Abstract
The conserved AmtB/Mep/Rh family of proteins mediate the transport of ammonium across cellular membranes in a wide range of organisms. Certain fungal members of this group are required to initiate filamentous growth. We have investigated the functions of two members of the AmtB/Mep/Rh family from the pathogenic basidiomycete Cryptococcus neoformans. Amt1 and Amt2 are low- and high-affinity ammonium permeases, respectively, and a mutant lacking both permeases is unable to grow under ammonium-limiting conditions. AMT2 is transcriptionally induced in response to nitrogen limitation, whereas AMT1 is constitutively expressed. Single and double amt mutants exhibit wild-type virulence in two models of cryptococcosis. Consistent with this, the formation of two C. neoformans virulence factors, cell wall melanin and the extracellular polysaccharide capsule, is not impaired in cells lacking either or both of the Amt1 and Amt2 permeases. Amt2 is, however, required for the initiation of invasive growth of haploid cells under low-nitrogen conditions and for the mating of wild-type cells under the same conditions. We propose that Amt2 may be a new fungal ammonium sensor and an element of the signaling cascades that govern the mating of C. neoformans in response to environmental nutritional cues.
Ammonium is an excellent nitrogen source for microorganisms, and studies on a variety of fungi suggest that each species contains at least one high- and one low-affinity ammonium transporter (2, 12, 26, 29, 31, 39). These belong to the AmtB/Mep/Rh family of proteins, which are conserved between bacteria and humans (13, 27). Structural studies with bacterial members of this family predict that these transporters form channels that allow the passive diffusion of ammonium to occur (1, 13, 48). Ammonium selectivity arises from the presence of a narrow hydrophobic channel that necessitates the deprotonation of the translocating ammonium ion to form ammonia gas that is reprotonated on the cytoplasmic side of the cell (13). The fungal ammonium permeases are transcriptionally regulated to ensure that appropriate pathways are expressed in response to particular nitrogen sources (reviewed in reference 5).
Some fungal members of the AmtB/Mep/Rh family are required to initiate a dimorphic transition from yeast to filamentous growth (2, 23, 39). This enables cells within a colony to grow away from an area of nutrient limitation in a coordinated manner, and in some pathogenic fungi this can contribute to virulence, if it is the filamentous form that is infectious (20). The molecular mechanisms that are involved in the ammonium permease-dependent aspect of this process are not understood. In particular, it is not known if these permeases act as sensors of ammonium availability and couple ammonium transport with the regulation of a signal transduction pathway. Interestingly, certain fungal homologues of the S. cerevisiae Mep2 permease can restore pseudohyphal growth in a strain that lacks the endogenous Mep2 (12, 39). This suggests that aspects of this process are conserved in different fungal species.
The ability and propensity of fungal pathogens to undergo sexual reproduction and its influence on the virulence of these organisms are fundamental. Studies on environmental populations of pathogenic fungi suggest that although sexual reproduction occurs, it is often limited and that predominantly clonal populations exist. This suggests that a clonal existence enables the successful colonization of a particular host or environment and that sexual reproduction may enable survival in response to adverse selective pressure (reviewed in reference 9). The balance between clonal and sexual populations of the fungal pathogen Cryptococcus neoformans is therefore of significant interest. C. neoformans is a saprophytic yeast that is predominantly a pathogen of immunocompromised individuals. It occurs as a haploid yeast of two mating types, α and a, although the vast majority of environmental and clinical samples are of the α type (15). An environmental population that exhibits evidence of recombination has been isolated within sub-Saharan Africa, which is consistent with the occurrence of geographically restricted sexual reproduction (19).
The discovery of a novel sexual cycle in C. neoformans has raised interesting questions relating to the balance between the sexual and clonal populations of this organism. Monokaryotic fruiting by C. neoformans has been known for some time and occurs predominately with haploid α cells when they are cultured under certain conditions (45). This developmental pathway results in morphological changes strikingly similar to those that occur during mating. Recently, monokaryotic fruiting has been found to be dependent upon conserved meiotic proteins and to involve the formation of a diploid intermediate that is resolved by a process involving recombination and the production of haploid spores (18).
Interest in the link between mating and virulence of C. neoformans extends beyond the maintenance of a clonal population as a pathogenic strategy. The spores that result from both traditional a-α mating and monokaryotic fruiting are the potential infectious agent of this organism (40, 49). In addition, several genes within the 100-kb region of the mating type locus are linked to the virulence of this yeast (4, 44). In certain strain backgrounds, α cells are more virulent than a cells (16). In the most common pathogenic serotype, there is no difference in virulence between the two mating types; however, in a murine model of coinfection, α cells are more successful at penetrating the blood-brain barrier (34, 35). Study of the environmental signals that initiate both traditional a-α mating and monokaryotic fruiting will allow a fuller understanding of the sexual process in C. neoformans. Nitrogen starvation induces both processes, which is consistent with spore formation contributing to survival during periods of nutrient deprivation.
The observation that C. neoformans mates or undergoes monokaryotic fruiting in response to low nitrogen levels prompted us to characterize the ammonium permease homologues that are evident following the sequencing of the C. neoformans genome (21). Although mating in C. neoformans is distinct from ammonium-dependent filamentous growth seen in other yeasts, both involve developmental processes, and in C. neoformans it involves morphological changes that include the formation of hyphae, basidia, and spores (6, 45). In this study, we show that C. neoformans expresses distinct low- and high-affinity ammonium transporters that we have designated Amt1 and Amt2, respectively. Haploid cells respond to limiting nitrogen conditions by undergoing invasive growth, and this process is dependent upon Amt2. In addition, Amt2 is also required for the induction of mating in response to low ammonium concentrations. This work, therefore, identifies a novel role for one member of the Amt/Mep/Rh family of proteins and extends the number of proteins that are involved in mating and spore formation in C. neoformans to include those involved in ammonium metabolism.
MATERIALS AND METHODS
Fungal strains.
