Abstract
In Fusarium fujikuroi, the production of gibberellins and bikaverin is repressed by nitrogen sources such as glutamine or ammonium. Sensing and uptake of ammonium by specific permeases play key roles in nitrogen metabolism. Here, we describe the cloning of three ammonium permease genes, mepA, mepB, and mepC, and their participation in ammonium uptake and signal transduction in F. fujikuroi. The expression of all three genes is strictly regulated by the nitrogen regulator AreA. Severe growth defects of ΔmepB mutants on low-ammonium medium and methylamine uptake studies suggest that MepB functions as the main ammonium permease in F. fujikuroi. In ΔmepB mutants, nitrogen-regulated genes such as the gibberellin and bikaverin biosynthetic genes are derepressed in spite of high extracellular ammonium concentrations. mepA mepB and mepC mepB double mutants show a similar phenotype as ΔmepB mutants. All three F. fujikuroi mep genes fully complemented the Saccharomyces cerevisiae mep1 mep2 mep3 triple mutant to restore growth on low-ammonium medium, whereas only MepA and MepC restored pseudohyphal growth in the mep2/mep2 mutant. Overexpression of mepC in the ΔmepB mutants partially suppressed the growth defect but did not prevent derepression of AreA-regulated genes. These studies provide evidence that MepB functions as a regulatory element in a nitrogen sensing system in F. fujikuroi yet does not provide the sensor activity of Mep2 in yeast, indicating differences in the mechanisms by which nitrogen is sensed in S. cerevisiae and F. fujikuroi.
The sensing and uptake of nitrogen are essential processes for fungal growth and development. Ammonium and glutamine are preferred nitrogen sources (48), as their presence results in the repression of genes that are involved in the acquisition and utilization of other sources of nitrogen. This regulatory system is known as nitrogen metabolite repression in filamentous fungi (53) or nitrogen catabolite repression (NCR) in Saccharomyces cerevisiae (6). A family of GATA-type transcription factors that activate gene expression when levels of preferred nitrogen sources become limiting mediates this regulation. Members of this family include AreA from Aspergillus nidulans and Fusarium fujikuroi, NIT2 from Neurospora crassa, and Gln3p from S. cerevisiae (12, 21, 26, 50).
Under nitrogen starvation conditions, the rice pathogenic fungus F. fujikuroi produces the red pigment bikaverin and gibberellins (GAs), mainly the gibberellic acids GA3, GA4, and GA7, the causative agents of the “bakanae” disease of rice seedlings. GAs are isoprenoid plant hormones used as plant growth regulators in agriculture and horticulture (40). Bikaverin is a polyketide which is responsible for the deep red color exhibited under certain growth conditions by cultures of some Fusarium species (23). Interestingly, neither of these secondary metabolites contains nitrogen, and they do not have any obvious function in nitrogen metabolism (reviewed in reference 51). Both the GA and bikaverin biosynthetic genes are strictly repressed by nitrogen. For the GA biosynthetic genes a direct dependence on AreA has already been shown (33, 46, 47, 50), whereas the role of AreA in regulation of bikaverin genes is not yet clear.
Additional components of nitrogen metabolism also influence the correct expression of the GA and bikaverin biosynthetic genes in F. fujikuroi. Glutamine synthetase (GS) is required for the wild-type expression of a variety of genes, including those involved in ribosome biogenesis and translation initiation, which are induced in a mutant strain lacking GS (46). Surprisingly, GS activity is also required for the expression of the GA and bikaverin biosynthetic genes, which is abolished, rather than induced, in a GS null mutant (46). This suggests an important role for GS in control of cellular metabolism and highlights the complexity of the regulatory networks that respond to nitrogen metabolism in F. fujikuroi.
The GA and bikaverin biosynthetic genes are also targets of the TOR signaling pathway (47). In yeast, the conserved TOR (target of rapamycin) kinases play a significant role in nutrient sensing and cell growth by affecting nuclear localization of transcription factors, such as Gln3, and thereby controlling genes subject to NCR (reviewed in reference 41). In F. fujikuroi, several AreA-regulated genes, e.g., GA and bikaverin biosynthetic genes, are only partially derepressed by rapamycin suggesting that additional regulatory pathways involved in nitrogen metabolism exist.
We are interested in identifying components that mediate nitrogen sensing and which act upstream of the transcription factor AreA in F. fujikuroi. In yeast and filamentous fungi, nitrogen permeases, such as ammonium and amino acid permeases, can form part of the nitrogen regulatory network (15). Ammonium transport in fungi is mediated by permeases that belong to the conserved AmtB/Mep family of proteins that are related to the mammalian rhesus blood group antigens. These proteins are highly conserved within bacteria, plants, and animals and share a similar protein structure (13, 16, 18, 29, 31, 49). Within certain fungal species, one of the permeases has evolved a regulatory function. Examples include Mep2 (S. cerevisiae and Candida albicans), Amt1 (Hebeloma cylindrosporum), and Ump2 (Ustilago maydis), which are required for the induction of filamentous growth under low-nitrogen conditions (3, 18, 24, 25, 44, 52). In S. cerevisiae, this dimorphic transition requires the cooperation of two signaling pathways, the mitogen-activated protein kinase and the cyclic AMP-dependent pathways (22).
It is not clear at present the extent to which regulatory ammonium permeases are conserved within fungal species. In this study we have characterized three members of the AmtB/Mep family from F. fujikuroi that we have designated MepA, MepB, and MepC. Phenotypic and methylamine uptake studies in S. cerevisiae confirm that they are functional ammonium permeases. The genes encoding these permeases are under AreA-mediated nitrogen metabolite repression control. MepA and MepC but not MepB fully restored pseudohyphal growth in the S. cerevisiae mep2 mutant. MepB is probably the major ammonium permease in F. fujikuroi, and its deletion results in derepression of the GA and bikaverin biosynthetic genes and other genes subject to nitrogen metabolite repression under ammonium-sufficient conditions. Furthermore, overexpression of mepC in the ΔmepB background partially suppresses the strong growth defect but not the derepression of AreA target genes on high ammonium concentrations. We suggest, therefore, a sensing or regulatory role of MepB in addition to its function as a permease.
MATERIALS AND METHODS
Fungal strains and culture conditions.
Strain IMI58289 (Commonwealth Mycological Institute, Kew, United Kingdom) is a GA-producing wild-type strain of F. fujikuroi. For regulation studies, the areA deletion strain T19 (50), the gdhA deletion mutant (B. Tudzynski, unpublished data), and the glnA mutant (46) were used. For all cultivations, F. fujikuroi strains were precultivated for 48 h in 300-ml Erlenmeyer flasks with 100 ml of Darken medium (DVK) (8) with 2.0 g/liter glutamine instead of (NH4)2SO4 on a rotary shaker; 1 ml of this culture was used as inoculum for cultivations in ICI (Imperial Chemical Industries Ltd., United Kingdom) medium.
For DNA isolation and protoplasting, F. fujikuroi strains were incubated in complete medium (39) at 28°C on a rotary shaker at 200 rpm for 3 days or 18 h, respectively. For analysis of GA production, the fungus was grown for 5 days at 28°C on a rotary shaker (190 rpm) in a liquid production medium containing 60 g/liter sunflower oil, 15 g/liter corn steep solids (Sigma, Germany), 1.0 g/liter glutamine, and 1.0 g/liter KH2PO4 or in ICI medium (14) with 20% of the ammonium nitrate concentration (20% ICI). The S. cerevisiae strains MLY40α (ura3-52 MATα), MLY131α (mep1::LEU2 mep2::LEU2 mep3::G418 ura3-52 MATα), MLY97a/α (ura3-52/ura3-52 leu2::hisG/leu2::hisG MATa/α), and MLY108a/α (mep2::LEU2/mep2::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG MATa/α) were used to analyze the function of the F. fujikuroi mep genes (24, 25).
Bacterial strains and plasmids.
Escherichia coli strain Top10 (Invitrogen, Groningen, The Netherlands) was used for plasmid propagation. Genomic DNA fragments carrying the F. fujikuroi mepA, mepB, and mepC genes or parts of the genes were cloned into the vector pUC19 (Fermentas, Germany). For the replacement of mepA, a 1.1-kb KpnI/SalI fragment of the 5′ noncoding region and a 0.9-kb HindIII/BamHI fragment of the 3′ noncoding region were cloned into the plasmid pUCH2-8 (1) carrying the hygromycin B resistance cassette. A KpnI/BamHI fragment of the resulting replacement vector, pΔmepA, carrying both flanks and the hygromycin resistance cassette, was used for gene replacement experiments. To construct the mepB gene replacement vector, a 0.8-kb SacII/XbaI fragment of the 5′ noncoding region and a 0.5-kb ClaI/SalI fragment from the 3′ noncoding region were cloned into the plasmid pNRI (28) carrying the nourseothricin resistance gene nat1. For targeted replacement of mepC, a 0.7-kb SacI/XbaI fragment of the 5′ noncoding region and a 0.8-kb HindIII/SalI right flank were cloned into vector pNR1.
For complementation of the mepB mutant with the wild-type mepB copy, a 4.8-kb genomic SacI fragment was cloned into the vector pPUCH2-8 (1) carrying the hygromycin resistance cassette. The circular vector pmepB-hyg was used to transform the mepB mutant strain T1 (mepB-T1, where T1 is for transformant 1). For constructing the mepC overexpression vector, the promoter of the F. fujikuroi glnA gene (46) was amplified by PCR using the primers glnA-prom-XbaI and glnA-prom-Bam. The coding region of the mepC gene was amplified with the primers MepC-Bam-F and MepC-Hind-R. The restricted promoter and mepC fragments were cloned by one step into the XbaI/HindIII-restricted vector pUCH2-8.
Screening of genomic library.
About 40,000 recombinant phages of the F. fujikuroi m567 genomic library (28) were plated with E. coli strain XII-Blue MRF′ and screened by plaque hybridization as described previously (42). Plaque lifts (Gene Screen nylon membranes; DuPont, Germany) were hybridized with [32P]dCTP-labeled 0.5-kb PCR fragments of the F. fujikuroi mepA, mepB, and mepC genes. Hybridization and washing steps were performed at 65°C. The blots were washed at 65°C (once with 2 × SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 0.1% sodium dodecyl sulfate [SDS]; once with 0.1 × SSC and 0.1% SDS) as described previously (46). Putative positive phages were purified in a second screening round. Phage DNA was isolated as described previously (42) and used for restriction analysis and subcloning.
