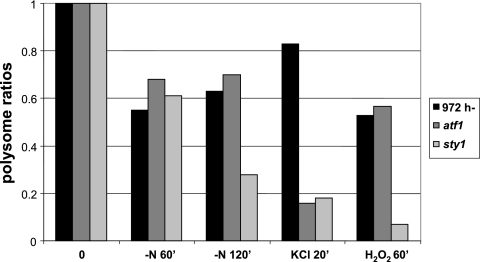
We wanted to see if the transcription factor Atf1, a major target of Sty1 in the fission yeast stress response, was involved also in regulation of global translation. Polysomal profiles from atf1Δ mutants showed that loss of translation activity after KCl was as profound as for sty1 mutants at 20 min, when polysomal levels had recovered in the wild type but were close to minimal in the two mutants (Fig. 7). By contrast, the polysomal content in atf1Δ mutants exposed to 1 mM H2O2 or to nitrogen starvation was similar to that in wild-type cells (Fig. 7).
FIG. 7.
Polysomal contents of atf1 mutant cells in different stress conditions. Cells were grown and lysates were prepared as for Fig. 4 to 6. Values for wild-type or sty1 cells are taken from Fig. 4 to 6 for comparison. Cells were exposed to nitrogen deprivation, 0.6 M KCl, or 1 mM H2O2 for the indicated times.
Phosphorylation and amount of eIF3a are reduced in sty1 mutant cells.
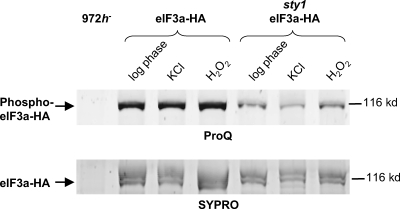
In view of the physical interactions between Sty1 and translation factors and their variation during stress conditions, we wanted to investigate the status of eIF3a in mutant cells lacking Sty1. As seen in Fig. 2C, the level of eIF3a protein is much lower in sty1Δ mutants than in the wild-type background; it should be noted that we observed the eIF3a level to vary considerably with the growth phase of the culture. We also compared phosphorylation of eIF3a in wild-type and sty1Δ mutant cells that were untreated or exposed to osmotic, oxidative, or nutrient stress (Fig. 8). Phosphorylation was found to be reduced in sty1Δ mutants relative to wild-type cells. This difference was seen in untreated cells and was accentuated after H2O2 treatment, where eIF3a phosphorylation was somewhat increased in wild-type cells (Fig. 8).
FIG. 8.
Phosphorylation of eIF3a in wild-type and sty1 mutant cells. Wild-type (EA128) or sty1 mutant (DN7) cells expressing eIF3a-HA were harvested before treatment or after treatment with stress agents as indicated. Nondenaturing protein lysates were prepared as for Fig. 1 to 3, and HA-tagged eIF3a was purified with HA probe. Approximately equal amounts of total eIF3a were loaded in each lane. The position of a protein size marker is shown to the right. The eIF3a band is indicated; the band visible just above it in the SYPRO staining corresponds to eIF3c copurifying under native conditions, which migrates slightly slower than eIF3a (46). Upper panel, phosphorylated eIF3a detected with ProQ Diamond phosphoprotein stain. Lower panel, total eIF3a detected with SYPRO Ruby protein stain.
Posttranslational regulation of eEF2 upon stress and nitrogen starvation in S. pombe.
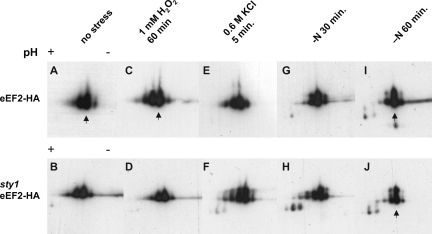
To test whether S. pombe eEF2 is posttranslationally modified after stress, sty1+ and sty1Δ strains tagged with eEF2-HA were grown to early log phase and subjected to 1 mM H2O2, 0.6 M KCl, or nitrogen deprivation as earlier. Proteins were separated on 2D gels and detected with anti-HA antibodies. As seen in Fig. 9A, at least four isoforms of eEF2-HA with the same apparent molecular weight but different pIs, in the interval from pI 6 to 6.5, were detected in undisturbed wild-type cells. The major form had the second highest pI. In the sty1Δ mutant strain, the appearance of eEF2 isoforms was similar (Fig. 9B). After nitrogen starvation, there was a relative increase in the amount of the most abundant isoform at the expense of spots with higher or lower pI. This shift started earlier (at 30 min) and was more pronounced in wild-type (Fig. 9G and I) than in sty1Δ (Fig. 9 H and J) cells. We examined HA-tagged eEF2 by 2D analysis in an mkp1Δ deletion strain but found no differences compared to the wild type (data not shown).
