Abstract
In Saccharomyces cerevisiae, Och1p and Mnn9p mannosyltransferases are localized in the cis-Golgi. Attempts to live image Och1p and Mnn9p tagged with green fluorescent protein or red fluorescent protein, respectively, using a high-performance confocal laser scanning microscope system resulted in simultaneous visualization of the native proteins in a living cell. Our observations revealed that Och1p and Mnn9p are not always colocalized to the same cisternae. The difference in the dynamics of these mannosyltransferases may reflect differences in the mechanisms for their retention in the cis-Golgi, since it has been reported that Mnn9p cycles between the endoplasmic reticulum and the cis-Golgi whereas Och1p does not (Z. Todorow, A. Spang, E. Carmack, J. Yates, and R. Schekman, Proc. Natl. Acad. Sci. USA 97:13643-13648, 2000). We investigated the localization of chimeric proteins of Mnn9p and Och1p in sec12 and erd1 mutant cells. A chimeric protein, M16/O16, which consists of the N-terminal cytoplasmic region of Mnn9p and the transmembrane and luminal region of Och1p, behaved like Mnn9p, suggesting that the N-terminal cytoplasmic region is important for the intracellular dynamics of Mnn9p. This observation is supported by results from subcellular-fractionation experiments. Mutational analysis revealed that two arginine residues in the N-terminal region of Mnn9p are important for the chimeric protein to cycle between the endoplasmic reticulum and the Golgi apparatus.
In eukaryotic cells, secretory proteins that are synthesized in the endoplasmic reticulum (ER) are transported to their final destinations through the Golgi apparatus. The Golgi apparatus is composed of cis, medial, and trans cisternae, and secretory proteins must traverse the Golgi apparatus in the cis-to-trans direction. It has long been debated how secretory cargo proteins pass through the cis-to-trans cisternae (26). Progress in live-imaging technology has contributed to resolving this issue. Recently, two groups have directly observed the replacement of one Golgi apparatus protein with another in individual cisternae (15, 16) and concluded that cargo proteins are transported as proposed by the cisternal-maturation model, in which secretory cargo proteins remain in the cisternae as the cisternae mature from cis to trans while resident Golgi apparatus enzymes, which variously modify the secretory proteins, move backwards.
Glycosyltransferases are major Golgi apparatus-resident enzymes. In Saccharomyces cerevisiae, secretory proteins are often modified with mannan, which is a long, branched polymer of approximately 200 mannoses. α-1,6-Mannosyltransferase activity is provided by Och1p (21, 23) and two enzyme complexes, M-Pol I (V complex) and M-Pol II (A complex) (10, 12). Och1p adds a single mannose to eight mannose-containing core oligosaccharides synthesized in the ER (24). The mannan backbone structure is synthesized by M-Pol I and M-Pol II. M-Pol I consists of Mnn9p and Van1p, while M-Pol II consists of Mnn9p, Anp1P, Hoc1p, Mnn10p, and Mnn11p. The M-Pol I complex adds approximately 10 mannoses to the mannose residue added by Och1p to form an α-1,6-linked mannan backbone, and then M-Pol II elongates the mannan backbone by adding more mannoses (9, 10, 12). As is often the case with Golgi apparatus-localized glycosyltransferases, Och1p and all components of M-Pol I and M-Pol II are type II integral membrane proteins, which are composed of a short cytoplasmic segment in the N-terminal region, a single transmembrane domain, and a large luminal region containing a catalytic domain and a stem region lying between the transmembrane and the catalytic domain. Many studies have been performed using deletion and fusion proteins to determine the domain critical for Golgi apparatus localization. These studies clarified that the lengths of the transmembrane domain (3, 19), the luminal domain in the C-terminal region (6, 33), and the cytoplasmic region (18) are important for the efficient Golgi apparatus localization of Golgi apparatus-localized glycosyltransferases, suggesting that there are different kinds of Golgi apparatus localization mechanisms. The cisternal-maturation model indicates the necessity for an intra-Golgi apparatus or Golgi apparatus-ER retrograde transport pathway for glycosyltransferases. Although it is presently unclear how Golgi apparatus enzymes move from each Golgi apparatus subcompartment, the involvement of COPI vesicles and tubules has been suggested (16). It has been reported that Och1p travels as far as the trans-Golgi network and then is transported back to the cis-Golgi (8). A study using sec12-4 mutant cells, in which vesicle budding from the ER is defective, showed that Mnn9p, Anp1p, and Van1p cycle back and forth between the Golgi apparatus and the ER, while Och1p does not (28, 32). On the other hand, another study investigated the retrieval mechanism of Och1p using sec23-1 mutant cells, which are also defective in vesicle budding from the ER, and showed that Och1p is transported back to the ER (11). It is currently unclear what causes the different results regarding Och1p dynamics between sec12-4 mutant and sec23-1 mutant cells.
Two ER retrieval signals involved in the cycling process from the Golgi apparatus to the ER, which are conserved from yeasts to mammals, have been characterized. One is the KDEL (mammal)/HDEL (yeast) sequence. Luminal ER-resident proteins harboring the KDEL/HDEL sequence at the C-terminal end are recognized by a specific receptor at the Golgi apparatus and then are returned to the ER (20). The other ER-retrieval signal is a dilysine motif (KKXX or KXKXX) that is usually found in the C-terminal cytosolic tail of type I membrane proteins (4). In yeast, the ER-resident subunit of oligosaccharyl transferase Wbp1p (5) and the lectin-binding protein ERGIC-53 homologue Emp47p (28) have been identified as the proteins harboring a dilysine motif. The KKXX motif has been reported to be recognized by COPI coat proteins (13). In mammalian cells, an arginine-based signal has also been reported to be involved in ER localization of multimeric membrane proteins (17). This signal was originally found in the cytoplasmic region of the invariant chain Ii p35 of the major histocompatibility complex class II, which is a type II membrane protein (29). Although several hallmarks of the arginine-based ER localization signal have been revealed, details of the recognition mechanism, such as whether one particular COPI subunit acts as the direct receptor for transport to the ER, remain obscure. It is also unclear if these ER retention/retrieval signals are involved in the correct localization of glycosyltransferases in the Golgi apparatus.