DNA isolation.
Lyophilized mycelium was ground into a fine powder with a mortar and pestle and dispersed (in the case of DNA for use in PCR) in extraction buffer as described by Cenis (5). DNA for Southern hybridization experiments was prepared following the protocol of Doyle and Doyle (9). Plasmid DNA was extracted using a plasmid extraction kit (Genomed, Germany).
Southern-blot analysis.
For Southern blot analysis, genomic, plasmid, or phage DNA was digested to completion with appropriate restriction enzymes (Fermentas, Germany), fractionated in 1.0% (wt/vol) agarose gels, and transferred to nylon N+ membranes (Amersham, Germany) by vacuum blotting. DNA probes were randomly labeled, and hybridizations were carried out overnight at 65°C. The blots were washed under hybridization conditions (2 × SSC-0.1% SDS at 65°C followed by 0.1 × SSC-0.1% SDS).
Northern blot analysis.
For RNA isolation, the fungal strains were grown in ICI medium with 20 mM glutamine as a nitrogen source instead of ammonium nitrate for 3 days on a rotary shaker at 28°C. After 24, 48, and 72 h, mycelia were harvested and used for RNA preparation. For shift experiments, the mycelium was grown for 3 days in ICI medium with 20 mM glutamine, washed, and transferred into synthetic ICI medium without nitrogen for 2 h to induce starvation; the mycelium was then shifted into medium without nitrogen or with 10 mM NH4NO3 or 10 mM glutamine, with (200 ng/ml) or without rapamycin (Calbiochem). Mycelia were harvested after 1 h. The irreversible inhibitor of the GS, l-methionine sulfoximine (MSX) (Sigma-Aldrich, Germany), was added in a concentration of 4.0 mM. Total F. fujikuroi RNA was isolated using an RNAgents total RNA isolation kit (Promega, Germany).
PCR and reverse transcription-PCR (RT-PCR).
PCR primers MepL 5′-CAATGGTTCTTCTGGGGCTACTC-3′ and MepR 5′-CGAACCAGCCGA ACCAGAGGAA-3′ were used for amplification of F. fujikuroi ammonium permease genes mepA and mepB. For cloning the third ammonium permease gene, mepC, primers MepC-F1 5′-CTTCAACATGTCTTATGTTATCCC-3′ and MepC-R1 5′- GCAAGTTTAAGCATGCTTCTCG-3′ were used. To construct gene replacement vectors pΔmepA, pΔmepB, and pΔmepC, the 3′ and 5′ noncoding regions were amplified with the following primers: epA-GR1-KpnI, 5′-GGAATTCGGTACCCTTGAGGAC-3′; MepA-GR2-SalI, 5′-TGTAAGGAGTGGTCATGTCGAGCAGACTGC-3′; MepA-GR3-Hind, 5′-TAAGCTTCGAGAAGAAGTGGAGCACC-3′; MepA-GR4-BamHI, 5′-CGGATCCTTGATAGGAGTATAAAGGACC-3′; MepB-GR1-SacII, 5′-CGAAAAAAACGTCACCGCGGTACCGG-3′; MepB-GR2-XbaI, 5′-CTAGAGGAACCATTAACAGCACTAGAGC-3′; MepB-GR3-ClaI, 5′-GATCGATCGATATCGATAACGAGTCCG-3′; MepB-GR4-SalI, 5′-CGTCGACTTCAGACTGCTGCTATTAGTCC-3′; MepC-GR1-SacI, 5′-TTGGGGAGCTCGTACTGATAAACATCCATGAGATGG-3′; MepC-GR2-XbaI, 5′-GCAATAAAATAACCCCTCTAGATCAAACAAGG-3′; MepC-GR3-HindIII, 5′-GGTGATTAACGAGAAGCATGCTTAAGCTTGC-3′; MepC-GR4-SalI, 5′-ATGTCGACGTTACTCTGGCTCCCAGATCAACACG-3′.
Derived fragments were first cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced. For constructing the yeast expression vectors, the full-length cDNA fragments of the three F. fujikuroi mep genes were generated by RT-PCR using the following primers: MepA-Sca, 5′-GCAGTACTGTAAAAATGACCACTCC-3′; MepA-Xba, 5′-CACTAATCTAGACCTTTAGACC-3′; MepB-Sca, 5′-GCAGCCTAGTACTATCGCAAAAATGCTCTCG-3′; MepB-Xba, 5′-GGTATCTAGAACTCGACAATTCC-3′; MepC-ScaI, 5′-CCACGAGTACTTATAAACACAATAATCAAAAATGTC-3′; and MepC-Xba, 5′-GCAATAAAATAACCCCTCTAGATCAAACAAGG-3′.
The cDNA fragments of mepA, mepB, and mepC were first cloned into the PCR cloning vector pCR2.1-TOPO (Invitrogen) and then subcloned into the ScaI/XbaI-restricted yeast expression vector pYES2.1 (Invitrogen) to yield complementation vectors pYes2.1-mepA, pYes2.1-mepB, and pYes2.1-mepC. These plasmids were sequenced and used for complementation of the yeast triple mep1 mep2 mep3 and mep2 mutants. Vector pYes2.1-mepB was also used for complementing the F. fujikuroi ΔmepB mutant.
For constructing the mepC overexpression vector, the following primers were used: MepC-Bam-F, 5′-ATGGATCCAACATGTCTTATGTTATCCCTGG-3′; MepC-Hind-R, 5′-ATAAGCTTCTTTGCGTGCTACTCCTAGCATCCCGC-3′; glnA-prom-Xba, 5′-AGTCTAGACGGAGCAAAGCGGTTTATATCCGCC-3′; and glnA-prom-Bam, 5′-TTGGATCCTGTGAATGTGGTTGTGATACGGGG-3′.
PCRs contained 25 ng of DNA, 50 ng of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, and 2 U of Taq polymerase (Red Taq; Sigma-Aldrich, Germany) in 50 μl. PCR was carried out at 94°C for 4 min followed by 30 cycles of 94°C for 1 min, 53 to 57°C for 30 s, and 72°C for 1.5 min. For RT-PCR 1 μg of total RNA of nitrogen-starved wild-type mycelium served as a template to create cDNA by using a One-step qRT-PCR kit (Invitrogen, Groningen, Germany).
Fungal transformations.
Preparation of protoplasts from F. fujikuroi mycelium was carried out as described previously (50). A total of 107 protoplasts of strain IMI58289 were transformed with 10 μg of the KpnI/BamHI, SacI/SalI, or SacI/SalI fragment of the replacement vectors pΔmepA, pΔmepB, or pΔmepC, respectively. For gene replacement, transformed protoplasts were regenerated at 28°C in a complete regeneration agar (0.7 M sucrose, 0.05% yeast extract, 0.1% Casamino Acids) containing 120 μg/ml hygromycin B (for pΔmepA) (Calbiochem, Germany) or 100 μg/ml nourseothricin (for pΔmepB and pΔmepC) (Werner-Bioagents, Germany) for 6 to 7 days. For construction of mepA mepB double mutants, strain ΔmepA-T10 was transformed with 10 μg of the SacI/SalI-fragment of vector pΔmepB. For construction of mepB mepC double mutants, strain ΔmepB-T1 was cotransformed with 10 μg of the SacI/SalI fragment of vector pΔmepC and 10 μg of the vector pUCH2-8 carrying the hygromycin resistance cassette. For complementation, the ΔmepB-T1 mutant was cotransformed with 10 μg (each) of the hygromycin B resistance-mediating vector pUCH2-8 and the pYES2.1 vector carrying the mepB cDNA fragment. The transformants were additionally selected by regeneration on a medium containing low ammonium concentrations (1 mM ammonium citrate) as the only nitrogen source. Single conidial cultures were established from hygromycin B- or nourseothrecin-resistant transformants and used for DNA isolation and Southern blot analysis.
Analysis of mepA, mepB, and mepC function in S. cerevisiae.
To test the function of the F. fujikuroi mep genes in S. cerevisiae, the vectors pYes2.1-mepA, pYes2.1-mepB, and pYes-mepC and control vectors pYes2.1-Mep2 and pYes2.1 were transformed into the haploid S. cerevisiae strain MLY131α (mep1 mep2 mep3). The wild-type MLY40α containing pYes2.1 served as a control strain. Growth of transformants under ammonium-limiting conditions was assayed by plating serial dilutions of washed overnight cultures onto SD plates (yeast nitrogen base with ammonium sulfate and without amino acid supplements plus 2% glucose) and SLADG plates (yeast nitrogen base without ammonium sulfate and without amino acids supplemented with 50 μM ammonium sulfate, 2% galactose, and 0.2% glucose). Pseudohyphal growth was analyzed by streaking transformants of the diploid MLY108a/α (mep2/mep2) S. cerevisiae strain containing the F. fujikuroi pYes2.1-mep vectors to single cells onto SLADG agar, which were then grown for 6 days at 30°C and photographed.
DNA sequencing and sequence homology searches.
DNA sequencing of recombinant plasmid clones was accomplished with the automatic sequencer Li-Cor 4000 (MWG, München, Germany). The two strands of overlapping subclones obtained from the genomic DNA clones were sequenced using the universal and the reverse primers or specific IRD-800-labeled oligonucleotides obtained from MWG Biotech (Munich, Germany). DNA and protein sequence alignments were done with DNA Star (Madison, WI). Sequence homology searches were performed using the NCBI database server. Protein homology was based on BlastX searches (2). For further investigations, the programs of DNA STAR Inc. (Madison, WI) were used.
[14C]methylamine uptake.
The analysis of methylamine uptake was carried out as previously described (44). Briefly, the S. cerevisiae diploid mep1 mep2 mep3 null strain was transformed with the yeast vector pYES2.1 that contained mepA, mepB, and mepC under the control of the GAL1 promoter. Individual transformants were grown in synthetic medium lacking uracil with galactose (3%) as the carbon source and proline as the nitrogen source (1 mM) for 6 h. The cells were pelleted, washed, and resuspended in phosphate buffer (20 mM; pH 7) to an optical density at 595 nm of 8 and incubated on ice. Aliquots were added to phosphate buffer (20 mM; pH 7) containing 0.1 mCi [14C]methylamine hydrochloride (MP Biomedicals, Inc) and increasing concentrations of methylamine. The cells were resuspended in a water bath at 30°C, and samples of 1 ml were removed at 1-min intervals and washed over GC Whatman filters. The level of [14C]methylamine was then quantified using liquid scintillation counting.