FIG. 9.
2D gel electrophoresis analysis of eEF2 isoforms after stress treatments. Wild-type (upper row) or sty1 mutant (lower row) cells expressing HA-tagged eEF2 were treated as indicated. Lysates were prepared and proteins separated on 2D gels, blotted, and probed with anti-HA antibodies. The arrows (A, C, I, and J) point to the predominant isoform.
DISCUSSION
We report physical interactions between two components of the translation machinery, eEF2 and eIF3a, and the stress-activated MAPK Sty1. eEF2 interacts with Sty1 in log-phase-growing cells, and the interaction is stable after oxidative or hyperosmotic stress. During nitrogen starvation, the interaction reaches a maximum after 30 min and then disappears. We also find eIF3a bound to Sty1 in log-phase cells; however, this interaction decreases after stress. In sty1 mutants, both the total amount and the degree of phosphorylation of eIF3a are reduced. The fraction of Sty1 engaged in binding to translation factors is significant. Since the eIF3a homolog in S. cerevisiae is about 10-fold more abundant than the Sty1 homolog (14), it is reasonable to estimate that on the order of 20% of Sty1 is engaged in associating with eIF3a. In contrast to the readily detected interaction of the translation factors with Sty1, we found no significant binding of either to Mkp1. Recovery of translation after stress is impaired in sty1 mutants but not in mkp1 mutants.
The impact of stress on translation efficiency and the involvement of the Sty1 pathway have been under investigation previously (12). The interpretation was that Sty1 reduces the phosphorylation level of eIF2α, which is phosphorylated and inactivated after various stresses in S. pombe by three different eIF2α kinases, Hri1, Hri2, and Gcn2 (45). As the effects on eIF2α phosphorylation are dependent on the transcription factor Atf1, this probably occurs through indirect transcriptional effects (12). Our findings verify the important role of Sty1, but with quantitative modifications. A plausible explanation for the differences is that we have used a sty1Δ deletion allele, whereas Dunand-Sauthier et al. (12) used spc1-m13 (13), which may be a hypomorph rather than a true null allele.
We have demonstrated a requirement for the transcription factor Atf1 for maintenance of polysome levels after hyperosmotic stress but not oxidative stress or nitrogen starvation. Atf1 has previously been shown to be required for transcriptional induction of the ctt1+ gene after treatment with 6 mM but not with 1 mM of H2O2 (31); Atf1 is also needed for induction of ctt1+ in hyperosmotic stress (44). This is consistent with the subdivision of the Sty1-dependent stress response on the translation level that we observe. The mechanism for this is not clear, as our data do not discriminate between a requirement for Atf1 for transcription of target genes essential for translation during stress on one hand and a requirement for the protein synthesis apparatus for translation of the atf1+ message during stress on the other.
In all stress conditions examined, we observe reduced polysome levels, with the amounts in sty1 mutants being far lower than those in wild-type cells. This fact, in combination with the reduction in protein synthesis measured by amino acid incorporation, indicates that a major effect is on the initiation step. We find the amount as well as the degree of phosphorylation of the initiation factor eIF3a to be reduced in sty1Δ cells. This could be due to destabilization of the unphosphorylated protein species or to the absence of an interaction partner. There are indications from mammalian cell phosphorylation (29) as well for wide variations in eIF3a levels. Thus, the amount of the human homolog of eIF3a, p170, is elevated in cancer cells. The level of p170 is cell cycle regulated, with a maximum in G1/S. Experimentally modulating p170 expression levels has little effect on the overall translation efficiency, but there is a subset of mRNA for which translation is highly dependent on eIF3a levels (10).