Advances in live-cell-imaging technology now enable the visualization of Golgi apparatus-localized mannosyltransferases without requiring overexpression of the target proteins. In this study, we used a newly developed confocal microscope system to clarify how the intracellular dynamics of Och1p and Mnn9p differ from each other. This confocal system combines the spinning-disk (Nipkow disk) confocal scanning method and a high-gain avalanche rushing amorphous photoconductor camera. We attempted to identify the regions of Mnn9p and Och1p that determine their dynamics. The results obtained with an Och1p/Mnn9p chimeric protein indicate that the N-terminal cytoplasmic region of Mnn9p is crucial for cycling the chimeric protein from the Golgi apparatus to the ER, further confirming the importance of two arginine residues in the N-terminal region.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in YPD (2% peptone, 1% yeast extract, and 2% glucose) or in SC (0.67% yeast nitrogen base without amino acids, 2% glucose, and required supplements) (30) medium. C-terminal tagging of Och1p with green fluorescent protein (GFP) and Mnn9p with monomeric red fluorescent protein (mRFP) were carried out by a PCR-based method using an integrative cassette derived from the plasmid pFA6a-GFP(S65T)-His3MX6 or pFA6a-mRFP5-His3MX6 as a template (14).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Origin or Reference |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | EUROSCARF |
| BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | EUROSCARF |
| YMO47 | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 OCH1-GFP::his5+ | This study |
| YMO50 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 MNN9-mRFP::his5+ | This study |
| YMO56 | MATα his3Δ1 leu2Δ0 ura3Δ0 OCH1-GFP::his5+MNN9-mRFP::his5+ | This study |
| YMO63 | MATahis3Δ1 leu2Δ0 ura3Δ0 erd1Δ::kanMX6 OCH1-GFP::his5+MNN9-mRFP::his5+ | This study |
| YMO65 | MATahis3Δ1 leu2Δ0 ura3Δ0 erd1Δ::kanMX6 MNN9-mRFP::his5+ | This study |
| YMO59 | MATahis3 leu2 ura3 (suc or SUC, gal2 or GAL2) sec12-4 OCH1-GFP::his5+MNN9-mRFP::his5+ | This study |
| YMO60 | MATahis3 leu2 ura3 (suc or SUC, gal2 or GAL2) sec12-4 MNN9-mRFP::his5+ | This study |
| YMO69 | MATahis3 leu2 ura3 (suc or SUC, gal2 or GAL2) sec12-4 mnn9(R9/11A)-mRFP::his5+ | This study |
| YS57-2C | MATaoch1::LEU2 leu2 ura3 trp1 his1 his3 | 21 |
| MBY10-7A | MATaleu2-3,112 ura-3-52 trp1-289 his3 his4 suc gal2 sec12-4 | 22 |
Yeast strain YMO56 was generated by crossing YMO47 with YMO50. YMO63 and YMO65 were generated by crossing YMO50 with erd1Δ::kanMX6 (EUROSCARF). YMO59 and YMO60 were segregants from a diploid generated by crossing MBY10-7A (22) with YMO56. YMO69 was constructed by chromosomal integration of pRS306-mnn9(R9/11A)ΔC into strain YMO50 after digestion by PmaCI.
Plasmid constructions.
To construct plasmid pFA6a-mRFP5-His3MX6, a DNA fragment coding for mRFP was amplified by PCR using plasmid pRSETB containing mRFP1 (kindly provided by Roger Tsien, University of California, San Diego) as a template and inserted into the PacI-AscI region of pFA6a-3HA-His3MX6 (14).
pMO36 (pRSOCH1-GFP) was constructed as follows. A fragment containing the open reading frame of OCH1 with a promoter and terminator was amplified from genomic DNA by a two-step PCR method and cloned between the SalI and HindIII sites in YEp352 (URA3 2μ). Next, the XbaI-XbaI fragment containing GFP was excised from pUC19-GFP and inserted into the XbaI site located just before the stop codon of OCH1 to generate pMO34 (YEpOCH1-GFP). The SalI-BglII fragment containing GFP-tagged OCH1 with a promoter and terminator was excised from pMO34 and inserted into the SalI-BamHI site of pRS316.
The plasmids carrying MNN9 and OCH1 chimeric genes were constructed as follows. An NheI site was inserted just before the initiation codon of OCH1 in pMO36 by using a Quick Change Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Next, the XbaI site located at the 3′ end of the GFP coding region in the pMO36 derivative was removed to generate pMO39. The fragments coding for the chimeric proteins M93/O80, M33/O37, and M16/O16 were amplified by a two-step PCR using genomic DNA as a template. To construct pRS316-M93/O80-GFP and pRS316-M33/O37-GFP, the amplified fragments were digested with NheI and XbaI and inserted into the NheI-XbaI site of pMO39. The fragment coding for M16/O16 was digested with NheI and BamHI, and inserted into the NheI-BamHI site of pMO39 to generate pRS316-M16/O16-GFP.
The pRS306-mnn9(R9/11A)ΔC plasmid was constructed as follows. The SalI-NheI region of pMO39 was replaced by a PCR fragment containing the promoter of MNN9, and the NheI-XbaI region of the plasmid was replaced by a PCR fragment containing the open reading frame of MNN9 to construct pRS316-MNN9-GFP. Next, the PCR fragment containing the coding sequence for the R9/11A mutation was cloned into the NheI-EcoRI region of pRS316-MNN9-GFP. A SalI-EcoRI fragment from this plasmid was inserted into pRS306 at SalI and EcoRI sites to generate pRS306-mnn9(R9/11A)ΔC.
Microscope observations.
Except for sec12-4 mutant cells, the cells were grown to exponential phase in SC medium at 25°C. The sec12-4 mutant cells were grown to exponential phase at 23°C, 100 μg/ml cycloheximide (CHX) (Nacalai Tesque, Kyoto, Japan) was added, and the culture was incubated at 23°C for a further 30 min. Then, the cells were incubated at 37.5°C for 40 min. Except for time-lapse observations, fluorescence images were obtained using a BX50 fluorescence microscope (Olympus, Tokyo, Japan) and photographed with a cooled charge-coupled-device camera (MicroMAX; Princeton Instruments, Trenton, NJ). The time-lapse images were observed using an IX71 fluorescence microscope (Olympus) equipped with a special high-performance confocal laser scanning unit (Yokogawa Electric, Tokyo, Japan), a beam splitter Dual-View Micro-Imager (Photometrics, Tucson, AZ), an image intensifier (Hamamatsu Photonics, Shizuoka, Japan), and a high-gain avalanche rushing amorphous photoconductor camera system (NHK Engineering Service and Hitachi Kokusai Electric, Tokyo, Japan). For simultaneous recording of green and red fluorescence, the Dual-View Micro-Imager was used to project green and red components side by side. The image intensifier was used to detect the weak fluorescent signals. The collected images were analyzed with IPLab software (Scanalytics, Fairfax, VA).
Subcellular fractionation.