GA determination by TLC.
Amounts of produced GA3 and GA4/7 were determined by thin-layer chromatography (TLC) on silica gel eluted with ethyl acetate-chloroform-acetic acid (60:40:5).
Nucleotide sequence accession numbers.
The sequences of the F. fujikuroi mepA, mepB, and mepC genes were deposited in the GenBank database under accession numbers AM168272, AM168273, and AM283470, respectively.
RESULTS
Cloning of the permease genes mepA, mepB, and mepC.
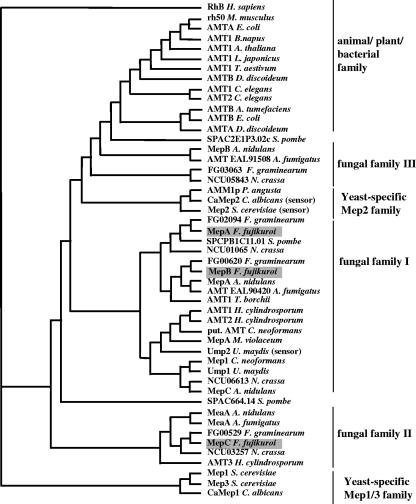
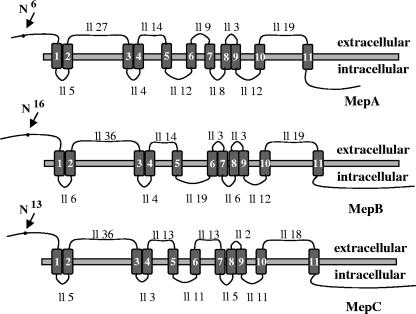
Primers based on the conserved MEP sequences from other fungi were used to isolate homologues from F. fujikuroi. Isolated PCR fragments were used to probe a genomic library and resulted in the isolation of two ammonium permease genes, mepA and mepB. During the course of this work, we noted that the genome of Fusarium verticillioides, a close relative of F. fujikuroi, contains a third mep gene. Primers based on the sequence of this gene were used to amplify the coding region and regulatory sequences of a third ammonium permease-encoding gene, mepC in F. fujikuroi. The three genes are predicted to encode proteins of 466 (MepA), 502 (MepB), and 513 (MepC) amino acids. All three permeases share a high degree of sequence similarity with other permeases of the AmtB/Mep family (Table 1). A phylogenetic tree of protein sequences of known ammonium transporters of the Mep/Amt family revealed a clear cluster of fungal ammonium permeases that is distinct from those of animals, plants, and bacteria (Fig. 1). F. fujikuroi MepC is most similar to MeaA from A. nidulans (fungal family II), and both MepA and MepB group together with MepA from A. nidulans in the main group of fungal ammonium permeases. The three F. fujikuroi permeases are predicted to contain 11 transmembrane helices with an Nout-Cin topology, based on the TMHMM and HMMTOP prediction programs (45), and this prediction is in agreement with predictions for other fungal ammonium transporters (32, 49) (Fig. 2). In addition, putative glycosylation sites are predicted for MepA (N6), MepB (N16), and MepC (N13) by the program PROSITE (43).
TABLE 1.
Sequence similarities of the F. fujikuroi Mep proteins with other permeases of the AmtB/Mep family
| F. fujikuroi Mep | AmtB/Mep family protein (accession no.) | Description | Organisma | % Sequence identity |
|---|---|---|---|---|
| MepA | FG02094 | F. graminearum | 91 | |
| MepA (CAD21326) | N. crassa | 64 | ||
| MepA (AAL73118) | High affinity | A. nidulans | 62 | |
| AMT1 (AAL11032) | High affinity | T. borchii | 63 | |
| Ump2 (XM756943) | High affinity; sensor | U. maydis | 56 | |
| MeaA (AAL73117) | High capacity | A. nidulans | 52 | |
| Mep2 (P41948) | High affinity, low capacity; sensor | S. cerevisiae | 50 | |
| MepB | FG00620 | F. graminearum | 97 | |
| AMT1 (AAL11032) | High affinity | T. borchii | 77 | |
| MepA (AAL73118) | High affinity | A. nidulans | 79 | |
| MepA (CAD21326) | N. crassa | 71 | ||
| MepC | F. fujikuroi | 64 | ||
| Ump2 (XM756943) | High affinity; sensor | U. maydis | 59 | |
| Mep2 (P41948) | High affinity, low capacity, sensor | S. cerevisiae | 50 | |
| MepC | FG00529 | F. graminearum | 94 | |
| MeaA (AAL73117) | High capacity | A. nidulans | 73 | |
| MepA (AAL73118) | High affinity | A. nidulans | 58 | |
| Mep1 (CAA97132) | High capacity, low affinity | S. cerevisiae | 57 | |
| Mep3 (P53390) | High capacity, low affinity | S. cerevisiae | 56 | |
| AMT1 (AAL11032) | High affinity | T. borchii | 56 | |
| MepA | High affinity | F. fujikuroi | 53 | |
| Mep2 (P41948) | Sensor | S. cerevisiae | 46 |
F. graminearum, Fusarium graminearum.
FIG. 1.
Dendrogram of aligned protein sequences of known mammalian, plant, bacterial, and fungal permeases of the MEP/AMT family or putative ammonium permeases chosen by homology. The following are accession numbers for the indicated proteins: Homo sapiens RH type B, AAG01086; Mus musculus rh50, AAC25155; Escherichia coli AMTA, AAA97110; Brassica napus AMT1, AAG28780; Arabidopsis thaliana AMT1, P54144; Lotus japonicus AMT1, AAG24944; Triticum aestivum AMT1, AAS19466; Dictyostelium discoideum AMTB, BAB39710; Caenorhabditis elegans AMT1, P54145; C. elegans AMT2, Q20605; Agrobacterium tumefaciens AMTB, AAL43739, E. coli AMTB, AAD14837; D. discoideum AMTA, BAB39709; S. cerevisiae Mep1, P40260; S. cerevisiae Mep3, P53390; H. cylindrosporum AMT3, AAK82417; Aspergillus nidulans MeaA, EAL73117; Aspergillus fumigatus MeaA, EAL87679; Schizosaccharomyces pombe hypothetical protein SPAC664.14, CAB65815; Cryptococcus neoformans Mep1, AAW40795; U. maydis Ump1, AAL08424; H. cylindrosporum AMT1, AAM21926; H. cylindrosporum AMT2, AAK82416; C. neoformans putative ammonium transporter, AAW45844; U. maydis Ump2, AAO42611; Microbotryum violaceum MepA, AAD40955; A. fumigatus ammonium transporter, EAL90420; A. nidulans MepA, AAL73118; F. fujikuroi MepB; T. borchii AMT1, AAL11032; N. crassa MepA, CAD21326; F. fujikuroi MepA; S. cerevisiae Mep2, P41948; Candida glabrata unnamed protein, XP_447968; C. albicans hypothetical protein CaO19.13117, XP_713400; Pichia angusta AMM1p, AAQ76838; A. fumigatus ammonium transporter EAL91508; Phytophthora infestans ammonium transporter, AAN31513.
FIG. 2.
Transmembrane structure of MepA, MepB, and MepC. Data were obtained from TMHMM2 (20). All three proteins consist of 11 transmembrane helices and 10 loops of denoted length (ll).
Analysis of mepA, mepB, and mepC gene expression.
The three F. fujikuroi mep genes are repressed when cells are grown in medium containing nitrogen sources such as ammonium, glutamine, arginine, and nitrate at concentrations of 10 and 100 mM, whereas weak expression of the mep genes is observed when 10 mM glutamate is used as a nitrogen source (data not shown). To determine if mep gene expression is dependent on AreA, we compared the expression pattern of the mep genes in wild-type cells and in the areA and glnA mutants. Both strains were initially grown for 5 days in a synthetic medium containing 20 mM glutamine, due to the inability of the areA mutant to use ammonium as a nitrogen source. Following a 5-h starvation in a nitrogen-free medium, the mycelia were transferred to various media containing no nitrogen at all or 10 mM ammonium nitrate or glutamine as nitrogen sources. To find out whether the expression of the mep genes is controlled by the TOR kinase via AreA as has been already shown for the GA and bikaverin biosynthesis genes (47), we added 200 ng/ml rapamycin to one set of flasks.
All three mep genes are strongly repressed by ammonium and glutamine and derepressed under nitrogen starvation conditions (Fig. 3). The three mep genes were not expressed in the areA deletion strain under all of the growth conditions tested, and several single and double GATA or TATC sequence elements were found in the 5′ noncoding regions of all three genes (data not shown), which is consistent with the idea that the mep genes are directly regulated by AreA. Interestingly, the mepA and mepB genes are also not expressed in the ΔglnA mutant whereas mepC is upregulated in this mutant except for the medium with glutamine (Fig. 3).
FIG. 3.
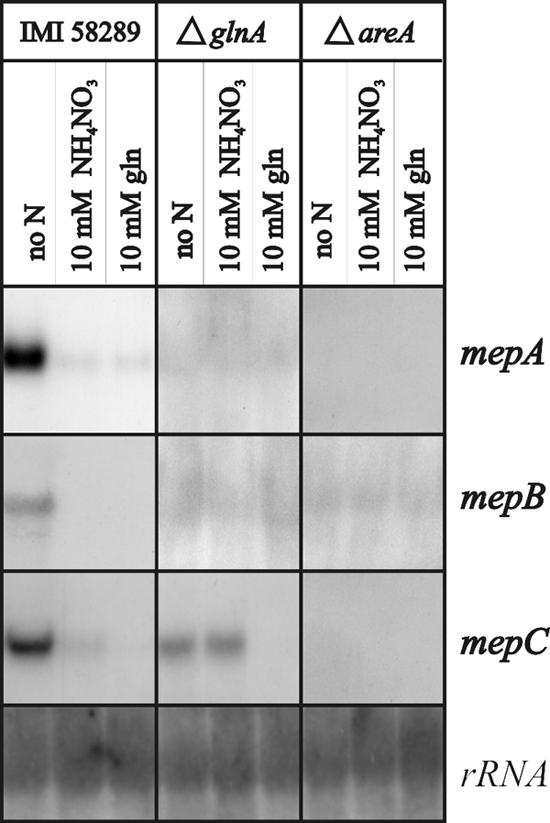
Expression analysis of the F. fujikuroi ammonium permease genes mepA, mepB, and mepC in the wild-type IMI58289 and the ΔglnA and ΔareA mutant strains. The fungal strains were cultivated for 5 days in ICI medium with 10 mM glutamine, and after a 2-h incubation in nitrogen-free ICI medium, the washed mycelia were shifted into medium without nitrogen, with 10 mM ammonium nitrate, or with 10 mM glutamine.