The strains used in this study are listed in Table 1. Derivative mutant strains of H99 MATα in which AMT1 and AMT2 were separately disrupted were generated by replacing the coding region of each gene with a cassette conferring resistance to, respectively, G418 (KAN) and nourseothricin (NAT). Each cassette consisted of the drug resistance gene, flanked by approximately 1 kb of the promoter and terminating sequences of the relevant AMT gene that was generated by overlap PCR. Transformation of each cassette was carried out using a biolistic particle delivery system (Bio-Rad model PDS-1000/He) (41). To obtain a double mutant MATα amt1Δ amt2Δ strain, a second transformation was carried out by replacing the coding region of AMT1 with the KAN resistance cassette in the H99 MATα amt2::NAT strain. PCR and Southern blotting confirmed integration at the correct locus and the absence of multiple integration events. The resulting H99 MATα amt1::KAN amt2::NAT strain was crossed with KN99 MATa, and basidiospores from the cross were analyzed to obtain multiple independent MATα and MATa amt1Δ, amt2Δ, and amt1Δ amt2Δ strains. Matings were carried out on MS (32, 46) medium for up to 3 weeks in the dark.
TABLE 1.
Strains used in this study
| Species and strain | Genotype | Reference |
|---|---|---|
| C. neoformans | ||
| Serotype A | ||
| H99α | MATα | 37, 41 |
| JR1 | amt1::NEO MATα | This study |
| JR3 | amt2::NAT MATα | This study |
| JR5 | amt1::NEO amt2::NAT MATα | This study |
| KN99a | MATa | 34 |
| JR7 | amt1::NEO MATa | This study |
| JR9 | amt2::NAT MATa | This study |
| JR11 | amt1::NEO amt2::NAT MATa | This study |
| Bt63 | MATa | 19 |
| JKH7 | MATα pka1Δ::URA5 ura5 | 10 |
| Serotype D | ||
| JEC20 | MATa | 16 |
| JEC21 | MATα | 16 |
| B3502 | MATa | 16 |
| B3501 | MATα | 16 |
| KN433a | MATa | 36 |
| KN433α | MATα | 36 |
| KN3501a | MATa | 36 |
| KN3501α | MATα | 36 |
| S. cerevisiae MLY40 | MATaura3-52 | 22 |
Phylogenetic analysis of protein sequences.
An unrooted tree was derived with heuristic maximum-likelihood analyses in PHYML using the WAG amino acid substitution model with a portion of invariable sites and gamma-distributed substitution rates. Ambiguously aligned regions were excluded from the analysis. A similar topology was recovered by neighbor joining in ClustalX using the Gonnet matrix and Bayesian analysis in MrBayes version 3. For the Bayesian analysis, the rtREV matrix with a portion of invariable sites and gamma-distributed substitution rates was used. The matrix and settings used for the Bayesian analysis were determined using Prottest. Branch support values were calculated from 500 replicates of nonparametric maximum-likelihood bootstrap analysis (first percent) from 1,000 replicates of distance bootstrap analysis (second percent) and Bayesian posterior probability (lower percent). Branches supported by two or more methods (≥75% bootstrap and ≥95% posterior probability) were considered supported and are shown as thickened branches on the tree.
In vitro phenotypic analysis.
The growth of strains was compared on yeast extract-peptone-dextrose (YPD), complete synthetic medium-dextrose containing added amino acids (CMD), and yeast nitrogen base medium supplemented with different nitrogen sources (YNB) (42). Melanin production was assayed on Niger seed agar and capsule formation induced by growth on Dulbecco's modified Eagle's (DME) agar. Sensitivity to methylamine was carried out using YNB-proline (1 mM) plates supplemented with 10 mM methylamine. All dilution series on agar plates were carried out with overnight yeast cultures that had been washed twice with water before plating. Invasive growth was assayed by spotting a 10-μl drop of a prewashed overnight culture onto synthetic low-ammonium dextrose (SLAD) agar (YNB without amino acids plus 50 μM ammonium sulfate), and the plates were incubated for 1 week at 30°C. Cells on the surface of the agar were carefully washed away with water to reveal the extent of invasive growth. To quantify the level of invasive growth, the degrees of invasion were compared by eye and scores from 0 (no visible invasive growth) to 4 (best invasive growth) assigned. Assays of this phenotype were repeated eight times, and the scores were averaged and used as trait values. Individual embedded cells were examined using a Zeiss Axiophot 2 Plus fluorescence microscope and photographed with an AxioCam MRM digital camera. Northern blot analysis was carried out as previously described (3) using DNA probes hybridizing to AMT1, AMT2, and ACT1 that had been amplified from genomic C. neoformans serotype A (H99) DNA using specific primers.
In vivo analysis of virulence.
For murine assays, wild-type and mutant strains of C. neoformans were used to infect female A/Jcr mice (NCI/Charles River Laboratories) by nasal inhalation as described previously (34). Ten animals per strain were inoculated with 50 μl phosphate-buffered saline containing 1 × 105 yeast cells of the wild-type H99 strain or the amt mutant strains. Animals were examined daily, and the protocol used was approved by the Duke University Animal Use Committee. P values were calculated using the Mann-Whitney U test. The wax moth virulence assay was carried out as previously described (33) using Galleria mellonella larvae (Vanderhorst Inc., St. Marys, OH). Each larva was inoculated with 1 × 105 yeast cells and incubated at 37°C. Larvae were examined daily and inviable larvae defined as those that did not respond to touch.
[14C]methylamine uptake.
The rate of [14C]methylamine uptake was determined as previously described (39, 43). Briefly, cells were grown in synthetic medium with proline (1 mM) as the nitrogen source to mid-log phase. The cells were pelleted, washed, resuspended in phosphate buffer (20 mM, pH 7) to an optical density at 595 nm of 8, and incubated on ice. Labeled methylamine (0.1 mCi; MP Biomedicals Inc., Irvine, CA) was added to each of a series of tubes that contained increasing concentrations of unlabeled methylamine in phosphate buffer. The total number of moles of methylamine in each tube was therefore equivalent to 0.1 mCi. Cells were taken from the ice and added to the first tube, which was then resuspended in a water bath at 30°C, and samples were taken at 1, 2, 3, and 4 min and added to an ice-cold nonlabeled methylamine solution (100 mM) to stop the reaction. The cells were vacuum filtered onto GC Whatman filters and washed with ice-cold nonlabeled methylamine solution (100 mM). This process was repeated for each methylamine concentration and the level of [14C]methylamine quantified using liquid scintillation counting. The measured 14C was used to calculate the number of moles of methylamine imported for each sample (with 0.1m Ci representing increasing concentrations of methylamine) and a rate of uptake (moles/min) calculated. For all experiments the rate of import increased with increasing external concentrations of methylamine until the experimental system became saturated. Methylamine uptake was linear and external labeled methylamine not exhausted over the 4-min sampling period. A Lineweaver-Burk plot was used to determine the maximal velocity (Vmax) and the Michaelis constant (Km) of the methylamine uptake reaction.