Cells were grown in SC−Ura medium to logarithmic phase (optical density at 600 nm = 0.4 to 0.8) at 23°C and treated with 100 μg/ml CHX. After 30 min, the culture was divided in two; one portion was kept at 23°C, while the other was shifted to 37.5°C. Both cultures were then incubated for a further 40 min. Then, 10 mM NaN3 was added, the cultures were incubated for 5 min at their respective temperatures, and the cells were collected. The cells were resuspended in 0.1 M Tris-HCl (pH 9.4) buffer containing 10 mM dithiothreitol. After incubation at 30°C for 10 min, the cells were collected and resuspended in spheroplasting buffer (1.35% yeast nitrogen base, 10% glucose, and 1 M sorbitol). Zymolyase 100T (250 μg/ml; Seikagaku-Kogyo, Tokyo, Japan) was added to the cell suspension, and incubation was continued for 10 to 20 min at 30°C. The Zymolyase-treated samples were overlaid on 2.0 ml of 1.4 M sorbitol solution and centrifuged at 1,600 × g in a swinging-bucket rotor. The resultant spheroplasted cells (1 × 109 cells) were suspended in 1 ml of ice-chilled lysis buffer (20 mM HEPES-KOH, pH 6.8, 150 mM potassium acetate, 200 mM sorbitol, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Roche, Basel, Switzerland]) and were homogenized with about five strokes for 1 min at 1-min intervals in a Dounce homogenizer while being kept on ice. This homogenization process was repeated five times. The lysates were centrifuged at 500 × g for 5 min to remove unbroken cells, the supernatants were subjected to centrifugation at 10,000 × g for 15 min at 4°C, and the pellet was recovered as the P10 fraction. The supernatant was further centrifuged at 100,000 × g for 60 min at 4°C to generate a high-speed pellet (P100) and supernatant (S100). Aliquots from the P10, P100, and S100 fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. GFP was detected by anti-GFP monoclonal antibody JL-8 (1:5,000; BD Biosciences Clontech, Palo Alto, CA), followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) (1:20,000; Cell Signaling Technology, Danvers, MA). mRFP was detected with anti-mRFP polyclonal antibody (1:5,000) (31), followed by HRP-conjugated anti-rabbit IgG (1:20,000; Cell Signaling Technology). Och1p was detected with anti-Och1p polyclonal antibody (1:3,000) (23), followed by HRP-conjugated anti-rabbit IgG (1:5,000). Dpm1p was detected with anti-Dpm1p monoclonal antibody (1:2,000; Invitrogen, Carlsbad, CA), followed by HRP-conjugated anti-mouse IgG (1:10,000).
RESULTS
Och1p and Mnn9p are not always colocalized to the same cis-Golgi cisternae.
In the N-linked-glycan synthetic pathway, Och1p transfers α-1,6-linked mannose to the core oligosaccharide in the cis-Golgi. The M-Pol I complex, which is composed of Mnn9p and Van1p, and the M-Pol II complex, which is composed of Mnn9p, Anp1p, Hoc1p, Mnn10p, and Mnn11p, also provide α-1,6-mannosyltransferase activity in the cis-Golgi. The cis-Golgi localization of these mannosyltransferases has been demonstrated by indirect immunofluorescence (9, 27). However, it is unclear whether these enzymes always colocalize to the cis-Golgi with each other. While Och1p is involved in both core-type oligosaccharide and mannan synthesis, M-Pol I and M-Pol II are mainly involved in mannan synthesis. These results raised the possibility that the localization pattern of Och1p may be different from that of M-Pol I and M-Pol II. Therefore, we attempted to detect the patterns of localization of these enzymes simultaneously in living cells using a high-performance confocal laser scanning microscope system. This system can detect weak fluorescent signals within living cells and, further, can detect two distinct fluorescent signals at the same time. To compare the intracellular dynamics of Och1p with those of M-Pol I and M-Pol II, we selected Mnn9p, which is common to both components of M-Pol I and M-Pol II, from among the proteins contained in the complexes.
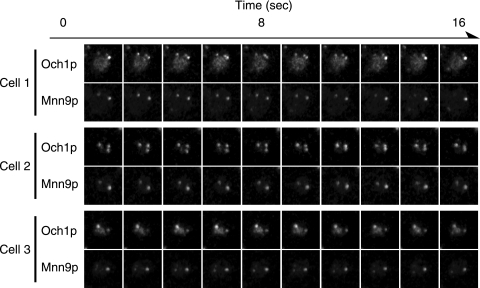
For live imaging, the coding sequences for GFP and mRFP were fused just upstream of the stop codon of the chromosomal OCH1 and MNN9 genes. This chromosome tagging had no obvious influence on the growth of the yeast, confirming that Och1p-GFP and Mnn9p-mRFP retained their original functions. Time-lapse observation revealed that the localization of Och1p-GFP is not always identical to that of Mnn9p-mRFP: some cis-Golgi cisternae contained Och1p-GFP but not Mnn9p-mRFP, and vice versa (Fig. 1; see Fig. S1 in the supplemental material). By microscope observation, we checked 201 Golgi apparatus cisternae and found that 38 cisternae (19% of the total counted) contained only Och1p and 12 cisternae (6%) contained only Mnn9p, while 151 cisternae (75%) contained both Och1p and Mnn9p. Therefore, we tentatively concluded that the localization mechanism of Och1p to the cis-Golgi is different from that of Mnn9p.
FIG. 1.
Dual-fluorescence observation of cis-Golgi-localized α-1,6-mannosyltransferases. After YMO56 cells were incubated at 25°C in SC medium, Och1p-GFP and Mnn9p-mRFP were observed at the same time with a high-performance confocal laser scanning microscope system. Images were taken at a rate of one frame per 1.6 seconds. Observations of three different cells are shown.
Chimeric protein M16/O16 behaves like Mnn9p.
In a previous report using sec12 mutant cells, in which the budding of COPII vesicles from the ER was blocked by using a restrictive temperature, Todorow et al. reported that Mnn9p cycles back and forth between the ER and the Golgi apparatus while Och1p cycles within the Golgi apparatus (32). We speculated that the difference between the observed dynamics of Och1p and those of Mnn9p might be caused by differences in retrograde transport mechanisms. Therefore, we further investigated the differences in cellular localization between Och1p and Mnn9p in sec12 mutant cells.