Functional complementation assays in yeast.
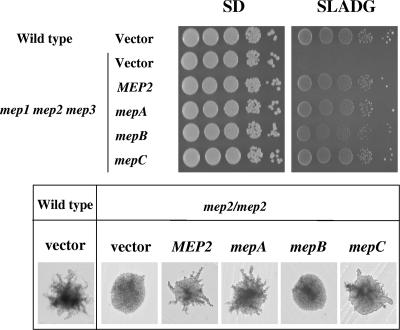
To determine whether the F. fujikuroi mep genes encode functional ammonium transporters, we tested the ability of the F. fujikuroi permeases to rescue the growth defect of an S. cerevisiae mutant (MLY131α) in which all three Mep-encoding genes had been deleted. All three F. fujikuroi mep genes were able to complement the yeast mep1 mep2 mep3 triple mutant to restore growth on low-ammonium medium, consistent with these encoding functional ammonium permeases (Fig. 4). The level of growth was equal to the same strain transformed with the S. cerevisiae MEP2 gene that was expressed from the same vector as the F. fujikuroi mep genes (Fig. 4).
FIG. 4.
Complementation of the yeast mep1 mep2 mep3 triple mutant with the F. fujikuroi mepA, mepB, and mepC cDNA fragments under the control of the S. cerevisiae GAL1 promoter. All three mep genes fully restored the growth of the mep1 mep2 mep3/mep1 mep2 mep3 mutant on SLADG minimal medium. Transformation of the diploid yeast mep2/mep2 mutant with the F. fujikuroi mepA and mepC genes, but not with the F. fujikuroi mepB gene, restored pseudohyphal growth on the nitrogen limited medium SLADG.
The most striking phenotype of a fungal ammonium permease mutant is the loss of filamentous growth by S. cerevisiae and C. albicans mutant strains that lack the permease Mep2 (3, 25). Certain members of the AmtB/Mep2/Ump2/Rh family are able to complement the pseudohyphal defect of the S. cerevisiae Mep2 diploid mutant, e.g., the U. maydis UMP2 and the H. cylindrosporum AMT1 genes (18, 44). Therefore, we analyzed the extent to which the F. fujikuroi mep genes were able to restore the pseudohyphal growth defect of the S. cerevisiae diploid mep2Δ/mep2Δ strain. Transformants containing the individual F. fujikuroi genes were grown on low-ammonium medium for 6 days. The mepA and mepC permease genes were able to complement the mep2Δ/mep2Δ mutant under these conditions, with MepA being the stronger inducer of pseudohyphal growth. The F. fujikuroi mepB gene did not complement the S. cerevisiae mutant, notwithstanding the fact that it was expressed from the same promoter as the other two permease genes.
[14C]methylamine uptake.
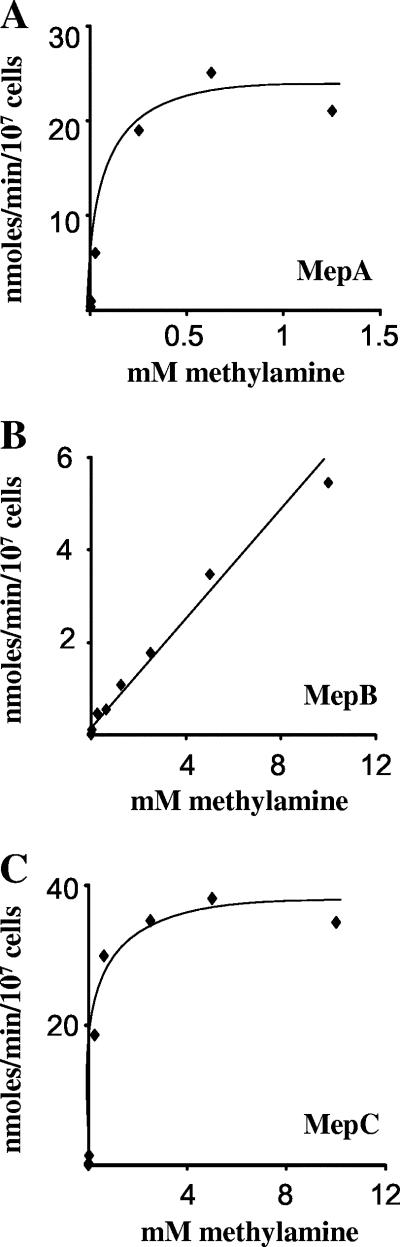
To confirm that the F. fujikuroi mep genes encode functional ammonium permeases, we assayed their ability to mediate [14C]methylamine uptake when expressed in a S. cerevisiae Mep-deficient mutant from a galactose-induced promoter. Cells were grown to mid-log phase using proline as a nitrogen source, and the rate of methylamine uptake over a range of methylamine concentrations was determined. All three F. fujikuroi Mep proteins mediated detectable methylamine uptake that increased with increasing concentrations of external methylamine. In the case of MepA and MepC, the permeases became saturated over the range of methylamine concentrations used (Fig. 5). MepA has the highest relative affinity for methylamine with an apparent Km of 140 μM. MepC exhibited a lower affinity for methylamine with an apparent Km of 1.37 mM and a higher Vmax than MepA. Under these experimental conditions, the highest external concentrations of methylamine did not saturate MepB and precluded the determination of the kinetic parameters of this particular permease. We note that it was also not possible to determine the Km of the low-affinity permease Ump1 from U. maydis using the same experimental system (44). We can conclude, therefore, that MepB has the lowest affinity and highest capacity for methylamine of the three F. fujikuroi ammonium permeases.
FIG. 5.
The Mep proteins of F. fujikuroi mediate [14C]methylamine uptake. Rate of uptake versus substrate concentration curves for MepA (A), MepB (B), and MepC (C) when proteins are expressed from the GAL1-10 promoter in a S. cerevisiae mep1 mep2 mep3 triple mutant strain (see Material and Methods).
Generation and analysis of mepA, mepB, and mepC single and double deletion strains.
Deletion of the three ammonium permease genes mepA, mepB, and mepC was performed by transforming the wild-type strain with the replacement cassettes of the vectors pΔmepA (hygromycin resistance marker), pΔmepB, and pΔmepC (both with the nourseothricin resistance cassette), respectively (Fig. 6) (see Materials and Methods). Altogether, three ΔmepA mutants (ΔmepA-T4, -T9, and -T10), three ΔmepB mutants (ΔmepB-T1, -T2, and -T10) and four ΔmepC mutants (ΔmepC-T4, -T11, -T17, and -T22) were obtained. Deletion mutants transformed with the same replacement cassette showed similar phenotypes whereas transformants with ectopic integrations of the replacement cassette behave as the wild type.
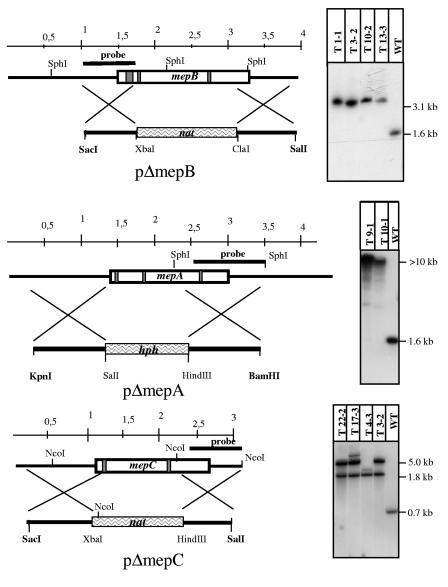
FIG. 6.
Strategy for generating the replacement vectors pΔmepA, pΔmepB, and pΔmepC which were used for deletion of the corresponding ammonium permease genes. mepB and mepA were replaced by the nourseothricin and mepA by the hygromycin resistance cassettes. Introns are shown as gray bars. One of the flanks of each replacement vector was used as a probe for hybridization in Southern blotting. For confirmation of homologous integration of the replacement cassette into the corresponding mep locus, the genomic DNA of the wild type and the deletion mutants was digested with SphI (mepA and mepB) or with EcoRI (mepC). In all three cases, the loss of the wild-type (WT) fragment is shown.
The growth of one deletion strain from each group was examined on plates with various ammonium concentrations (0 to 100 mM) and with 10 mM glutamine. The ΔmepA strain exhibited wild-type levels of growth on all ammonium concentrations. This is consistent with our data from [14C]methylamine uptake experiments showing that this permease seems to have a minor role in ammonium acquisition not only in S. cerevisiae but also in F. fujikuroi and that the lack of MepA is fully compensated by MepB and MepC. The ΔmepB strain, and to a lesser extent the ΔmepC strain, exhibited a decreased growth rate on medium with decreasing levels of ammonium, which was most severe on plates that completely lacked ammonium. Under these latter conditions, the ΔmepA and ΔmepC strains were still able to form a thin mycelium, similar to that on nutrient-free water agar, in contrast to the ΔmepB strains. (Fig. 7A). This is consistent with the idea that MepB is the major ammonium transporter in F. fujikuroi under these growth conditions, with the elimination of this permease causing a strong starvation-induced growth inhibition, probably due to a significant defect in ammonium transport. Surprisingly, 100 mM ammonium revealed a rather inhibitory effect on growth of the wild type and the mutants; the reasons for this growth inhibition are not yet clear.
FIG. 7.