RESULTS
C. neoformans expresses homologues of low- and high-affinity ammonium transporters.
The complete genome of the C. neoformans serotype A strain H99 was searched for homologues of the Mep2 ammonium permease from Saccharomyces cerevisiae, which is a high-affinity transporter that is required for the initiation of pseudohyphal growth (23, 26). Two open reading frames (CNAG00235.1 and CNAG04758.1) that encode proteins with significant similarity to Mep2 were identified. Based on these similarities, we have designated these genes AMT1 (for ammonium transporter) (CNAG00235.1) and AMT2 (CNAG04758.1). Mep2 exhibits 51% identity (over 429 residues) and 45% identity (over 420 residues) with Amt1 and Amt2, respectively.
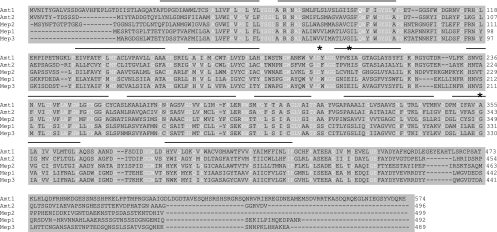
Sequence alignments of all three ammonium permeases from S. cerevisiae (Mep1 to -3) with the two Amt permeases from C. neoformans reveal residues that are conserved throughout the entire proteins, including those within predicted membrane helices (Fig. 1). Residues that are required, or predicted to be essential, for ammonium transport are also conserved between Amt1, Amt2, and the S. cerevisiae Mep proteins. Residue D186 of Mep2 (equivalent to D195 of Amt1 and D183 of Amt2) is positioned at the outer pore of the ammonium channel based on the structure of the bacterial AmtB permease (13). An asparagine substitution at position 186 in Mep2 results in the loss of Mep2-dependent ammonium transport without the loss of Mep2 membrane localization (25). The AmtB structure also revealed two histidine residues (H168 and H318) that are positioned within the ammonium channel and that are required for ammonium transport (11). The AmtB His-318 residue is conserved in all of the S. cerevisiae Mep and C. neoformans Amt proteins. Fungal homologues of AmtB contain either a histidine (Amt2 and Mep2) or a glutamic acid (Amt1, Mep1, and Mep3) residue at the equivalent position of the AmtB H168 (Fig. 1). Replacement of the equivalent histidine residue with a glutamic acid residue within AmtB reduces but does not abolish AmtB activity (11).
FIG. 1.
Alignment of Amt1 and Amt2 with the three ammonium permeases from S. cerevisiae (Mep1 to -3) using ClustalW. Conserved residues are in white, and lines above the alignment indicate the positions of the 11 transmembrane helices based on the structure of bacterial AmtB and alignment of representative members of the AmtB/Mep/Rh family of proteins (13). Asterisks show the positions of three key residues that are, or are predicted to be, ligands for ammonium/ammonia as it is transported through the permease.
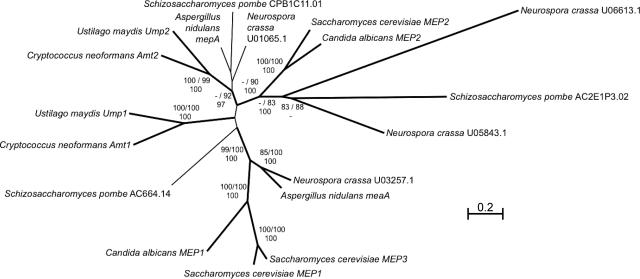
A phylogenetic tree showing the evolutionary relationship between various established and putative members of the AmtB/Mep/Rh family in fungi revealed that Amt1 and Amt2 cluster with other basidiomycete fungal ammonium permeases (Fig. 2). The conservation of residues essential for ammonium transport and the evolutionary relationship of Amt1 and Amt2 to other ammonium permeases are consistent with Amt1 and Amt2 being ammonium permeases. Interestingly, the C. neoformans Amt proteins exhibit a close evolutionary relationship with the Ump proteins from Ustilago maydis. However, these data do not allow a prediction to be made as to whether Amt1 and Amt2 are high- or low-affinity permeases or whether they have an additional signaling function equivalent to that of Mep2 or Ump2 from S. cerevisiae and U. maydis, respectively.
FIG. 2.
Tree representing the phylogenetic relationship between selected fungal ammonium transporters. The evolutionary relationship of representative members of the AmtB/Mep/Rh family was determined for those that are known or predicted to be ammonium permeases (see Materials and Methods). The gene or open reading frame and the name of the organism are indicated.
Amt1 and Amt2 are required for growth under ammonium-limiting conditions.
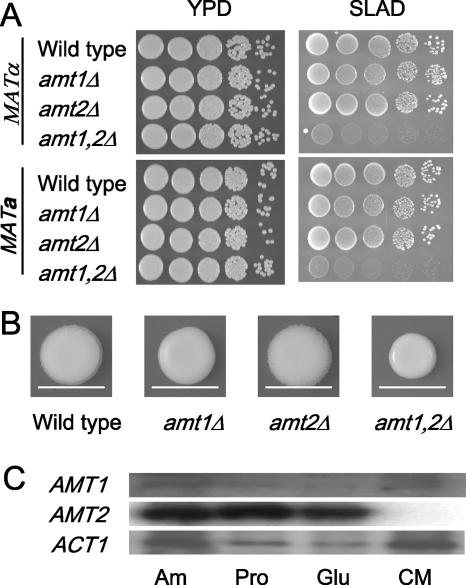
To assess the functions of Amt1 and Amt2, single and double mutant strains were generated in which the coding regions of the AMT1 and AMT2 genes were deleted from the serotype A, MATα wild-type strain H99. Isogenic MATa strains were isolated following a meiotic cross of the MATα amt1Δ amt2Δ strain and the KN99 MATa wild-type strain. The resulting wild-type and single and double mutant strains were inoculated on low-ammonium agar at 30°C for 5 days to examine their ability to grow under ammonium-limiting conditions. This growth assay did not reveal any obvious difference between the wild-type and single amt1Δ and amt2Δ mutant strains (Fig. 3A). The amt1Δ amt2Δ double mutant strains, however, exhibited a severe growth defect on low-ammonium agar, which is consistent with Amt1 and Amt2 having overlapping functions under these conditions. Identical phenotypes were observed in MATa mutant strains that had been derived from a cross between the wild-type MATa cells and the MATα amt1Δ amt2Δ double mutant strain (Fig. 3A).