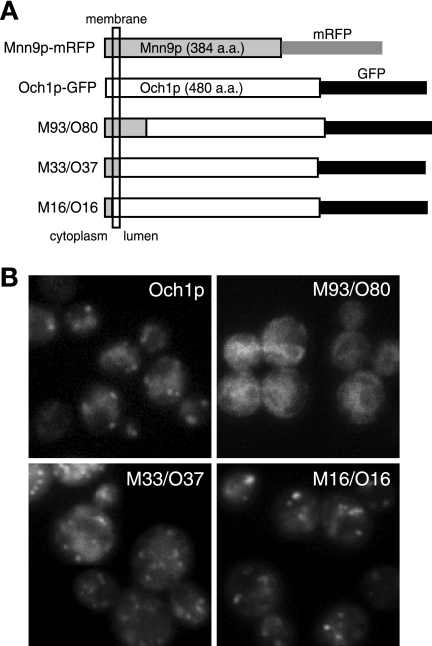
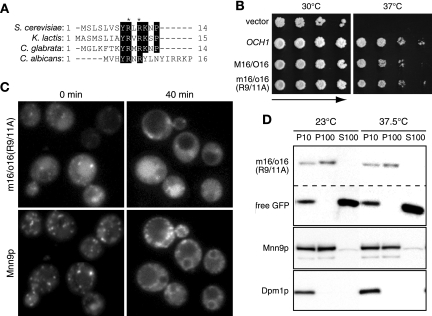
To investigate which region determines the recycling pathway of each protein, we constructed several chimeric proteins consisting of Och1p and Mnn9p. The plasmids expressing these chimeric genes, which were fused with the gene encoding GFP under the OCH1 promoter, were introduced into wild-type cells, and the localization of the chimeric proteins was observed. The chimeric protein M93/O80, consisting of the N-terminal stem region of Mnn9p and most of the catalytic region of Och1p, was mislocalized to the ER, suggesting that it is not folded correctly. Another chimeric protein, M33/O37, consisting of the N-terminal cytosolic and transmembrane regions of Mnn9p and the entire luminal region of Och1p, showed weak punctate staining patterns that are characteristic of Golgi apparatus localization. The chimeric protein M16/O16, in which the N-terminal cytosolic region of Och1p is substituted for that of Mnn9p but the native luminal region of Och1p is retained, showed a much clearer Golgi apparatus localization pattern (Fig. 2). Therefore, we focused on characterizing the M16/O16 chimeric protein further.
FIG. 2.
Visualization of Mnn9/Och1 chimeric proteins. (A) Schematic representation of chimeric constructs. Och1p was constructed from 480 amino acids (a.a.) and tagged with GFP at the C terminus. Mnn9p (384 amino acids) was tagged with mRFP at the C terminus. All chimeric proteins were tagged at their C termini and expressed under the OCH1 promoter on a low-copy-number (CEN) plasmid. The chimeric proteins were named for the region containing Och1p and Mnn9p. For example, M16/O16 means that the protein was constructed by fusing a peptide from the first to the 16th amino acids of Mnn9p with a polypeptide from the 16th to the 480th amino acids of Och1p. (B) BY4741 cells expressing each chimeric protein were incubated at 25°C in SC−Ura medium and observed under a fluorescence microscope.
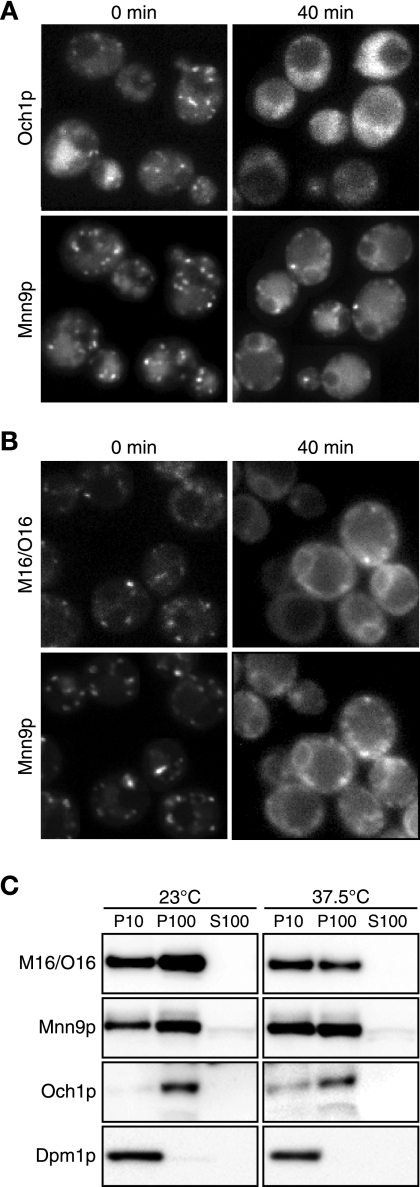
To investigate the intracellular dynamics of the M16/O16 chimeric protein, we employed a procedure using sec12 mutant cells. This procedure allowed investigation of whether the proteins preexisting in the Golgi apparatus are cycled back to the ER. The sec12 mutant cells were used, since they were treated with CHX before the restrictive temperature shift, thus preventing contributions from newly synthesized proteins. As reported previously, Och1p-GFP and Mnn9p-mRFP are distributed throughout cells with a punctate structure that is characteristic of the Golgi apparatus at the permissive temperature before being shifted to the restrictive temperature (Fig. 3A, 0 min). Due to the inhibition of protein synthesis by CHX, the Golgi apparatus-localized signals for Och1p-GFP and Mnn9p-mRFP appeared weaker than those without inhibition. The high background fluorescence when Och1p-GFP was detected was caused by strong excitation light to detect weak GFP signals. After the shift to 37.5°C, Mnn9p-mRFP exhibited a central ring structure and strings at the peripheries of the cells, indicating a typical ER distribution. On the other hand, although Och1p-GFP with a punctate structure observed at the permissive temperature was decreased, Och1p-GFP did not show the ER localization pattern (Fig. 3A, 40 min). We then examined the behavior of the M16/O16 chimeric protein in the sec12 mutant cells. Before the temperature shift, the M16/O16-GFP fusions were localized to the Golgi apparatus (Fig. 3B). However, after the temperature shift, they accumulated in the ER, like Mnn9p-mRFP. This result suggests that M16/O16-GFP followed the Mnn9p recycling pathway, even though the M16/O16 chimeric protein consists primarily of the Och1p sequence, except for the short cytoplasmic region derived from Mnn9p.
FIG. 3.