After 1 h without external nitrogen, the polysomal ratio in wild-type cells is roughly halved. At this time, eIF3a almost disappears and protein synthesis resumes even though no nitrogen has been added. This is suggestive of a redirection of the translation apparatus to other mRNA sets, driven by intracellular signaling events, and with a different setup of translation factors. The findings with mammalian cells and our results with fission yeast show that it is possible to maintain general translation without eIF3a. S. pombe eIF3 exists in two different forms, both consisting of the five core subunits plus eIF3f but containing either eIF3h and eIF3m or eIF3d and eIF3e. The first complex is required for global protein synthesis, whereas the second interacts with a small subset of mRNAs, thereby regulating their translation (46). Other changes in eIF3 composition may occur during nutrient stress; whether another protein replaces eIF3a under nitrogen starvation is unknown.
In addition to regulation of the initiation step, there are arguments why regulation of translation elongation is desirable. A temporary change in nutrient status or an environmental insult can be dealt with by decreasing elongation rates, preserving polysomes intact, and allowing protein synthesis to resume quickly as soon as conditions improve (6). This may be an important aspect of regulation of translation upon transient stress that is defective in sty1 mutants, as the main defect seems to be in the recovery phase. The eEF2-Sty1 interaction could be central in this regard, placing Sty1 near critical targets during the elongation phase. After nitrogen deprivation, this interaction peaks in the same time interval as the Mkp1-Sty1 interaction (Fig. 3) and Sty1 kinase activity (35), indicating that this is a pivotal time for S. pombe cells adapting to nutrient stress.
We find changes in eEF2 isoforms after stress. Phosphorylation of mammalian eEF2 T56 by eEF2 kinase inhibits elongation by preventing binding to the ribosome. S. cerevisiae eEF2 is subject to Rck2-dependent phosphorylation at T57 after osmotic shock (39). Fission yeast eEF2 lacks T57, but an additional potential phosphorylation site at T59 is conserved between S. cerevisiae, S. pombe, and mammals. Isoforms of mammalian eEF2 have been detected on 2D gels using specific antibodies (32). The pattern of eEF2 isoforms with the same apparent molecular weight but different pIs (horizontally shifted) is virtually identical to what we detect for S. pombe eEF2 (Fig. 9), and using antibodies specific for eEF2 phosphorylated at T56, it was inferred that they correspond to multiply phosphorylated forms. The previous results do not suggest direct phosphorylation of eEF2 by the MAPK extracellular signal-regulated kinase (ERK) but rather suggest an indirect regulation, since phosphorylated T56 increases, rather than decreases, after ERK inhibition.
On a 2D gel detected with anti-HA against tagged eEF2, we identify four to five horizontally shifted spots, which we interpret as multiply phosphorylated forms. After nitrogen withdrawal, the amounts of the less abundant, horizontally shifted isoforms decrease, and after 60 min the major form accounts for the majority (Fig. 9I and J). We judge it likely that, as for the ERK dependence in mammalian cells (32), the Sty1 dependence of these changes between horizontally shifted forms is indirect and not the result of direct phosphorylation of eEF2 by Sty1, since there are no highly phosphorylated forms completely absent in sty1 mutants.
While we find rather strong interactions between Sty1 and translation factors and a major impact of the sty1 mutation on recovery of translation after stress, we see no corresponding physical or functional links between Mkp1 and translation in fission yeast. In mammalian cells, a role has been demonstrated for the p38-activated kinase MAPKAPK-2 both in posttranscriptional control of translation and mRNA stability (22, 28) and in cell cycle G2/M control through inhibitory phosphorylation of the Cdc2-activating phosphatase Cdc25 (24). Rck1 and Rck2, the budding yeast homologs of Mkp1 and Mkp2, were identified as suppressors of cell cycle checkpoint mutations in S. pombe, indicating a role in cell cycle control (7), and indeed Mkp1 was later shown to phosphorylate Cdc25 in a way analogous to that in mammalian cells (23). In budding yeast, Rck2 is implicated in translation control (37, 39). On the other hand, Rck2 has not been shown to exert a comparable effect on the G2/M transition, possibly because phosphorylation of Mih1, the Cdc25 homolog, does not cause a significant G2/M delay (34). It thus appears as though whereas both tasks are fulfilled by mammalian MAPKAPKs, the budding yeast homolog is specialized for translation control and the fission yeast homolog Mkp1 for cell cycle control. This might be explained by the above-mentioned absence of the otherwise conserved threonine phosphorylation site (T57) in fission yeast eEF2.