FIG. 3.
The AMT1 and AMT2 genes are required for growth under low-ammonium conditions and are differently regulated in response to nitrogen availability. (A) Wild-type MATα (H99) and MATa (KN99) and isogenic amt1Δ, amt2Δ, and amt1Δ amt2Δ strains were grown overnight in YPD medium, washed, serially diluted and plated on YPD and low-ammonium (50 μM) medium, and grown for 2 days at 30°C. (B) Overnight cultures of H99 wild-type cells and isogenic amt1Δ, amt2Δ, and amt1Δ amt2Δ cells were washed and spotted onto complete synthetic medium (CMD) and grown for 2 weeks at room temperature. The white lines represent the diameter of the wild-type colony for comparison purposes. (C) Northern analysis of the levels of AMT1 and AMT2 transcripts in wild-type (H99) cells grown CMD or synthetic medium with limiting quantities of glutamine (Glu), proline (Pro), or ammonium (Am) (50 μM). Levels of ACT1 transcripts served as the loading control.
During the course of this work we noted that wild-type strains and the amt mutant strains developed different colony morphologies when grown for long periods. To analyze this in a systematic way, washed cells were spotted onto complete synthetic medium and grown for 2 weeks. Although this medium contains a high level of ammonium sulfate, the lack of Amt1 resulted in slower growth of the colony, which was exacerbated in the amt1Δ amt2Δ double mutant (Fig. 3B). Interestingly, the cells lacking Amt1 also developed colonies with a smooth surface, whereas the wild-type colonies appeared wrinkled. Under these conditions, the amt2Δ mutant strain grew to the same extent and developed the same colony morphology as wild-type cells (Fig. 3B).
As Amt1 and Amt2 are together required for growth under ammonium-limiting conditions, we were interested in determining the extent to which the AMT1/2 genes are regulated by ammonium availability. Wild-type cells were grown in synthetic medium containing limiting concentrations of ammonium sulfate, glutamine, or proline. Northern analysis revealed that AMT1 is constitutively expressed at the same low level under all of the growth conditions tested. In contrast, AMT2 is not expressed under ammonium-sufficient conditions but is induced to high levels when ammonium levels are limiting or when low levels of alternative nitrogen sources are present (Fig. 3C). The induced level of AMT2 expression is considerably higher than the constitutive level of expression of AMT1. Control experiments using MATα amt1Δ amt2Δ cells confirmed the specificity of the RNA probes used (data not shown).
Amt1 and Amt2 are low- and high-affinity ammonium transporters, respectively.
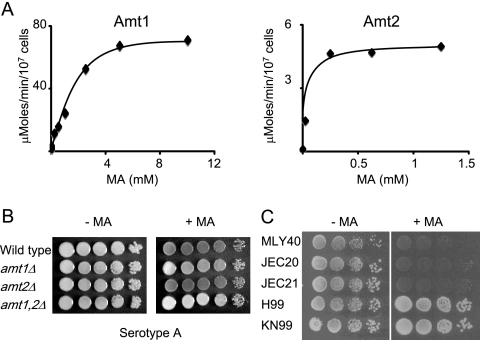
To confirm that Amt1 and Amt2 are ammonium transporters and to quantify the level of transport, we separately assessed the activities of Amt1 and Amt2 by assaying the uptake of [14C]methylamine in amt1Δ mutants expressing only Amt2 and in amt2Δ mutants expressing only Amt1. This ammonium analogue has previously been used to quantify the activities of members of the AmtB/Mep/Rh family of permeases (39). Cells were grown in synthetic media to mid-log phase with proline as the nitrogen source. The rate of [14C]methylamine uptake over a range of external methylamine concentrations was determined. Analysis of the rate of uptake by Amt1 or Amt2 revealed a kinetic profile similar to that described for other fungal ammonium permeases (Fig. 4A) (31, 39). Cells expressing either Amt1 or Amt2 became saturated for [14C]methylamine uptake over the range of external methylamine concentrations used. Amt1 exhibited the greater rate of transport, at 63 μmol moles/min/107 cells compared to 5.1 μmol/min/107 cells for Amt2. The determination of the Km for transport for [14C]methylamine revealed apparent Kms of 1.25 mM and 104 μM for Amt1 and Amt2, respectively. Control experiments identified no [14C]methylamine uptake in the amt1Δ amt2Δ mutant strain, consistent with Amt1 and Amt2 being the only ammonium transporters in C. neoformans (data not shown). Together, these data are consistent with Amt1 being a low-affinity, high-capacity ammonium permease and with Amt2 being a high-affinity, low-capacity ammonium permease.
FIG. 4.
Amt1 and Amt2 mediate the transport of methylamine. (A) Rate of [14C]methylamine uptake versus substrate concentration for the amt2Δ (left) and amt1Δ (right) deletion mutants. (B) Wild-type and isogenic amt1Δ, amt2Δ, and amt1Δ amt2Δ strains were grown overnight in synthetic dextrose (SD)-proline medium, washed, serially diluted and plated on SD-proline plates with and without methylamine (10 mM), and grown for 4 days at 30°C. (C) Wild-type S. cerevisiae (MLY40) and C. neoformans (JEC20a, JEC21α, H99α, and KN99a) strains grown under the same conditions as for panel B.
Methylamine is toxic to yeast cells in the absence of appropriate resistance mechanisms, and therefore methylamine resistance has been used as a qualitative assay for ammonium permease function (39). To test the sensitivities of the C. neoformans wild-type and mutant strains to methylamine, mid-log-phase cultures of cells were serially diluted and plated onto medium containing proline as the nitrogen source and increasing concentrations of methylamine. The wild-type and amt2Δ strains exhibited a partial reduction in growth on plates containing 10 mM methylamine. Cells lacking Amt1 were modestly more resistant to this level of methylamine, with the double amt1Δ amt2Δ strain being more resistant than the single amt1Δ strain (Fig. 4B). Higher concentrations of methylamine did not accentuate the difference between the resistances of wild-type and mutant strains to methylamine, as all cells became sensitive at these concentrations (data not shown). Therefore, the loss of Amt1 results in cells exhibiting a partial resistance to methylamine. To test the relative sensitivities of different strains to methylamine, we grew the serotype A strains (H99α and KN99a), the serotype D strains (JEC20a and JEC21α), and the S. cerevisiae strain MLY40 on medium containing 10 mM methylamine. The C. neoformans serotype A strains (H99 and KN99) grew significantly more robustly than the other strains (Fig. 4C). On close inspection the C. neoformans serotype D strains appeared to be slightly more resistant to methylamine than the S. cerevisiae strain. All cells were sensitive to 20 mM methylamine (data not shown).