Intracellular dynamics of M16/O16 chimeric protein in sec12 cells. YMO59 cells (A) and YMO60 cells harboring pRS316-M16/O16-GFP (B) were treated with 100 μg/ml CHX for 30 min at 23°C, incubated at 37.5°C for 40 min, and then observed. (C) Subcellular fractionation of sec12 mutant cells expressing M16/O16-GFP and Mnn9p-mRFP. YMO60 cells harboring pRS316-M16/O16-GFP were treated as described for panel B, lysed, and fractionated by centrifugation. Equal amounts of P10, P100, and S100 were electrophoresed and then immunoblotted using anti-GFP, anti-mRFP, anti-Och1p, and anti-Dpm1p antibodies.
Next, we performed subcellular-fractionation and immunoblot analyses to confirm the results of the above-mentioned M16/O16-GFP microscope analyses. sec12 mutant cells expressing M16/O16-GFP were incubated in the presence of CHX at either the permissive or nonpermissive temperature and then were harvested and lysed. The lysates were fractionated by centrifugation as described in Materials and Methods (Fig. 3C). In this experiment, the ER was fractionated into P10, as shown by the localization of the ER marker Dpm1p, and the cis-Golgi was fractionated mainly into P100 (with a smaller amount in P10), as shown by the localization of the cis-Golgi marker Och1p. At 23°C, the P100 fraction contained larger amounts of Mnn9p-mRFP and M16/O16-GFP than the P10 fraction; the relative amounts of Mnn9p-mRFP in the P100 and P10 fractions were 57% and 37% of the total lysate, respectively, while those of M16/O16-GFP in the P100 and P10 fractions were 66% and 33%, respectively, indicating that these proteins are mainly localized in the cis-Golgi. On the other hand, at 37.5°C, larger amounts of Mnn9-mRFP and M16/O16-GFP were observed in the P10 fraction than in the P100 fraction; the relative amounts of Mnn9p-mRFP in the P100 and P10 fractions were 47% and 49% of the total lysate, respectively, while those of M16/O16-GFP in the P100 and P10 fractions were 40% and 60%, respectively. This observation indicates that some Mnn9p-mRFP and M16/O16-GFP are relocated from the cis-Golgi to the ER due to the inhibition of the ER-to-Golgi apparatus anterograde transport system. This result again shows that the M16/O16 chimeric protein cycles back and forth between the ER and the Golgi apparatus in the same manner as Mnn9p and that the cytoplasmic N-terminal region of Mnn9p determines the intracellular dynamics of the M16/O16 chimeric protein.
The intracellular dynamics of M16/O16 chimeric protein is not affected by deletion of ERD1.
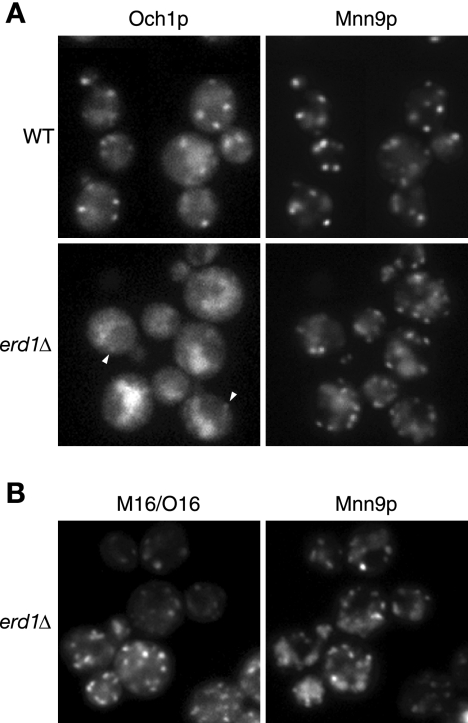
The erd1 mutant secretes endogenous resident ER proteins containing the HDEL signal sequence, which is required for the retrieval of the resident ER proteins from early Golgi apparatus compartments. Cells lacking the ERD1 gene have been reported to be defective in the Golgi apparatus-dependent modification of several glycoproteins (7, 25). We suspected that the defect in Golgi apparatus-dependent glycosylation in the erd1 deletion mutant was caused by the mislocalization of several glycosyltransferases, such as Och1p and Mnn9p. To address this possibility, we investigated the localization of Och1p and Mnn9p in the erd1Δ cells.
In wild-type cells, both Och1p-GFP and Mnn9p-mRFP showed a punctate structure, indicating a typical Golgi apparatus localization (Fig. 4A, top). In the erd1Δ cells, Mnn9p-mRFP was primarily localized to the Golgi apparatus, and weak signals due to mRFP were also detected in the vacuole. On the other hand, Och1p-GFP was mainly localized to the vacuole, and only very weak signals due to GFP were observed in the Golgi apparatus (Fig. 4A, bottom), indicating that the localization of Och1p to the Golgi apparatus is severely affected by the deletion of ERD1 while the localization of Mnn9p is not appreciably affected. We thought that the difference in the localization of Och1p and Mnn9p in erd1Δ cells might be caused by differences in the recycling patterns of the proteins. Thus, we further investigated the localization pattern of the M16/O16 chimeric protein in erd1Δ cells and found that M16/O16-GFP showed a punctate structure, similar to Mnn9p (Fig. 4B). Although the physiological function of ERD1 remains to be elucidated, this result indicates that the M16/O16 chimeric protein behaves like Mnn9p even in erd1Δ cells, suggesting again that the N-terminal region of Mnn9p in the M16/O16 chimeric protein plays an important role in determining the pathways used for relocation to the cis-Golgi compartment.
FIG. 4.
Localization of M16/O16 chimeric protein in erd1Δ cells. (A) YMO56 (WT) and YMO63 (erd1Δ) cells were grown at 25°C in SC medium. The arrowheads indicate the weak dot signal arising from the Golgi apparatus localization. (B) YMO65 cells harboring pRS316-M16/O16-GFP were grown at 25°C in SC−Ura medium.
The M16/O16 chimeric protein shows intracellular dynamics similar to those of Mnn9p in wild-type cells.
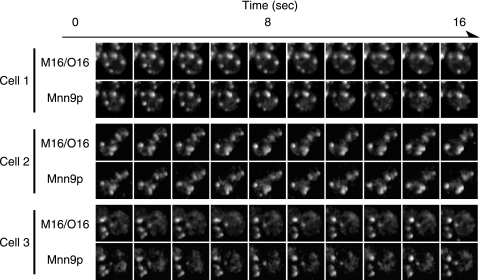
If the N-terminal cytoplasmic region of Mnn9p has an important role in determining the pathways used for the relocation to the cis-Golgi, M16/O16 chimeric protein should behave like Mnn9p, even in wild-type cells. To address this possibility, we tried time-lapse imaging of M16/O16-GFP and Mnn9p-mRFP simultaneously in wild-type living cells with the high-performance confocal laser scanning microscope system. The time-lapse observation revealed that the intracellular dynamics of M16/O16-GFP are similar to that of Mnn9p; almost all M16/O16-GFP colocalized with Mnn9p-mRFP (Fig. 5; see Fig. S2 in the supplemental material). We also checked 150 Golgi apparatus cisternae under the microscope and found that 5 cisternae (3% of the total counted) contained only Och1p while 145 cisternae (97%) contained both Och1p and Mnn9p. We could not find the cisternae that contained only Mnn9p. These results show again that the cytoplasmic region determines the localization pattern as with Mnn9p and that the difference in the intracellular dynamics of Och1p and Mnn9p reflects the difference in the recycling pathway to the cis-Golgi compartment.