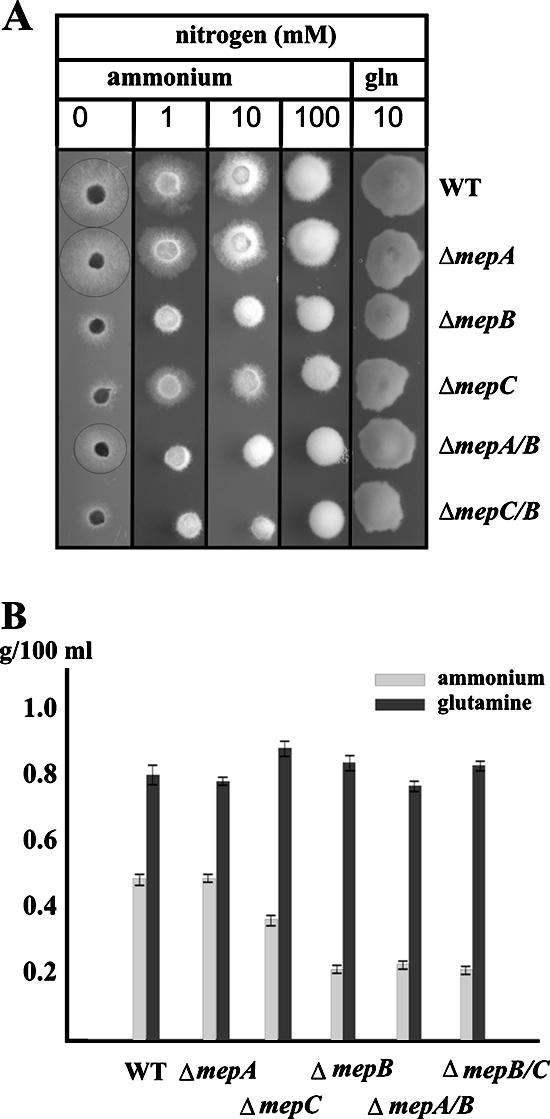
Plate assays for monitoring the growth of the wild-type and mutant strains on ICI medium with different ammonium citrate concentrations. (A) Growth of single and double ammonium permease deletion strains on different ammonium citrate concentrations (0, 1, 10, and 100 mM). The circles around the wild type (WT), mepA, and mepC mutants show the borders of the colonies on plates without any nitrogen. mepB single and ΔmepA ΔmepB (ΔmepA/B) and ΔmepB ΔmepC (ΔmepB/C) double mutants are not able to grow on this medium. (B) Biomass production of the wild type (WT) and the five mutant strains in ICI medium with 10 mM ammonium sulfate or 10 mM glutamine after 72 h of incubation.
To test the combinational effects of the loss of multiple ammonium permeases, we generated ΔmepA ΔmepB and ΔmepB ΔmepC double mutants. Both the ΔmepA ΔmepB and the ΔmepB ΔmepC strains exhibited no further reduction in growth rate under low-ammonium conditions compared with the ΔmepB single mutant. The lack of increased sensitivity to low-ammonium growth by the additional deletion of either ΔmepA or ΔmepC confirmed the major role of MepB. The minimal growth of the ΔmepB ΔmepC strain under these conditions is probably attributed to nonspecific ammonium uptake and the presence of MepA. We were not able to construct a triple mepA mepB mepC mutant due to the lack of a suitable third resistance marker for F. fujikuroi. Therefore, the extent of nonspecific ammonium uptake cannot be precisely determined.
To quantify the growth defect of the mutants lacking one or two Mep permeases, we grew the wild type and the single and double mutants in liquid synthetic medium with ammonium sulfate (10 mM) or glutamine (10 mM) for 48 h and determined the dry weight of the mycelia. While all strains produce almost the same biomass when grown with glutamine as the nitrogen source, the ΔmepB strain and ΔmepA ΔmepB and ΔmepB ΔmepC double mutants produced significantly fewer mycelia when grown on ammonium (Fig. 7B). To link the phenotype of the ΔmepB mutants with the deletion of the mepB permease gene, we complemented the mutant ΔmepB-T10 with the mepB wild-type copy cloned into the vector pGPC1. Hygromycin-resistant transformants were screened for their ability to grow on low-ammonium-containing medium (1 mM ammonium citrate) and for the integration of the complementing mepB gene. Five of the 12 transformants tested revealed the expected PCR fragment for the mepB gene, and these transformants exhibited almost the same growth as the wild type and showed wild-type-like expression of the mepB gene (data not shown). Thus, the growth defect of ΔmepB mutants on low ammonium is clearly linked to the deletion of the mepB ammonium permease gene.
Deletion of mepB but not mepA or mepC confers highly deregulated secondary metabolism.
To date, the impact of single ammonium transporters on secondary metabolism has not been studied in filamentous fungi. In F. fujikuroi, two different nitrogen-free metabolites, the GAs and the red pigment bikaverin, are strictly regulated by nitrogen metabolite repression. To test if the F. fujikuroi ammonium permeases influence GA and bikaverin biosynthesis, we analyzed the production of these secondary metabolites in the mep deletion strains. In wild-type cells, the initiation of bikaverin biosynthesis is a marker for nitrogen depletion, and the production of the pink pigment is accompanied by induction of GA biosynthesis (23, 50).
Wild-type and mutant strains were grown for 48 h in a medium containing 10 mM glutamine to support equal growth for all strains. Aliquots were inoculated into synthetic ICI medium containing 20 mM ammonium or glutamine. The color of the cultures was monitored at 15, 24, 48, and 72 h. The color of the wild-type and ΔmepA cultures changed from white (up to 24 h) to light pink (30 h to 48 h) to yellowish pink (72 h) (Fig. 8). In contrast, the single ΔmepB and the double ΔmepA ΔmepB and ΔmepB ΔmepC mutant strains displayed a red pigmentation already 15 h after inoculation that increased with longer incubation (Fig. 8). The pigmentation of the mepC deletion strain was also apparent after 15 h of cultivation but was not as intense as in strains that lacked mepB. Surprisingly, a slightly earlier pigmentation of ΔmepB, ΔmepA ΔmepB, and ΔmepC mutants was also observed in cultures with glutamine as nitrogen source (Fig. 8).
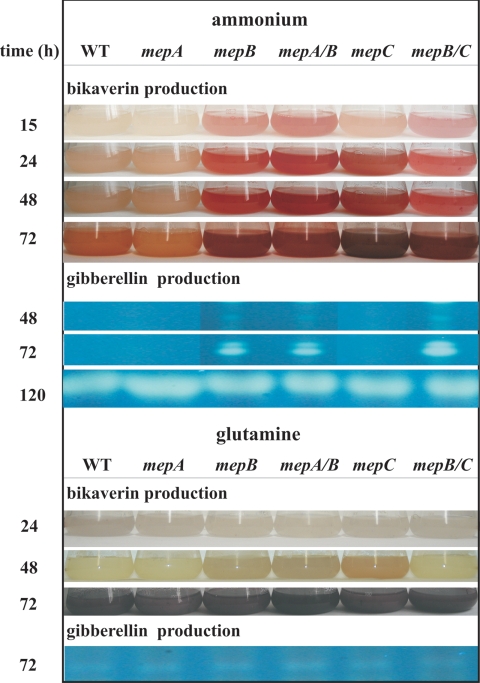
FIG. 8.
Production of bikaverin and GA3 by the wild-type strain IMI58289; the ΔmepA, ΔmepB, and ΔmepC single mutants; and the ΔmepA ΔmepB (mepA/B) and ΔmepB ΔmepC (mepB/C) double mutants. The strains were cultivated in a time course for 3 days in medium containing 12 mM ammonium sulfate or 12 mM glutamine. The pigmentation of the culture fluids demonstrates the much earlier start of the bikaverin production in the ΔmepB single and double mutants and also in the ΔmepC mutant with ammonium as the nitrogen source. The GA3 production under the same conditions is shown by TLC. As for the bikaverin production, the GAs were produced much earlier (at 30 h) in the ΔmepB single and double mutants, whereas the production starts only at 90 h in the wild type and ΔmepA mutant.
To identify the extent to which GA production in F. fujikuroi is also regulated by the ammonium permeases, we monitored the accumulation of GA3 in the wild-type and mutant strains. In a synthetic medium with 20 mM ammonium, the wild-type produced no GAs up to the full depletion of nitrogen sources in the medium but accumulated high amounts of these products under starvation conditions (33). The ΔmepA and mepC mutants behaved as the wild type: these strains did not produce visible amounts of GAs at 30 and 48 h. However, the ΔmepB strain and the ΔmepA ΔmepB and ΔmepB ΔmepC double mutants had already initiated GA3 production by 30 h (Fig. 8). With glutamine, all strains started to produce GA3 only at 72 h (Fig. 8). Thus, deletion of mepB affects the biosynthesis of both secondary metabolites in a similar fashion. The early bikaverin and GA formation in the medium containing ammonium confirms the importance of MepB in ammonium uptake and cellular nitrogen homeostasis.
Gene expression of secondary metabolism genes is deregulated in mepB deletion strains.
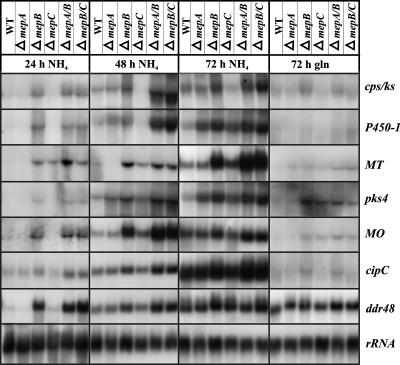
We analyzed the expression of GA and bikaverin biosynthesis genes using the same cultivation conditions that had been used for monitoring bikaverin and GA production. In medium containing 20 mM ammonium, the bikaverin biosynthetic genes pks4, MT, and MO (encoding the polyketide synthase, a methyltransferase, and a monooxygenase from the bikaverin gene cluster) (23; also P. Wiemann and B. Tudzynski, unpublished data) and the GA biosynthetic genes (cps/ks and P450-1) (reviewed in reference 51) were expressed by 24 h in the mepB single and double mutants. The same transcripts were detectable only at 48 or 72 h in the wild-type and the mepA and mepC mutant strains (Fig. 9). This pattern of expression has also been observed for other areA target genes, e.g., the peptide transporter gene mtd1 and the amino acid permease gene aap8 (B. Schönig and B. Tudzynski, unpublished data; also data not shown) as well as a set of GS target genes, e.g., ddr48 and cipC (Fig. 9), which are not expressed in the glnA mutant (46). As expected, nitrogen deregulation is most significant when the cultures contain ammonium as the nitrogen source. However, the expression of the GA and bikaverin biosynthetic genes is also partially deregulated in single and double mepB mutants and, for some genes, also in mepC mutants with glutamine as a nitrogen source, although the effect is not as clear as with ammonium (Fig. 9).