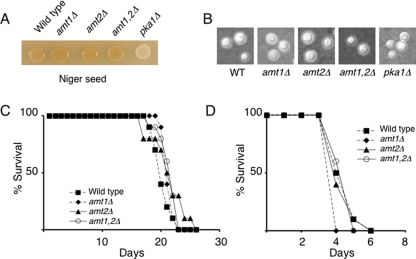
Loss of Amt1 and Amt2 does not influence the pathogenicity of C. neoformans.
The importance of Amt1 and Amt2 in the provision of an important nitrogen source led us to test the extent to which these permeases might be required for the virulence of C. neoformans. Initially, the wild-type and mutant strains were tested for differences in production of two known virulence factors. Cells were assayed for their ability to produce melanin on Niger seed medium and for capsule production in DME medium. In all cases, the production of both virulence factors was induced to the same extent in wild-type and mutant cells (Fig. 5A and B). To assess the importance of these ammonium permeases during in vivo infection, two virulence assays using established model systems were used to test the survival of wild-type and mutant cells lacking the Amt permeases. In the murine inhalation model of cryptococcosis, the wild-type and mutant strains had equivalent virulence (P ≥ 0.07 for comparisons of wild type versus mutant) (Fig. 5C). In addition, when the wild-type and amt mutant strains were also tested in the wax moth larva model of C. neoformans infection, no differences in virulence were exhibited (P ≥ 0.06 for comparisons of wild type versus mutant) (Fig. 5D).
FIG. 5.
Amt1 and Amt2 are not required for the virulence of C. neoformans. Wild-type (WT) and isogenic amt1Δ, amt2Δ, and amt2Δ strains were plated on Niger seed plates to induce melanin production (A) or on DME plates to induce the polysaccharide capsule (B). Capsules were visualized by staining cells using India ink. A mutant lacking Pka1 was used as a negative control (10). (C) Virulence of the wild-type and isogenic amt1Δ, amt2Δ, and amt1Δ amt2Δ strains was assayed by intranasally infecting mice with 1 × 105 cells and monitoring progression to severe morbidity. P values were 0.0753, 0.2799, 0.0892, 0.9118, 0.5787, and 0.9705 for wild-type/amt1Δ, wild-type/amt2Δ, wild-type/amt1Δ amt2Δ, amt1Δ/amt2Δ, amt1Δ/amt1Δ amt2Δ, and amt2Δ/amt1Δ amt2Δ strains, respectively. (D) To determine virulence in the wax moth Galleria mellonella, larvae were injected with yeast cells (1 × 105) and survival was monitored daily. P values were 0.0630, 0.79759, 0.9118, 0.1431, 0.0232, and 0.6305 for wild-type/amt1Δ, wild-type/amt2Δ, wild-type/amt1Δ amt2Δ, amt1Δ/amt2Δ, amt1Δ/amt1Δ amt2Δ, and amt2Δ/amt1Δ amt2Δ strains, respectively.
The high-affinity Amt2 permease is required for induction of invasive growth in response to low ammonium levels.
The established role of fungal high-affinity ammonium permeases in filamentous growth led us to test whether C. neoformans Amt1 and Amt2 serve a similar function. The Mep2 ammonium permease is required for pseudohyphal growth in both diploid S. cerevisiae and Candida albicans (2, 23). Haploid S. cerevisiae cells undergo a similar but more limited dimorphic switch that leads to the invasive growth of cells into low-ammonium and rich medium agar (38, 43). Colonies of C. neoformans haploid cells grown on low-ammonium plates did not exhibit the filamentous growth at the edges of the colony that is a hallmark of pseudohyphal growth. The colonies did, however, undergo invasive growth into the agar as revealed by washing away surface cells followed by microscopy (Fig. 6). The strains lacking Amt2 did not undergo invasive growth under these conditions, whereas the amt1Δ strains exhibited wild-type levels of invasive growth. Microscopic inspection of the area of wild-type invasive growth revealed that the invasive phenotype consists of stochastic invasions of the agar that give rise to small groups of embedded cells (Fig. 6D). Interestingly, the elongated cell phenotype that is associated with S. cerevisiae diploid pseudohyphal growth is not observed with haploid C. neoformans cells undergoing invasive growth.
FIG. 6.
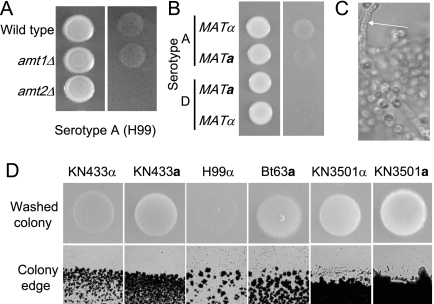
The extent of invasive growth is variable among different C. neoformans strains and is dependent on Amt2. Wild-type H99 and isogenic amt1Δ and amt2Δ strains (A), H99α/KN99a (serotype A) and JEC20a/JEC21α (serotype D) strains (B), and other strains as indicated (D) were grown overnight, washed and inoculated on low ammonium medium (50 μM), and grown for 1 week at 30°C. Invasive growth was visualized by washing away surface yeast cells with water (A, B, and D). The morphology of invading cells was visualized with a Zeiss Axiophot 2 Plus fluorescence microscope and filaments were detected in JEC21 MATα cells (C, arrow).
The difference in methylamine sensitivity exhibited by serotype A and D strains is consistent with there being a difference in ammonium metabolism between these strains. We were therefore interested in testing the extent to which different C. neoformans strains undergo invasive growth. Initially, wild-type MATα and MATa cells (serotypes A and D) were analyzed for their ability to undergo invasive growth under ammonium-limiting conditions. The serotype A cells exhibited a more robust invasive phenotype than the serotype D cells, with the H99 MATα strain being modestly more efficient at invasive growth than the congenic MATa strain KN99a (Fig. 6B). Only a few sites of invasion by the serotype D strains could be identified, although interestingly, the microscopic examination of the sites of serotype D MATα invasion revealed that a few cells had undergone a morphological switch from the yeast to the filamentous form (Fig. 6C). An equivalent dimorphic switch was not observed within the areas of serotype A MATα invasive growth.