FIG. 5.
Dual-fluorescence observation of M16/O16-GFP and Mnn9p-mRFP. After YMO50 cells harboring pRS316-M16/O16-GFP were incubated at 25°C in SC−Ura medium, M16/O16-GFP and Mnn9p-mRFP were observed at the same time with a high-performance confocal laser scanning microscope system. Images of three different cells were taken as the same rate as for Fig. 1.
Two arginine residues located in the cytoplasmic region of Mnn9p are important but not essential for its retrograde transport from the Golgi apparatus to the ER.
We attempted to identify the sequence located in the N-terminal cytoplasmic region of the M16/O16 chimeric protein required for cycling Mnn9p from the Golgi apparatus to the ER. Several amino acid residues were found to be conserved in multiple alignments of N-terminal sequences of Mnn9p homologues from S. cerevisiae, Kluyveromyces lactis, Candida glabrata and Candida albicans (Fig. 6A). We focused on the two arginine residues in the conserved region because double-arginine motifs (RR or RXR) located near the cytoplasmic N terminus have been reported to direct the retrieval of ER-resident type II membrane proteins to the ER (29). To investigate if the arginine residues in positions 9 and 11 in the N terminus are important for the cycling of Mnn9p from the Golgi apparatus to the ER, we constructed the mutant m16/o16(R9/11A)-GFP, in which the arginine residues in positions 9 and 11 were replaced with alanine. The m16/o16(R9/11A)-GFP mutant protein suppressed the temperature sensitivity of och1Δ cells, indicating that it is indeed a functional protein (Fig. 6B). The m16/o16(R9/11A)-GFP mutant protein was expressed in the sec12 mutant cells, and then the localization of this mutant protein was observed under the same conditions described in Fig. 3B. Before the temperature shift, m16/o16(R9/11A)-GFP was barely localized to the Golgi apparatus, in addition to the vacuole, although its fluorescent signal was much weaker than that of M16/O16-GFP (Fig. 6C, 0 min). Since m16/o16(R9/11A)-GFP was not accumulated in the ER, we consider that the amino acid substitutions (R9/11A) do not influence the folding of m16/o16(R9/11A)-GFP. After the shift to 37.5°C, m16/o16(R9/11A)-GFP was not observed to localize to the ER, whereas Mnn9p-mRFP did (Fig. 6C, 40 min), indicating that m16/o16(R9/11A)-GFP was no longer being cycled to the ER. To confirm this result, we performed subcellular fractionation of m16/o16(R9/11A)-GFP (Fig. 6D). The relative amount of Mnn9p-mRFP in the P100 fraction was greater than that in the P10 fraction at 23°C, while that in the P100 fraction was less than that in the P10 fraction at 37.5°C, indicating that the Mnn9-mRFP moved to the ER after the temperature shift. When the cells were treated with CHX for 30 min, the m16/o16(R9/11A)-GFP tended to be degraded and a large amount of free GFP was detected in the P10 and S100 fractions at 23°C and 37.5°C. Under our experimental conditions, carboxypeptidase Y, a marker protein of the vacuole, was distributed into the P10 and S100 fractions (data not shown). Therefore, we speculate that the free GFP arose from the degradation of m16/o16(R9/11A)-GFP in the vacuole. At both 23°C and 37.5°C, the amount of intact m16/o16(R9/11A)-GFP in the P100 fraction was larger than that in the P10 fraction, suggesting that intact m16/o16(R9/11A)-GFP did not move to the ER. These results correspond to the microscopic observation of the intracellular localization and suggest that considerable amounts of m16/o16(R9/11A)-GFP are transported from the Golgi apparatus to the vacuole, not to the ER, and subsequently degraded. Therefore, we concluded that these two arginine residues are important for the retrieval of M16/O16 from the Golgi apparatus to the ER.
FIG. 6.
Intracellular dynamics of mutant m16/o16(R9/11A) chimeric protein in sec12 cells. (A) Multiple alignments of the N-terminal cytoplasmic regions of S. cerevisiae Mnn9p homologues in the yeast species K. lactis (accession number CAG99616), C. glabrata (CAG62299), and C. albicans (AAB05924). Conserved amino acid residues are boxed. The asterisks indicate the mutated arginine residues in m16/o16(R9/11A). (B) YS57-2C cells, which were transformed with pRS316, pRSOCH1-GFP, pRS316-M16/O16-GFP, or pRS316-m16/o16(R9/11A)-GFP, were incubated at the indicated temperature for 3 days. (C) YMO60 harboring pRS316-m16/o16(R9/11A)-GFP was incubated at 23°C in SC−Ura medium and treated with 100 μg/ml CHX for 30 min. m16/o16(R9/11A)-GFP and Mnn9p-mRFP were observed 0 or 40 min after being shifted to 37.5°C. (D) Subcellular fractionation of sec12 mutant cells expressing m16/o16-GFP(R9/11A)-mRFP. The cells were treated as described in the legend to Fig. 3C.
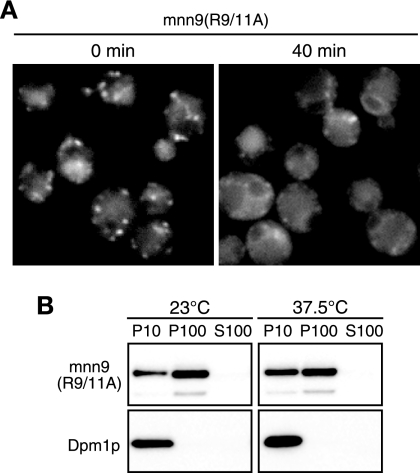
We further investigated whether the arginine residues in positions 9 and 11 are critical for recycling of authentic Mnn9p to the ER. In sec12-4 cells expressing mnn9(R9/11A)-mRFP, which have R9/11A mutations in Mnn9p, we observed the localization pattern of mnn9(R9/11A)-mRFP. mnn9(R9/11A)-mRFP was located in the Golgi apparatus before the temperature shift (Fig. 7A, 0 min), while it showed ER localization after the temperature shift (Fig. 7A, 40 min). We also performed subcellular fractionation of mnn9(R9/11A)-mRFP. The relative amount of mnn9(R9/11A)-mRFP in the P10 fraction was increased after the temperature shift (Fig. 7B), indicating that mnn9(R9/11A)-mRFP is transported from the Golgi apparatus to the ER. These results show that the mutations R9/11A do not have severe effects on the cycling of Mnn9p to the ER, implying that other factors may be involved in the ER-Golgi apparatus cycling of Mnn9p, in addition to the arginine residues in positions 9 and 11.