FIG. 9.
Northern blot analysis with the F. fujikuroi wild type (WT); the ΔmepA, ΔmepB, and ΔmepC single mutants; and the ΔmepA ΔmepB (ΔmepA/B) and ΔmepB ΔmepC (ΔmepB/C) double mutants. Strains were cultivated in a time course for 3 days in medium containing 12 mM ammonium sulfate or glutamine. For the glutamine cultures, only one time point (72 h) is shown due to the low expression levels of the GA and bikaverin genes at 24 and 48 h. The filters were probed with the cDNA fragments of the genes.
Is ammonium incorporation by metabolism required for nitrogen metabolite repression?
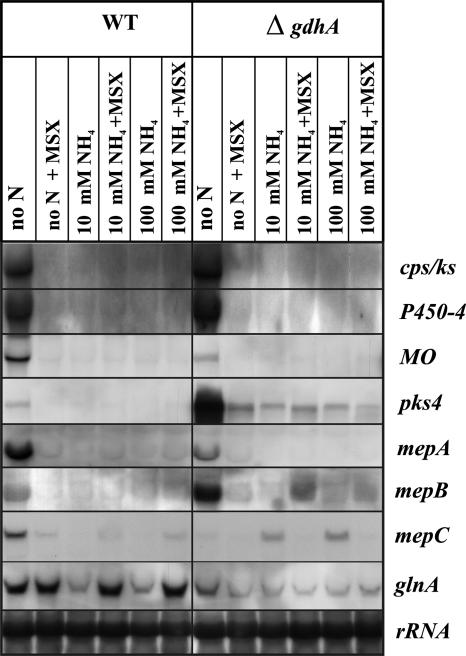
Fungal ammonium assimilation occurs via its incorporation into glutamate and glutamine by glutamate dehydrogenase A (GdhA) and GS, respectively (38). To answer the question whether ammonium itself acts as an effector of nitrogen metabolite repression, or if repression of GA and bikaverin biosynthesis genes (and other NCR genes) by ammonium is due to its conversion to glutamate and/or glutamine, we completely abolished ammonium incorporation into metabolism. For this, we used a gdhA deletion mutant (B. Tudzynski, unpublished data) which is unable to assimilate ammonium through reductive synthesis of glutamate from 2-oxoglutarate, and we inhibited GS with the specific inhibitor MSX to abolish the second route of ammonium assimilation by GS. We compared the expression of the three mep genes and the GA and bikaverin biosynthesis genes in the gdhA mutant with that in the wild type in medium without nitrogen and with ammonium, with or without MSX. Under these conditions, MSX did not prevent strong repression of gene expression by ammonium either in the wild type or in the gdhA mutant (Fig. 10). However, under starvation conditions (no nitrogen), several genes were upregulated in the gdhA mutant, suggesting that they are inhibited by both ammonium and glutamate/glutamine in the wild type. Interestingly, mepC is slightly induced by intracellular ammonium in the mutant, and this induction is abolished by MSX (Fig. 10). On the other hand, the expression of mepB is slightly induced in the mutant when MSX is added to the ammonium-containing medium, suggesting a functional link between MepB and the GS. In summary, these results demonstrate that ammonium itself causes strong nitrogen metabolite repression by transducing the signal of nitrogen availability.
FIG. 10.
Northern blot analysis with the F. fujikuroi wild type (WT) and ΔgdhA mutant. Strains were cultivated in a time course for 3 days in medium containing 10 mM ammonium sulfate. The mycelia were washed and shifted for 2 h into nitrogen-free medium or medium containing 10 or 100 mM ammonium sulfate, with or without the GS inhibitor MSX (2 mM). The filters were probed with the cDNA fragments of the GA (cps/ks and P450-4) and bikaverin (MO and pks4) biosynthetic genes as well as the three mep genes and the GS-encoding gene glnA.
Does MepB play a regulatory role?
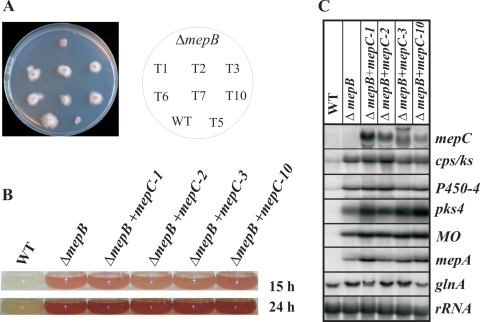
The strong effect of mepB deletion on expression of secondary metabolite and some other NCR genes (e.g., mtd1 and aap8) can be explained by its role as the major ammonium permease or by a suggested additional role as a sensor-like regulatory component transducing the signal of nitrogen availability via a yet unknown signaling pathway to AreA. To answer this question, we overexpressed mepC (which was shown to be a high-capacity permease beside MepB) in the mepB mutant to rescue the poor growth of mepB mutants on ammonium. We fused the mepC gene with the strong promoter of the F. fujikuroi glnA gene (glnAprom) (46) and cloned the chimeric fragment into vector pUCH2-8 containing the hygromycin resistance cassette. The resulting vector pglnA::mepC was used to transform the mepB mutant T1. Hygromycin-resistant transformants were evaluated by PCR for the presence of the glnAprom-mepC fusion product and by growth assays on medium with 1 mM ammonium. Six out of 12 transformants contained the PCR product and exhibited enhanced growth compared to the mepB mutant, whereas the other transformants (e.g., T5) did not contain the fusion product and grew as poorly on 1 mM ammonium as the ΔmepB recipient strain (Fig. 11A). Statistical analysis of the growth rate of the mepC-overexpressing strains compared with that of the mepB mutant and the wild type is shown in Table 2.
FIG. 11.
Overexpression of mepC (glnAprom::mepC) in the ΔmepB deletion mutant. (A) Growth of several transformants carrying the overexpression cassette on 1 mM ammonium sulfate. Transformants T1, T2, T3, and T10 were chosen for detailed analyses (see below). Transformant T5 does not contain the whole promoter-gene fusion fragment and shows the same growth defect as the ΔmepB deletion mutant (negative control). (B) Bikaverin production in the wild type, ΔmepB mutant, and four mepC-overexpressing transformants, which still show a much earlier pigment production than the wild type. (C) Northern blot analysis with the F. fujikuroi wild type, the ΔmepB mutant, and four mepC-overexpressing transformants. The strains were cultivated for 24 h in medium containing 12 mM ammonium sulfate. The early pigmentation (as shown in panel B) corresponds very well with early expression of several AreA target genes (for an explanation, see the legend of Fig. 10) in the mepC-overexpressing transformants. WT, wild type.
TABLE 2.
Partial restoration of vegetative growth of the ΔmepB mutant by overexpression of MepC
| Strain | Colony diam (mm) ata:
|
|||
|---|---|---|---|---|
| 48 h | 72 h | 96 h | 168 h | |
| Wild type | 11 ± 1 | 14 ± 0 | 16 ± 1 | 22 ± 2 |
| ΔmepB-T1 | 6 ± 1 | 8 ± 1 | 9 ± 1 | 11 ± 1 |
| ΔmepB+mepC-1 | 9 ± 1 | 12 ± 1 | 14 ± 2 | 21 ± 2 |
| ΔmepB+mepC-2 | 9 ± 1 | 12 ± 1 | 13 ± 1 | 21 ± 2 |
| ΔmepB+mepC-3 | 9 ± 1 | 12 ± 1 | 14 ± 1 | 22 ± 2 |
| ΔmepB+mepC-10 | 9 ± 1 | 13 ± 1 | 13 ± 2 | 21 ± 2 |
Data are the average of three independent measurements ± standard deviation.
If the strong phenotype of mepB mutants regarding the derepression of secondary metabolism genes is due to ammonium limitation, we would expect a loss of this deregulation by overexpressing mepC. We cultivated the wild type, the mepB mutant, and four mepC-overexpressing transformants (designated ΔmepB+mepC-1, -2, -3, and -10) in ICI medium with 20 mM ammonium sulfate. Surprisingly, the mepC-overexpressing mutants already showed a strong pigmentation after 15 h, similar to the mepB mutant despite having a wild-type growth pattern (Fig. 11B). The mycelia were harvested after 24 h and used for expression studies by Northern blot analysis. The mepC gene was strongly up-regulated in the mutants, as expected. The GA and bikaverin genes are highly expressed at 24 h as is the mepB mutant, despite the almost full restoration of growth (Fig. 11C). These data suggest that MepB plays an additional role in nitrogen regulation in F. fujikuroi, probably by sensing and transducing the signal of ammonium availability.
DISCUSSION
Transport capacity of MepA, MepB, and MepC.
In this study, we report the cloning and characterization of three genes, mepA, mepB, and mepC, which encode ammonium permeases in F. fujikuroi. All three proteins share a high level of sequence identity to other ammonium permeases of the AmtB/Mep family and are able to complement a S. cerevisiae mep1 mep2 mep3 triple mutant despite the evolutionary distance between S. cerevisiae and F. fujikuroi. Our data indicate that they differ in ammonium transport capacity and affinity toward ammonium ions. MepB appears to be the major ammonium transporter in F. fujikuroi, followed by MepC, as their deletion resulted in substantially reduced growth on medium containing 1 mM or 10 mM ammonium. These data correspond to the [14C]methylamine uptake data in S. cerevisiae by which MepB was shown to have the lowest affinity and highest capacity for methylamine of the three F. fujikuroi ammonium permeases. A similar phenotype has been described for meaA deletion mutants of A. nidulans, which exhibit a significantly reduced growth rate on medium with low concentrations of ammonium, are resistant to methylamine, and play a physiological role in retaining the ammonium derived from the metabolism of other nitrogen sources (34). Increased methylamine resistance has not been observed for F. fujikuroi mep mutants due to the extremely high resistance of the wild type. Our analysis of the relative affinities of the F. fujikuroi Mep permeases clearly shows that these proteins are able to mediate methylamine uptake. This suggests that F. fujikuroi contains an efficient mechanism to detoxify imported methylamine. Consistent with this, the F. fujikuroi genome contains at least three genes that code for homologues of fungal amine oxidases which are known to catalyze the oxidative deamination of amines and can therefore confer resistance to methylamine (11). The differential number, expression levels, and/or specificity of amine oxidases could result in a range of sensitivity to methylamine among different fungi species.