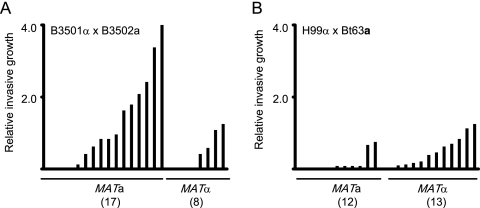
The differential invasive patterns of the two extensively used and laboratory-defined serotype A H99 and serotype D JEC21 strains led us to test the invasive phenotypes of a larger range of strains when grown under ammonium-limiting conditions. Under identical conditions, additional strains exhibited wide variation in the extent of invasive growth, with strains such as H99α and KN433α being considerably less invasive than strains such as KN3501α and KN3501a (Fig. 6D). In contrast to the finding that the serotype A MATα strain is more invasive than the congenic MATa strain, the serotype D MATa strains KN433 and KN3501 were more invasive than the congenic strains of the opposite mating type (KN433α and KN3501α). To test if invasive growth is linked to the MAT locus, the levels of invasion by the progeny of crosses of two nonisogenic pairs (B3501α × B3502a and H99α × Bt63a) were assessed. The progeny of a cross between B3501α and B3502a are recombinant based on genotyping using microsatellite and restriction fragment length polymorphism genetic markers (28). The MATa progeny of the B3501α × B3502a cross showed better invasive growth than the MATα progeny (Fig. 7A), which is consistent with the pattern of invasion of the KN433 and KN3501 strains. In contrast, the MATα progeny of the H99α × Bt63a cross were more invasive than the MATa strains (Fig. 7B).
FIG. 7.
Quantification of the level of invasive growth in the F1 progeny of two genetic crosses. The progeny of crosses between the serotype D strains B3501α and B3502a (A) and the serotype A strains H99α and Bt63a (B) were grown on SLAD medium for 6 days and the extent of invasive growth measured and plotted.
C. neoformans mating in response to ammonium-limiting conditions is Amt2 dependent.
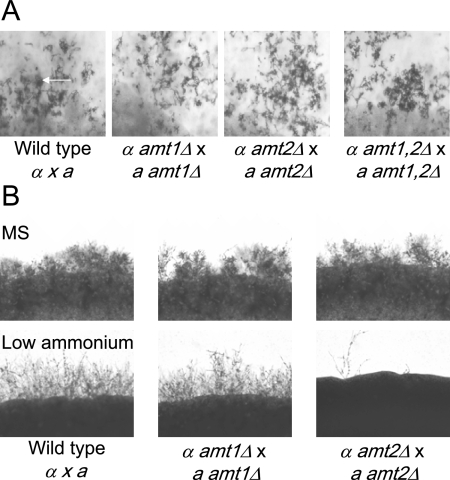
The initiation of mating in response to nutrient deprivation by C. neoformans and the influence of Amt2 on invasive growth led us to hypothesize that mating in C. neoformans may be dependent on this ammonium permease. To test this hypothesis, wild-type and mutant cells were cocultured on MS medium and SLAD medium. MS medium is a plant tissue culture medium (32) that supports significant mating of C. neoformans strains, is not nitrogen limiting, and thus supports growth of the amt1Δ amt2Δ double mutant strain. Under these growth conditions, mating is induced by the presence of inositol and not by a lack of ammonium (46). The wild-type and mutant strains produced mating filaments to the same extent when grown on MS medium (Fig. 8A and B). All of these matings resulted in the production of basidia and long chains of basidiospores. Therefore, under these growth conditions, Amt1 and Amt2 do not influence C. neoformans mating.
FIG. 8.
Amt2 is required for mating under low-nitrogen conditions. (A) Bilateral matings of wild-type cells (α × a) and amt mutant strains (α amt1Δ × a amt1Δ, α amt2Δ × a amt2Δ, and α amt1Δ amt2Δ × a amt1Δ amt2Δ) were carried out on MS medium, resulting in the formation of basidia (arrow in left panel). Chains of basidiospores were produced in each mating, including the mating of the amt1Δ amt2Δ double mutant strains (α amt1Δ amt2Δ × a amt1Δ amt2Δ). (B) The mating of the wild-type and single amt mutant strains was compared on MS medium and low-ammonium sulfate SLAD medium.
We then tested the mating of wild-type serotype A cells and the isogenic amt1Δ, amt2Δ, and amt1Δ amt2Δ mutants under low-ammonium conditions. Wild-type mating mixtures grown on SLAD medium produced significant numbers of mating filaments. A similar phenotype was observed with a bilateral cross of amt1Δ strains (α amt1Δ × a amt1Δ). However, mating between cells lacking amt2 (α amt2Δ × a amt2Δ) was severely impaired, indicating that Amt2 promotes mating under nitrogen-limiting conditions (Fig. 8B). Equivalent bilateral matings of all strains on medium containing higher concentrations of ammonium sulfate (500 μM) did not result in the production of mating filaments (data not shown). Together, these data indicate that low-ammonium conditions stimulate mating in an Amt2-dependent manner.
To confirm that ammonium-responsive invasive growth and mating are linked to the AMT2 locus, we generated reconstituted amt2Δ strains in which the AMT2 gene was reintegrated into the C. neoformans genome. The resulting strains did not, however, exhibit wild-type phenotypes (data not shown). Analysis of Mep2 function in C. albicans has shown that a reduction in MEP2 gene expression can detrimentally influence the induction of filamentous growth (2). It is possible that the role of Amt2 in C. neoformans development is also dependent on its expression levels, which may have been adversely influenced by integration of AMT2 into a nonnative locus. We therefore undertook a second approach and performed linkage analysis to analyze the phenotypes of 24 progeny from a cross between a wild-type strain and an amt2Δ mutant strain. In all cases, those progeny that contained the amt2 deletion exhibited reduced invasive growth and no mating when grown on ammonium-limiting medium. Conversely, all wild-type progeny displayed invasive growth and mating. These results support the conclusion that reduced invasive growth and reduced mating in response to low ammonium levels are caused by AMT2 deletion.