FIG. 7.
Intracellular dynamics of mutant mnn9(R9/11A) protein in sec12 cells. (A) YMO69 cells were incubated at 23°C in SC−Ura medium and treated with 100 μg/ml CHX for 30 min. mnn9(R9/11A)-mRFP was observed 0 or 40 min after being shifted to 37.5°C. (B) YMO69 cells were treated as described for Fig. 3C, and subcellular fractionation was performed.
DISCUSSION
The progress of live-imaging technology has led to many new findings, among them that yeast Golgi apparatus cisternae change the distribution of resident membrane proteins from a cis nature to trans over time, as proposed by the cisternal-maturation model (16). We applied live-imaging technology to the visualization of Golgi apparatus-localized mannosyltransferases. It is quite important to express genes encoding the transferases at a native level, without overexpression, to investigate the correct localization of these proteins, because overexpression often causes mislocalization of the transferases. For example, Anp1p, a subunit of M-Pol II, was first reported to be localized in the ER (2). However, this observation was in fact an artifact due to overexpression of ANP1 (9). It has been difficult to obtain live images of mannosyltransferases because of their low endogenous expression levels, but progress in live-imaging technology enabled us to overcome this hurdle. We utilized a high-performance confocal laser scanning microscope system to live image the cis-Golgi-localized mannosyltransferases, Och1p and Mnn9p. Such systems have already made great contributions to the visualization of cell wall proteins (31) and Golgi apparatus cisternae (16). We succeeded in the visualization of live images of GFP-tagged Och1p and mRFP-tagged Mnn9p without any artificial overexpression. The images of Och1p-GFP and Mnn9p-mRFP, which were taken simultaneously without any time lag using a Dual-View beam splitter, enabled precise analyses of the intracellular dynamics of both mannosyltransferases. Live imaging showed that the localization of Och1p-GFP is not always identical to that of Mnn9p-mRFP, even though both Och1p and Mnn9p are cis-Golgi-localized proteins. The results indicate that the cis-Golgi cisternae are neither uniform nor static but change their contents occasionally. To our knowledge, this is the first report describing live imaging of Golgi apparatus-localized glycosyltransferases without overexpression.
We considered that the difference in the intracellular dynamics of Och1p and Mnn9p might reflect differences in the mechanism for cis-Golgi retention of these enzymes. As reported previously, Och1p travels as far as the trans-Golgi and is transported back to the cis-Golgi (8), while Mnn9p cycles between the ER and cis-Golgi (32). Our results are consistent with these previous reports. During mannan synthesis, Mnn9p is involved downstream of Och1p in the synthetic pathway. Therefore, the order of function does not always match the localization dynamics of the proteins responsible for a synthetic step. M-Pol I elongates the α-1,6-mannan backbone at the mannose residue added by Och1p. Since M-Pol I cannot elongate the mannan backbone without Och1p, it is conceivable that the order of reactions mediated by these mannosyltransferases is regulated not by the localization of the enzymes, but by the strict substrate specificity of the enzymes.
Observation of chimeric proteins of Och1p and Mnn9p revealed that the N-terminal cytoplasmic region is important for the ER cycling of Mnn9p. It is known that arginine-based ER localization signals are sorting motifs involved in the transport of multimeric membrane proteins (17). The arginine-based signals consist of the consensus sequence Φ/Ψ/R-R-X-R, in which Φ/Ψ denotes an aromatic or bulky hydrophobic residue and X represents any amino acid (17). The N-terminal cytoplasmic region of Mnn9p has the sequence YRLR (from the 8th to the 11th amino acid residues), which shows good correspondence with the consensus sequence of the arginine-based ER localization signals. An important issue is that the arginine residues in positions 9 and 11 in the N terminus of Mnn9p indeed act as an ER localization signal. Our results suggest that the mutant m16/o16(R9/11A) protein is synthesized in the ER, transported to the cis-Golgi, and degraded in the vacuole without being transported back to the ER. Therefore, it is possible that the arginine residues in positions 9 and 11 in the N terminus of Mnn9p may act as a signal for M16/O16 chimeric protein to be retrieved from the Golgi apparatus to the ER and that the degradation of the mutant m16/o16(R9/11A) protein in the vacuole might be caused due to the lack of the ER localization signal.
The arginine-based signals have been reported to be recognized by COPI-coated vesicles (1, 34). It has also been reported that Mnn9p is incorporated into COPI vesicles by a coatmer-dependent vesicle budding assay (32). These reports suggest that Mnn9p may be transported back to the ER by COPI vesicles through the recognition of the sequence that contains two arginine residues. However, mutant mnn9(R9/11A) protein was till transported back to the ER. Considering that the arginine residues in positions 9 and 11 are required for the retrieval of M16/O16 from the Golgi apparatus to the ER, the behavior of mnn9(R9/11A) seems to be strange. One explanation that could overcome this apparent inconsistency is that Mnn9p may have another unidentified ER relocation signal in its transmembrane or luminal region. The inconsistency may be also explainable by the ability of Mnn9p to form protein complexes, M-Pol I and M-Pol II. It has been reported that the luminal region of Mnn9p interacts with that of Van1p, a component of M-Pol I (12), and that Van1p and Anp1p, a component of M-Pol II, also recycle between the ER and the Golgi apparatus (32). Therefore, we speculate that Van1p may have its own mechanism for ER-Golgi apparatus recycling and that some of the mutant mnn9(R9/11A) protein may be brought to the ER by interacting with Van1p through its C-terminal luminal region. We tried time-lapse observation of Van1p that was tagged with GFP and mRFP but failed to visualize them due to their low fluorescence intensities.