Interestingly, the MepB permease of F. fujikuroi does not group together with MeaA or yeast Mep1, despite their functional similarity as high-capacity permeases. Instead, MepB groups with the A. nidulans and Tuber borchii high-affinity permeases MepA and TbAMT1, respectively (34, 37). MeaA shows high sequence similarity to the F. fujikuroi MepC permease, which is also a high-capacity permease according to the reduced growth on medium containing 1 to 100 mM ammonium. In contrast to mepB and mepC mutants, mepA mutants exhibit no obvious phenotype. They grow at the same rate as the wild type and produce colonies with thin mycelia, even on plates without nitrogen. Analysis of methylamine uptake confirmed that MepA is a high-affinity and low-capacity permease. MepA groups into an AmtB/Mep subclass together with the A. nidulans MepA and the F. fujikuroi high-capacity permease MepB. Therefore, the two F. fujikuroi permeases MepA and MepB are phylogenetically highly related but differ in their affinity for ammonium and their transport capacity, demonstrating that a high degree of sequence similarity cannot be strictly correlated with functional inferences.
The expression of mepA, mepB, and mepC is highly dependent on AreA.
The expression of all three mep genes is uniformly regulated by AreA. In the ΔareA mutant, no transcript levels were detected for mepA, mepB, and mepC, consistent with the finding that the F. fujikuroi ΔareA mutant does not grow on medium with ammonium as the only nitrogen source. This contrasts with A. nidulans where disruption of the zinc finger region of areA does not result in loss of ability to grow on ammonium but prevents utilization of nitrogen sources other than ammonium and glutamine (21).
The expression of several Mep2-type permeases, such as the S. cerevisiae MEP genes (30) and the four permeases in A. nidulans (34, 35, 36), is nitrogen regulated and depends on activation by the transcription factors Gln3 and AreA, respectively. Genes encoding permeases of the Mep1/Mep3 type, such as Mep1 in C. albicans (3) and MeaA in A. nidulans (34, 35), are expressed at much lower levels than the high-affinity Mep2-type transporters. In F. fujikuroi, mepA shows the highest expression level similar to other Mep2-like permeases. Interestingly, the expression of mepC is upregulated in the glnA mutant in medium without nitrogen or with 10 mM ammonium, whereas glutamine overcomes this effect. In contrast, the mepA and mepB genes are repressed in both the areA and glnA mutants. Further studies will be required to identify the mechanism of GS-dependent expression of the mep genes.
Does MepB participate in nitrogen regulation?
Some fungal ammonium permeases have evolved a regulatory function. In S. cerevisiae, several plasma membrane-localized permeases are able to sense the environmental availability of amino acids, ammonium, and sugars (10, 52). Among the three Mep proteins, Mep2 is a receptor for ammonium and is required for pseudohyphal growth under nitrogen starvation conditions (25). In C. albicans, CaMep2, but not CaMep1, also induces filamentous growth under limiting nitrogen conditions via the activation of both mitogen-activated protein kinase and the Ras1-cyclic AMP-dependent pathways (3). The maize pathogen U. maydis expresses two ammonium permease-encoding genes, ump1 and ump2 (44). Ump2 is required to trigger filamentous growth on low-nitrogen medium and complements the pseudohyphal growth defect of the S. cerevisae mep2 mutant, consistent with a sensor role (44). Similarly, it has been proposed that in the mycorrhiza fungus H. cylindrosporum, the high-affinity ammonium transporters Amt1 and Amt2 induce, via an as yet unknown signal transduction cascade, a switch in the fungal growth mode during the formation of mycorrhiza (17).
The molecular mechanisms involved in transporter-mediated ammonium regulation are not presently known although some important aspects have been identified in yeast that may be conserved within fungi. The Mep2-like permeases are high-affinity transporters and are the most highly transcribed in response to ammonium limitation. Promoter exchange and reporter gene experiments show that CaMep2 needs to be expressed at very high levels to mediate filamentous growth induction under low-ammonium conditions (3). A mutant strain lacking such a regulatory permease would also be expected to exhibit a signaling phenotype, and no phenotype that could be attributed to a reduction in nitrogen availability, such as a reduction in growth rate. Other characteristics of the yeast ammonium permeases are less predictive with regard to a role as regulatory proteins. Sequence alignments of the currently available Meps in the database show that fungal Meps contain a putative protein kinase A (PKA) phosphorylation site [RRX(S/T), where X is any residue] that is not present in those from other organisms (44). Point mutations in these sites in Ump2 (Ser288-Ala) and Mep2 (Thr288-Ala) do not abolish ammonium transport, but both mutant Meps fail to complement the pseudohyphal growth defect of the S. cerevisiae mep2 mutant (44). However, nonsensing homologues of Mep2 (e.g., Mep1 and Mep3 of S. cerevisiae) also contain a similar potential PKA phosphorylation site. In addition, the S. cerevisiae Mep2 is glycosylated at the fourth amino acid. However, Mep2N4Q mutation of the S. cerevisiae glycosylation site does not prevent pseudohyphal growth (32). Many of the Mep2-specific features, such as glycosylation and putative PKA phosphorylation sites, are found in all three F. fujikuroi Mep proteins. In addition, all Fusarium Meps are predicted to have the same overall structure as Mep2, including the putative sensing loop (25). It is therefore challenging to identify conserved characteristics within the ammonium permeases of filamentous fungi that define a regulatory function.
The data presented in this study are most consistent with MepA having the characteristics of a regulatory permease: the gene is highly expressed, the mepA mutants show no severe growth defect on low ammonium, MepA (but also MepB and MepC) is predicted to be glycosylated, and MepA is a high-affinity transporter that is able to induce pseudohyphal growth in a mep2-deficient S. cerevisiae mutant. MepC also restores the pseudohyphal growth in the yeast mep2 mutant, but the deletion of mepC conferred an obvious growth phenotype in F. fujikuroi. However, the expression patterns of the AreA target genes argue against a role of MepA or MepC as extracellular ammonium sensors that link ammonium availability to the regulation of AreA activity. If MepA or MepC were a sensor permease, then the signal for nitrogen sufficiency would be transduced in ΔmepB mutants, resulting in strong repression of AreA target genes, but this is not the case.
In contrast, MepB does not restore pseudohyphal growth of the S. cerevisiae mep2 mutant, and mepB mutants do not grow under low-ammonium conditions, making it appear to be a less likely candidate for a regulatory permease. However, MepB is the only permease affecting the regulation of AreA target genes, such as the GA and bikaverin biosynthesis genes, which are derepressed in the ΔmepB mutant under ammonium-sufficient conditions. In addition, a partial derepression of GA and bikaverin genes in the mepB mutants was observed also on glutamine, similar to C. albicans where the Mep2 orthologue is required for nitrogen limitation-induced filamentation under conditions of both ammonium and amino acid limitation (3).
There are two possible explanations for the strong impact of MepB on regulation. First, the effects on gene expression may be due to reduced ammonium transport, in agreement with the growth defect of the mepB mutant and with methylamine uptake experiments demonstrating that MepB is the major ammonium transporter in F. fujikuroi. Second, MepB might play an additional role in nitrogen regulation despite not exhibiting a regulatory function in the heterologous S. cerevisiae model. If the derepressing effect on nitrogen-regulated genes in the ΔmepB mutant is the result of ammonium limitation due to the loss of the major transporter, overexpression of the second high-capacity permease, MepC, should restore not only the wild-type growth but also the wild-type expression levels of these genes. However, despite the significantly increased growth of the mepC-overexpressing mutants (Fig. 11), the deregulation of GA and bikaverin biosynthesis genes as well as other AreA-dependent genes is the same as in the ΔmepB mutant. These data support our suggestion that MepB might play an important role in transport and sensing of ammonium although this permease does not complement the pseudohyphal growth defect in yeast. However, due to the evolutionary distance between F. fujikuroi and S. cerevisiae, heterologous expression of F. fujikuroi mep genes need not necessarily mimic all of the regulatory events or expression levels that occur in the natural host.
However, more definitive evidence will be required before a sensing function can be attributed to any of the F. fujikuroi ammonium permeases. Such evidence includes the isolation of mutants, such as those recently identified in Mep2 of S. cerevisiae (J. Rutherford et al., personal communication), that separate the transport and sensing function of the permease, a demonstration of interactions with signal transduction proteins, or the identification of downstream physiological events such as altered transcriptional programs. This will also include the identification of mutants analogous to the hyperactive Mep2G349C mutant of S. cerevisiae that restores dimorphic growth in a pseudohyphal-deficient mutant (npr1Δ) and thereby establishes a clear correlation between Mep2 dependent transport and signaling (4).
Is the metabolism of ammonium essential for triggering nitrogen repression?
In S. cerevisiae, the stimulus for the expression of nitrogen-regulated genes by the AreA homologue Gln3 is a drop in the intracellular level of glutamine (7, 27). Therefore, we sought to establish whether the reduced intracellular glutamine level, resulting from the compromised ammonium transport capacity in the mepB mutants, or the decreased ammonium availability itself triggers the activation of AreA via a yet unknown signaling cascade and the derepression of AreA target genes. To answer this question, we inactivated both the GdhA and GS pathways for ammonium incorporation into metabolism. For this, we used a gdhA deletion mutant and treated the mycelium with the specific GS inhibitor MSX. Ammonium-induced strong repression of GA and bikaverin biosynthetic genes was not prevented under these conditions, demonstrating that the intracellular level of glutamine is not the only signal sensed by the fungal cells that activates/inactivates signaling pathways to AreA and that ammonium itself is an effector of nitrogen metabolite repression in F. fujikuroi.
Conclusions.
We identified one of the three ammonium permeases, MepB, as the major transporter and as a potential element in the F. fujikuroi nitrogen regulation network. Although the inability of MepB to restore the pseudohyphal growth defect of S. cerevisiae mep2 mutants argues against a direct role of MepB as a regulator, at least in a heterologous organism, several other features support such a role. The strongest argument for a regulatory role is the overexpression of mepC encoding the second high-capacity transporter in F. fujikuroi, which resulted in significantly improved growth but did not prevent the deregulated expression pattern of the nitrogen-regulated secondary metabolism genes.