DISCUSSION
The ability to sense nutrients and acquire them from the environment is fundamental to an organism's ability to survive. Fungi can utilize a wide range of nitrogen sources and many of the molecular mechanisms of nitrogen acquisition identified in model fungi are likely to be conserved throughout this kingdom. The study of the equivalent systems in an organism such as C. neoformans is of interest as they relate to the ability of this organism to cause disease. C. neoformans is able to survive both as a saprophytic yeast and as an opportunistic human pathogen, which involves survival within two diverse environments that provide different nutritional resources. Nitrogen availability also indirectly influences the virulence of C. neoformans, as this organism undergoes mating and monokaryotic fruiting in response to limiting nitrogen, which leads to the production of potentially infectious spores.
The genome of C. neoformans contains two genes that encode proteins with high similarity to members of AmtB/Mep/Rh family of ammonium permeases (21). Amt1 and Amt2 contain conserved residues that have been predicted or shown to be crucial for ammonium transport in other AmtB/Mep/Rh permeases. The role of Amt1 and Amt2 in ammonium metabolism was confirmed by the isolation of C. neoformans mutants in which the relevant genes had been deleted. Cells lacking either Amt1 or Amt2 were viable on low-ammonium medium; however, a double mutant lacking both permeases was unable to grow under these conditions. This phenotype was observed in multiple isolates of the double mutant of both MATa and MATα strains. The role of Amt1 and Amt2 in ammonium transport was confirmed by analyzing the rate of methylamine uptake. Amt1 is a low-affinity, high-capacity ammonium permease, and Amt2 is a high-affinity, low-capacity ammonium permease. All fungal species studied to date contain at least two members of the AmtB/Mep/Rh family of proteins, although some species contain additional paralogs. Cells that lack both Amt1 and Amt2 do not import methylamine, which is consistent with Amt1 and Amt2 being the only C. neoformans ammonium transporters.
The available evidence suggests that the ammonium permease-encoding genes are differentially regulated within a particular fungal species. The three MEP genes of S. cerevisiae are under nitrogen catabolite control and are induced under nitrogen-limiting conditions by the Gln3 and Gat1 transcription factors (26). The extent of induction, however, varies considerably, with MEP2 being approximately 20- and 50-fold more highly expressed than MEP1 and MEP3, respectively, when proline is used as a nitrogen source (26). In Aspergillus nidulans, nitrogen-responsive transcription is mediated by the AreA factor and the meaA and mepA genes encode low- and high-affinity permeases, respectively (30, 31). The meaA gene is constitutively expressed and does not respond to changes in external ammonium concentrations. In contrast, the mepA gene is induced to a high level under low-nitrogen conditions but is not expressed during nitrogen sufficiency. The C. neoformans AMT1 and AMT2 genes show a pattern of regulation similar to that of the A. nidulans genes that encode MeaA and MepA, with AMT1 being constitutively expressed and AMT2 induced by nitrogen limitation.
The uptake of methylamine by C. neoformans suggested that there would be an Amt-dependent phenotype associated with growth in the presence of this ammonium analogue. Methylamine is toxic to S. cerevisiae, and mutant strains lacking the Mep ammonium permeases exhibit resistance to it (7). In contrast, strains lacking Amt1 exhibited only weak resistance to methylamine, with the double amt1Δ amt2Δ mutant being more resistant than the single amt1Δ mutant. The amt2Δ strain grew like the wild type in the presence of methylamine. Analysis of the relative sensitivities of an S. cerevisiae strain and of C. neoformans serotype A and D strains revealed that S. cerevisiae is more sensitive to methylamine than C. neoformans. This suggests that the mechanisms of uptake and/or detoxification of methylamine are different in these two fungi and may account for the clear methylamine phenotype that has been described for the S. cerevisiae mep-deficient mutants. It is interesting to note that the genomes of C. neoformans serotype A and D strains contain homologues of amine oxidase, an enzyme that catalyzes the oxidative deamination of amines, which may confer resistance to methylamine. These enzymes are found throughout biology, including in fungal species (8), although a homologue is lacking in S. cerevisiae.
The loss of the Amt permeases does not influence the virulence of C. neoformans. First, we analyzed the influence of the ammonium permeases on the ability of C. neoformans to induce the formation of melanin and the polysaccharide capsule. We reasoned that ammonium availability, as a signal of nutrient deprivation, might influence the production of mechanisms that afford protection within this organism's natural environment. The lack of both ammonium permeases had no influence on the production of these virulence factors under standard laboratory conditions. We then analyzed directly a role for the Amt permeases in the virulence of C. neoformans using the murine and wax moth insect models of cryptococcosis. Mutants lacking one or both Amt permeases were as virulent as wild-type cells in both experimental models. This suggests that ammonium is not a source of nitrogen during infection and/or that in the absence of ammonium uptake other mechanisms of nitrogen acquisition, such as the uptake of amino acids, are sufficient to maintain wild-type growth within the host. This is consistent with the finding that the utilization of ammonium as the only source of nitrogen is not essential for the survival of S. cerevisiae as a pathogen in vivo (14).
Morphological change can be initiated in response to ammonium limitation in certain fungi, and this dimorphic switch is dependent on certain members of the AmtB/Mep/Rh family. In those fungi in which this has been studied, a role for the protein kinase A and mitogen-activated protein kinase signaling pathways in development has been demonstrated (17). However, the precise role of the ammonium permeases in this process is not yet known. Haploid S. cerevisiae undergoes invasive growth when grown for extended periods on rich medium or in the presence of fusel alcohols (24). Recently, this phenotype has also been observed in response to ammonium limitation and is dependent on one of the S. cerevisiae Mep proteins (43). This led us to analyze the influence of ammonium limitation on the growth of C. neoformans, which is predominately a haploid organism. Under these conditions, wild-type cells undergo invasive growth, which appears to consist of stochastic events that give rise to individual clusters of embedded cells. For serotype A cells of the H99 background, this process is dependent on the presence of Amt2. The extent of nitrogen responsive invasion varies considerably between different strains of C. neoformans. Quantification of the degree of invasive growth in the progeny of crosses revealed an intriguing relationship between the MAT locus and invasive growth in the serotype D background. The MAT locus of C. neoformans contains homologues of the mitogen-activated protein kinase signaling pathway of S. cerevisiae that have a dual role in mating and pseudohyphal growth in that organism (47). This suggests a complex relationship between the MAT locus and the levels of ammonium uptake in different C. neoformans strains, and consequently signaling, that is evidenced by the different sensitivities of C. neoformans strains to methylamine. The analysis of the congenic serotype D strains KN433 and KN3501 revealed that MATa cells were more invasive than MATα cells. The analysis of the congenic serotype A strain pair H99α and KN99a and of the progeny from a genetic cross of two genetically distinct serotype A parental strains (H99α and Bt63a) instead provided more limited evidence that enhanced invasive growth can be associated with the MATα locus. This suggests an interesting relationship between the MAT locus allele and other genomic interactions that may differ between the two divergent serotypes and which should form the basis for future studies to investigate these differences.