Och1p follows a recycling pathway different from that of Mnn9p. In erd1Δ cells, Och1p is mainly localized to the vacuole and only slightly to the Golgi apparatus, suggesting that Och1p may be transported to the vacuole through the Golgi apparatus without cycling to the cis-Golgi. On the other hand, M16/O16 chimeric protein is normally localized to the Golgi apparatus even in erd1Δ cells. These results suggest that the cytoplasmic region of Och1p is important for the recycling of Och1p, similar to Mnn9p. Erd1p might be involved in the recognition of the cytoplasmic region in the Och1p recycling pathway. We tried to investigate whether O15/M17-mRFP, a chimeric protein that was constructed by swapping the N-terminal 16 amino acids of Mnn9p with the N-terminal 15 amino acids of Och1p, cycles between the ER and the Golgi apparatus. In the system using sec12 mutant cells, O15/M17-mRFP seemed to be transported back to the ER less effectively than Mnn9p-mRFP, but the result was quite obscure (data not shown). This result would be reasonable if the N-terminal cytoplasmic region of Och1p possesses its own localization signal; some O15/M17 proteins behave like Och1p due to its localization signal in the cytoplasmic region, while some O15/M17 proteins behave like Mnn9p and are transported back to the ER due to its localization signal in the transmembrane or luminal region or due to interaction with Van1p. Further work will be necessary to reveal the recycling pathway of Och1p. The live-imaging system we utilized in the present study will continue to make contributions to revealing the cis-Golgi-retention mechanisms of mannosyltransferases.
Supplementary Material
Acknowledgments
We thank all the members of the Nakano group of the Dynamic-Bio Project for technical contributions to the advanced microscope system. We are grateful to Roger Y. Tsien for providing the mRFP gene and to Akihiko Nakano for providing strain MBY10-7A. We thank Takuji Oka, Hiroto Hirayama, Xiao-Dong Gao, Morihisa Fujita, Mariko Umemura, Yasunori Chiba, and Yoh-ichi Shimma for helpful discussions.
This study was supported by a national fund from the New Energy and Industrial Technology Development Organization.
Footnotes
Published ahead of print on 14 December 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Brock, C., L. Boudier, D. Maurel, J. Blahos, and J. P. Pin. 2005. Assembly-dependent surface targeting of the heterodimeric GABAB receptor is controlled by COPI but not 14-3-3. Mol. Biol. Cell 165572-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman, R. E., and S. Munro. 1994. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 134896-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colley, K. J. 1997. Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 71-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 2631629-1631. [DOI] [PubMed] [Google Scholar]
- 5.Gaynor, E. C., S. te Heesen, T. R. Graham, M. Aebi, and S. D. Emr. 1994. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham, T. R., M. Seeger, G. S. Payne, V. L. MacKay, and S. D. Emr. 1994. Clathrin-dependent localization of alpha 1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J. Cell Biol. 127667-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardwick, K. G., M. J. Lewis, J. Semenza, N. Dean, and H. R. Pelham. 1990. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J. 9623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, S. L., and M. G. Waters. 1996. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell Biol. 132985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jungmann, J., and S. Munro. 1998. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 17423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jungmann, J., J. C. Rayner, and S. Munro. 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 2746579-6585. [DOI] [PubMed] [Google Scholar]
- 11.Karhinen, L., and M. Makarow. 2004. Activity of recycling Golgi mannosyltransferases in the yeast endoplasmic reticulum. J. Cell Sci. 117351-358. [DOI] [PubMed] [Google Scholar]
- 12.Kojima, H., H. Hashimoto, and K. Yoda. 1999. Interaction among the subunits of Golgi membrane mannosyltransferase complexes of the yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 631970-1976. [DOI] [PubMed] [Google Scholar]
- 13.Letourneur, F., E. C. Gaynor, S. Hennecke, C. Demolliere, R. Duden, S. D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 791199-1207. [DOI] [PubMed] [Google Scholar]
- 14.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 15.Losev, E., C. A. Reinke, J. Jellen, D. E. Strongin, B. J. Bevis, and B. S. Glick. 2006. Golgi maturation visualized in living yeast. Nature 4411002-1006. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura-Tokita, K., M. Takeuchi, A. Ichihara, K. Mikuriya, and A. Nakano. 2006. Live imaging of yeast Golgi cisternal maturation. Nature 4411007-1010. [DOI] [PubMed] [Google Scholar]
- 17.Michelsen, K., H. Yuan, and B. Schwappach. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milland, J., S. G. Taylor, H. C. Dodson, I. F. C. McKenzie, and M. S. Sandrin. 2001. The cytoplasmic tail of alpha 1,2-fucosyltransferase contains a sequence for Golgi localization. J. Biol. Chem. 27612012-12018. [DOI] [PubMed] [Google Scholar]
- 19.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 144695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48899-907. [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi-Shindo, Y., K. Nakayama, A. Tanaka, Y. Toda, and Y. Jigami. 1993. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 26826338-26345. [PubMed] [Google Scholar]
- 22.Nakano, A., D. Brada, and R. Schekman. 1988. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama, K., T. Nagasu, Y. Shimma, J. Kuromitsu, and Y. Jigami. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 112511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama, K., Y. Nakanishi-Shindo, A. Tanaka, Y. Haga-Toda, and Y. Jigami. 1997. Substrate specificity of alpha-1,6-mannosyltransferase that initiates N-linked mannose outer chain elongation in Saccharomyces cerevisiae. FEBS Lett. 412547-550. [DOI] [PubMed] [Google Scholar]
- 25.Pelham, H. R., K. G. Hardwick, and M. J. Lewis. 1988. Sorting of soluble ER proteins in yeast. EMBO J. 71757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelham, H. R., and J. E. Rothman. 2000. The debate about transport in the Golgi—two sides of the same coin? Cell 102713-719. [DOI] [PubMed] [Google Scholar]
- 27.Rossanese, O. W., J. Soderholm, B. J. Bevis, I. B. Sears, J. O'Connor, E. K. Williamson, and B. S. Glick. 1999. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 14569-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroder, S., F. Schimmoller, B. Singer-Kruger, and H. Riezman. 1995. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in alpha-COP. J. Cell Biol. 131895-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schutze, M. P., P. A. Peterson, and M. R. Jackson. 1994. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 131696-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 31.Sumita, T., T. Yoko-o, Y. Shimma, and Y. Jigami. 2005. Comparison of cell wall localization among Pir family proteins and functional dissection of the region required for cell wall binding and bud scar recruitment of Pir1p. Eukaryot. Cell 41872-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todorow, Z., A. Spang, E. Carmack, J. Yates, and R. Schekman. 2000. Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 9713643-13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vowels, J. J., and G. S. Payne. 1998. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol. Biol. Cell 91351-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan, H., K. Michelsen, and B. Schwappach. 2003. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 13638-646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.