We have demonstrated that physical interactions between Sty1 and translation factors do take place, but not where the interaction occurs. This could be in the context of ongoing protein synthesis but also in cytoplasmic complexes of translationally inactive mRNA and proteins, stress granules, or processing bodies. Structures reminiscent of the stress granules described in plant or mammalian cells (21) have been reported for S. pombe (11). Mammalian stress granules have been shown to contain eIF3 (20), opening the possibility that Sty1-mediated regulation of translation through eIF3a occurs through the process of transferring mRNA between translationally active polysomes and inactive granules. In mammalian cells it has been shown that the drug emetine, which blocks dissociation of polysomes during stress, also blocks formation of stress granules (19). In budding yeast, rck2-kd, encoding a dominant-negative allele of the MAPKAPK Rck2, blocks dissociation of polysomes in a way reminiscent of emetine action (37).
The mechanisms by which the Sty1 pathway influences translation in fission yeast require additional analysis. However, our results emphasize the need of the cell to redirect the translational machinery to recover after stress and that Sty1 is required for this process.
Supplementary Material
Acknowledgments
Thanks are due to Malin Hult for technical advice on polysome separation.
This work was supported by the Swedish Research Council (2003-3189) and the Swedish Cancer Fund (2163-B05-16XAB).
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alemany, V., M. Sanchez-Piris, O. Bachs, and R. Aligue. 2002. Cmk2, a novel serine/threonine kinase in fission yeast. FEBS Lett. 52479-86. [DOI] [PubMed] [Google Scholar]
- 2.Anand, M., K. Chakraburtty, M. J. Marton, A. G. Hinnebusch, and T. G. Kinzy. 2003. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 2786985-6991. [DOI] [PubMed] [Google Scholar]
- 3.Asp, E., and P. Sunnerhagen. 2003. Mkp1 and Mkp2, two MAPKAP-kinase homologues in Schizosaccharomyces pombe, interact with the MAP kinase Sty1. Mol. Genet. Genomics 268585-597. [DOI] [PubMed] [Google Scholar]
- 4.Bilsland, E., C. Molin, S. Swaminathan, A. Ramne, and P. Sunnerhagen. 2004. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 531743-1756. [DOI] [PubMed] [Google Scholar]
- 5.Bilsland-Marchesan, E., J. Ariño, H. Saito, P. Sunnerhagen, and F. Posas. 2000. Rck2 kinase is a substrate for the osmotic-stress activated MAP kinase Hog1. Mol. Cell. Biol. 203887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, G. J., and C. G. Proud. 2002. Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 2695360-5368. [DOI] [PubMed] [Google Scholar]
- 7.Dahlkvist, A., G. Kanter-Smoler, and P. Sunnerhagen. 1995. The RCK1 and RCK2 protein kinase genes from Saccharomyces cerevisiae suppress cell cycle checkpoint mutations in Schizosaccharomyces pombe. Mol. Gen. Genet. 246316-326. [DOI] [PubMed] [Google Scholar]
- 8.Dahlkvist, A., and P. Sunnerhagen. 1994. Two novel deduced serine/threonine protein kinases from Saccharomyces cerevisiae. Gene 13927-33. [DOI] [PubMed] [Google Scholar]
- 9.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108545-556. [DOI] [PubMed] [Google Scholar]
- 10.Dong, Z., L. H. Liu, B. Han, R. Pincheira, and J. T. Zhang. 2004. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene 233790-3801. [DOI] [PubMed] [Google Scholar]
- 11.Dunand-Sauthier, I., C. Walker, C. Wilkinson, C. Gordon, R. Crane, C. Norbury, and T. Humphrey. 2002. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell 131626-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunand-Sauthier, I., C. A. Walker, J. Narasimhan, A. K. Pearce, R. C. Wek, and T. C. Humphrey. 2005. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell 41785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaits, F., K. Shiozaki, and P. Russell. 1997. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J. Biol. Chem. 27217873-17879. [DOI] [PubMed] [Google Scholar]
- 14.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425737-741. [DOI] [PubMed] [Google Scholar]
- 15.Grallert, B., S. E. Kearsey, M. Lenhard, C. R. Carlson, P. Nurse, E. Boye, and K. Labib. 2000. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell Sci. 1131447-1458. [DOI] [PubMed] [Google Scholar]
- 16.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 61099-1108. [DOI] [PubMed] [Google Scholar]