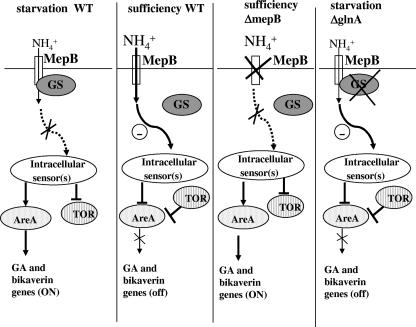
Recently, we showed that GA and bikaverin biosynthetic genes are strongly repressed in a ΔglnA mutant even under nitrogen-limiting conditions, despite the fact that the intracellular glutamine pool is drastically reduced. These results are in contrast to our expectation and suggest a regulatory role of GS in addition to its enzymatic function, e.g., by protein-protein interactions with other components of the nitrogen regulation network (46). Summarizing our current knowledge of the role of GS and other components involved in nitrogen regulation of secondary metabolism in F. fujikuroi and the postulated regulatory role of MepB, we propose a model for nitrogen sensing and signaling (Fig. 12). In this model, MepB is the major ammonium transporter which is able to sense ammonium (nitrogen) availability and to transduce a repressing signal to AreA by an as yet unknown signaling pathway. Under ammonium limitation conditions, GS binds to MepB, thereby blocking the transduction of this repressing signal and resulting in the induced expression of glnA and probably increased GS levels. The missing signal leads to an activation of AreA and derepression of AreA target genes, such as the mep and the GA and bikaverin biosynthetic genes. This model can also explain why GS is needed for the expression of the GA and bikaverin biosynthesis genes under nitrogen starvation conditions (46). The inability of GS to bind to MepB in the glnA deletion mutant would enable the putative sensor MepB to constitutively activate signaling, resulting in strong repression of AreA target genes. In support of this model, a close metabolic coupling of the membrane-bound GS to the ammonium channel AmtB has been recently shown for E. coli (19). This model will provide the basis of future work to identify the molecular mechanisms that link nitrogen availability to the regulation of secondary metabolism in F. fujikuroi.
FIG. 12.
Model of the nitrogen regulation network in F. fujikuroi. MepB is postulated as a key component in this model, acting as the major ammonium permease. In addition, mepB might also act as a nitrogen sensor either by mediating the signal of nitrogen availability or indirectly by causing a drop in the intracellular glutamine level. In the latter case we suggest an intracellular sensor transducing the glutamine signal to AreA via TOR and/or additional signaling pathways. WT, wild type.
Acknowledgments
Research in the Tudzynski laboratory was supported by the DFG (Tu1245/7), and research in the Heitman laboratory was supported by NIH/NIAID R01 grant AI39115-09.
We thank Sabine Huber (University Münster) for excellent technical assistance and Sabrina Wemhoff for cloning and sequencing the F. fujikuroi mepC gene. We also thank Mike Perlin for supplying us with the pYes2.1-Mep2 plasmid.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Alexander, N. J., S. P. McCormick, and T. M. Hohn. 1999. TRIII gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, K., and J. Morschhäuser. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56649-669. [DOI] [PubMed] [Google Scholar]
- 4.Boeckstaens, M., B. Andre, and A. M. Marini. 2007. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64534-546. [DOI] [PubMed] [Google Scholar]
- 5.Cenis, J. L. 1993. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 202380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, T. G., and R. A. Sumrada. 1983. What is the function of nitrogen catabolite repression in Saccharomyces cerevisiae? J. Bacteriol. 155623-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 996784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darken, M. A., A. A. L. Jensen, and P. Shu. 1959. Production of gibberellic acid by fermentation. Appl. Microbiol. 7301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 1213-15. [Google Scholar]
- 10.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 4091-109. [DOI] [PubMed] [Google Scholar]
- 11.Frébort, I., K. Matsushita, H. Toyama, K. Lemr, M. Yamada, and O. Adachi. 1999. Purification and characterization of methylamine oxidase induced in Aspergillus niger AKU 3302. Biosci. Biotechnol. Biochem. 63125-134. [DOI] [PubMed] [Google Scholar]
- 12.Fu, Y. H., and G. A. Marzluf. 1990. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc. Natl. Acad. Sci. USA 875331-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazzarrini, S., L. Lejay, A. Gojon, O. Ninnemann, W. B. Frommer, and N. von Wiren. 1999. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geissman, T. A., A. J. Verbiscar, B. O. Phinney, and G. Cragg. 1966. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry 5933-947. [Google Scholar]
- 15.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van de Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29556-564. [DOI] [PubMed] [Google Scholar]
- 16.Howitt, S. M., and M. K. Udvardi. 2000. Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta 1465152-170. [DOI] [PubMed] [Google Scholar]
- 17.Javelle, A., B. André, A. M. Marini, and M. Chalot. 2003. High-affinity ammonium transporters and nitrogen sensing in mycorrhizas. Trends Microbiol. 1153-55. [DOI] [PubMed] [Google Scholar]
- 18.Javelle, A., M. Morel, B. R. Rodriguez-Pastrana, B. Botton, B. Andre, A. M. Marini, A. Brun, and M. Chalot. 2003. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 47411-430. [DOI] [PubMed] [Google Scholar]
- 19.Javelle, A., G. Thomas, A. M. Marini, R. Krämer, and M. Merrick. 2005. In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem. J. 390215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 21.Kudla, B., M. X. Caddick, T. Langdon, N. M. Martinez-Rossi, C. F. Bennett, S. Sibley, and R. W. Davies. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 91355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linnemannstöns, P., J. Schulte, M. del Mar Prado, R. H. Proctor, J. Avalos, and B. Tudzynski. 2002. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet. Biol. 37143-148. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 167008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 171236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2901-18. [DOI] [PubMed] [Google Scholar]
- 27.Magasanik, B. 2005. The transduction of the nitrogen regulation signal in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 10216537-16538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malonek, S., M. C. Rojas, P. Hedden, P. Gaskin, P. Hopkins, and B. Tudzynski. 2004. The NADPH-cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem. 27925075-25084. [DOI] [PubMed] [Google Scholar]
- 29.Marini, A. M., S. Vissers, A. Urrestarazu, and B. Andre. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 133456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marini, A. M., S. Soussi-Boudekou, S. Vissers, and B. Andre. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 174282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marini, A. M., B. Andre, J. P. Cartron, and B. Cherif-Zahar. 2000. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat. Genet. 26341-344. [DOI] [PubMed] [Google Scholar]
- 32.Marini, A. M., and B. Andre. 2000. In vivo N-glycosylation of the Mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. Mol. Microbiol. 38552-564. [DOI] [PubMed] [Google Scholar]
- 33.Mihlan, M., V. Homann, T. W. Liu, and B. Tudzynski. 2003. AreA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol. Microbiol. 47975-991. [DOI] [PubMed] [Google Scholar]
- 34.Monahan, B. J., J. S. Fraser, M. J. Hynes, and M. A. Davis. 2002a. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 185-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monahan, B. J., S. E. Unkles, I. T. Tsing, J. R. Kinghorn, M. J. Hynes, and M. A. Davis. 2002b. Mutation and functional analysis of the Aspergillus nidulans ammonium permease MeaA and evidence for interaction with itself and MepA. Fungal Genet. Biol. 3635-46. [DOI] [PubMed] [Google Scholar]
- 36.Monahan, B. J., M. C. Askin, M. J. Hynes, and M. A. Davis. 2006. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell 5226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montanini, B., N. Moretto, E. Soragni, R. Percudani, and S. Ottonello. 2002. A high-affinity ammonium transporter from the mycorrhizal ascomycete Tuber borchii. Fungal Genet. Biol. 3622-34. [DOI] [PubMed] [Google Scholar]
- 38.Noor, S., and N. S. Punekar. 2005. Allosteric NADP-glutamate dehydrogenase from aspergilli: purification, characterization and implications for metabolic regulation at the carbon-nitrogen interface. Microbiology 1511409-1419. [DOI] [PubMed] [Google Scholar]
- 39.Pontecorvo, G. V., J. A. Roper, L. M. Hemmonns, K. D. Mac Donald, and A. W. J. Buften. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 141141-238. [DOI] [PubMed] [Google Scholar]
- 40.Rademacher, W. 1997. Gibberellins, p. 193-205. In T. Anke (ed.), Fungal biotechnology. Chapman and Hall, London, United Kingdom.
- 41.Rohde, J. R., and M. E. Cardenas. 2004. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 27953-72. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Sigrist, C. J. A., L. Cerutti, N. Hulo, A. Gattiker, L. Falquet, M. Pagni, A. Bairoch, and P. Bucher. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3265-274. [DOI] [PubMed] [Google Scholar]
- 44.Smith, D. G., M. D. Garcia-Pedrajas, S. E. Gold, and M. H. Perlin. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50259-275. [DOI] [PubMed] [Google Scholar]
- 45.Sonnhammer, E. L., G. Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6175-182. [PubMed] [Google Scholar]
- 46.Teichert, S., B. Schönig, S. Richter, and B. Tudzynski. 2004. Deletion of the Gibberella fujikuroi glutamine synthetase gene has significant impact on transcriptional control of primary and secondary metabolism. Mol. Microbiol. 531661-1675. [DOI] [PubMed] [Google Scholar]
- 47.Teichert, S., M. Wottawa, B. Schönig, and B. Tudzynski. 2006. Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell 51807-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ter Schure, E. G., N. A. van Riel, and C. T. Verrips. 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2467-83. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, G. H., J. G. I. Mullins, and M. Merrick. 2000. Membrane topology of the Mep/Amt family of ammonium transporters. Mol. Microbiol. 37331-344. [DOI] [PubMed] [Google Scholar]
- 50.Tudzynski, B., V. Homann, B. Feng, and G. Marzluf. 1999. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol. Gen. Genet. 261106-114. [DOI] [PubMed] [Google Scholar]
- 51.Tudzynski, B. 2005. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 66597-611. [DOI] [PubMed] [Google Scholar]
- 52.Van Nuland, A., P. Vandormael, M. Donaton, M. Alenquer, A. Lourenco, E. Quintino, M. Versele, and J. M. Thevelein. 2006. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 591485-1505. [DOI] [PubMed] [Google Scholar]
- 53.Wiame, J.-M., M. Grenson, and H. N. Arst, Jr. 1985. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv. Microbiol. Physiol. 261-87. [DOI] [PubMed] [Google Scholar]