- 17.Hinnebusch, A. G. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31553-562. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 122-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 1511257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1471431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. D. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 194-97. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Aviles, S., M. Grande, M. Gonzalez, A. L. Helgesen, V. Alemany, M. Sanchez-Piris, O. Bachs, J. B. Millar, and R. Aligue. 2005. Inactivation of the Cdc25 phosphatase by the stress-activated Srk1 kinase in fission yeast. Mol. Cell 1749-59. [DOI] [PubMed] [Google Scholar]
- 24.Manke, I. A., A. Nguyen, D. Lim, M. Q. Stewart, A. E. Elia, and M. B. Yaffe. 2005. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G(2)/M transition and S phase progression in response to UV irradiation. Mol. Cell 1737-48. [DOI] [PubMed] [Google Scholar]
- 25.Martin, V., M. A. Rodriguez-Gabriel, W. H. McDonald, S. Watt, J. R. Yates III, J. Bähler, and P. Russell. 2006. Cip1 and Cip2 are novel RNA-recognition-motif proteins that counteract Csx1 function during oxidative stress. Mol. Biol. Cell 171176-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mita, K., M. Morimyo, K. Ito, K. Sugaya, K. Ebihara, E. Hongo, T. Higashi, Y. Hirayama, and Y. Nakamura. 1997. Comprehensive cloning of Schizosaccharomyces pombe genes encoding translation elongation factors. Gene 187259-266. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 28.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 2773065-3068. [DOI] [PubMed] [Google Scholar]
- 29.Pincheira, R., Q. Chen, Z. Huang, and J. T. Zhang. 2001. Two subcellular localizations of eIF3 p170 and its interaction with membrane-bound microfilaments: implications for alternative functions of p170. Eur. J. Cell Biol. 80410-418. [DOI] [PubMed] [Google Scholar]
- 30.Prendergast, G. C. 2003. Signal transduction: putting translation before transcription. Cancer Cell 4244-245. [DOI] [PubMed] [Google Scholar]
- 31.Quinn, J., V. J. Findlay, K. Dawson, J. B. Millar, N. Jones, B. A. Morgan, and W. M. Toone. 2002. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 13805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, E. C., K. Hammond, A. M. Traish, K. A. Resing, and N. G. Ahn. 2006. Identification of G2/M targets for the MAP kinase pathway by functional proteomics. Proteomics 64541-4553. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Gabriel, M. A., G. Burns, W. H. McDonald, V. Martin, J. R. Yates III, J. Bähler, and P. Russell. 2003. RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 226256-6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell, P., S. Moreno, and S. I. Reed. 1989. Conservation of mitotic controls in fission and budding yeasts. Cell 57295-303. [DOI] [PubMed] [Google Scholar]
- 35.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 102276-2288. [DOI] [PubMed] [Google Scholar]
- 36.Smith, D. A., W. M. Toone, D. Chen, J. Bähler, N. Jones, B. A. Morgan, and J. Quinn. 2002. The Srk1 protein kinase is a target for the Sty1 stress-activated MAPK in fission yeast. J. Biol. Chem. 27733411-33421. [DOI] [PubMed] [Google Scholar]
- 37.Swaminathan, S., T. Masek, C. Molin, M. Pospisek, and P. Sunnerhagen. 2006. Rck2 is required for reprogramming of ribosomes during oxidative stress. Mol. Biol. Cell 171472-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tasto, J. J., R. H. Carnahan, W. H. McDonald, and K. L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18657-662. [DOI] [PubMed] [Google Scholar]
- 39.Teige, M., E. Scheikl, V. Reiser, H. Ruis, and G. Ammerer. 2001. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. USA 985625-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uesono, Y., and E. A. Toh. 2002. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 27713848-13855. [DOI] [PubMed] [Google Scholar]
- 41.Vasudevan, S., N. Garneau, D. Tu Khounh, and S. W. Peltz. 2005. p38 mitogen-activated protein kinase/Hog1p regulates translation of the AU-rich-element-bearing MFA2 transcript. Mol. Cell. Biol. 259753-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 71191-1200. [DOI] [PubMed] [Google Scholar]
- 43.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24437-440. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 102289-2301. [DOI] [PubMed] [Google Scholar]
- 45.Zhan, K., J. Narasimhan, and R. C. Wek. 2004. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics 1681867-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, C., F. Arslan, S. Wee, S. Krishnan, A. R. Ivanov, A. Oliva, J. Leatherwood, and D. A. Wolf. 2005. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.