The established role of ammonium in the initiation of mating in C. neoformans led us to analyze the influence of the Amt permeases on this process. Cells lacking Amt1 underwent mating to the same extent as wild-type cells under ammonium-limiting conditions. A bilateral cross between mutant amt2Δ strains, however, revealed that this strain has a severe mating defect under these conditions. All of the mutant strains, including the double amt1Δ amt2Δ mutant, were able to undergo mating and produce filaments and spores when grown on MS medium. Therefore, the amt2Δ strain is not sterile but lacks the ability to initiate mating in response to low-ammonium conditions. Importantly, analysis of the phenotypes of progeny from a cross between a wild-type and amt2Δ strain established linkage between AMT2 and ammonium-responsive invasive growth and mating.
The role of Amt2 in development suggests that it has evolved a function that is analogous to those of the Mep2 proteins of S. cerevisiae and C. albicans and Ump2 of U. maydis. Similar to Mep2 and Ump2, Amt2 is a high-affinity permease and is the most highly expressed ammonium permease within its species. The ability of the C. albicans Mep2 to initiate signaling is dependent on its high expression levels (2). It is presently unknown how the Mep2-like proteins regulate development at the molecular level. It is possible that this group of proteins act as sensors of ammonium availability that link signaling to the transport function of the protein. This hypothesis will remain speculative until the signaling partners of these putative signaling permeases are identified. Notwithstanding the precise mechanism of Amt2 function, this study extends the number of fungal developmental processes that depend on ammonium transport. This encourages the prediction that other homologues exist elsewhere in the fungal kingdom and that they link a variety of biological processes to ammonium availability. The challenge is to identify the mechanisms by which these permeases link ammonium availability to fungal development and the extent to which these processes are conserved between different fungi. This will reveal the molecular mechanisms that link species-specific responses to the insufficiency of an essential nutrient.
Acknowledgments
We thank Wei-Chiang Shen and Andy Alspaugh for contributions to early stages of this project. We also thank Alex Idnurm for assistance with the wax moth experiments, Charles Hall for advice on construction of the evolutionary trees, and Anna Floyd for excellent technical assistance.
This research was funded by NIH/NIAID grants AI39115 and AI50113 to J.H.
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Andrade, S. L., A. Dickmanns, R. Ficner, and O. Einsle. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 10214994-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas, K., and J. Morschhäuser. 2005. The Mep2 ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56649-669. [DOI] [PubMed] [Google Scholar]
- 3.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 133271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. C., G. F. Miller, and K. J. Kwon-Chung. 2003. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect. Immun. 714953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper, T. G. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, H., and W. Courchesne. 1998. A novel quantitative mating assay for the fungal pathogen Cryptococcus neoformans provides insight into signalling pathways responding to nutrients and temperature. Microbiology 1441691-1697. [DOI] [PubMed] [Google Scholar]
- 7.Dubois, E., and M. Grenson. 1979. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol. Gen. Genet. 17567-76. [DOI] [PubMed] [Google Scholar]
- 8.Frébort, I., K. Matsushita, H. Toyama, K. Lemr, M. Yamada, and O. Adachi. 1999. Purification and characterization of methylamine oxidase induced in Aspergillus niger AKU 3302. Biosci. Biotechnol. Biochem. 63125-134. [DOI] [PubMed] [Google Scholar]
- 9.Heitman, J. 2006. Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 16R711-R725. [DOI] [PubMed] [Google Scholar]
- 10.Hicks, J. K., Y.-S. Bahn, and J. Heitman. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 41971-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javelle, A., D. Lupo, L. Zheng, X.-D. Li, F. K. Winkler, and M. Merrick. 2006. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J. Biol. Chem. 28139492-39498. [DOI] [PubMed] [Google Scholar]
- 12.Javelle, A., M. Morel, B. R. Rodriguez-Pastrana, B. Botton, B. Andre, A. M. Marini, A. Brun, and M. Chalot. 2003. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 47411-430. [DOI] [PubMed] [Google Scholar]
- 13.Khademi, S., J. O'Connell III, J. Remis, Y. Robles-Colmenares, L. J. W. Miercke, and R. M. Stroud. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 3051587-1594. [DOI] [PubMed] [Google Scholar]
- 14.Kingsbury, J. M., A. J. Goldstein, and J. H. McCusker. 2006. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot. Cell 5816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of a and α mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108337-340. [DOI] [PubMed] [Google Scholar]
- 16.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genectic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lengeler, K. B., R. C. Davidson, C. D'souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, X., C. M. Hull, and J. Heitman. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 4341017-1021. [DOI] [PubMed] [Google Scholar]
- 19.Litvintseva, A. P., R. E. Marra, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 21162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, H.-J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 21.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 3071321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 167008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 171236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz, M. C., N. S. Cutler, and J. Heitman. 2000. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell 11183-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marini, A. M., M. Boeckstaens, F. Benjelloun, B. Chérif-Zahar, and B. André. 2006. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49364-374. [DOI] [PubMed] [Google Scholar]
- 26.Marini, A. M., S. Soussi-Boudekou, S. Vissers, and B. André. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 174282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marini, A. M., A. Urrestarazu, R. Beauwens, and B. André. 1997. The Rh (rhesus) blood group polypeptides are related to NH4+ transporters. Trends Biochem. Sci. 22460-461. [DOI] [PubMed] [Google Scholar]
- 28.Marra, R. E., J. C. Huang, E. Fung, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2004. A genetic map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsuzawa, H. 2006. Ammonium transporter genes in the fission yeast Schizosaccharomyces pombe: role in ammonium uptake and a morphological transition. Genes Cells 111183-1195. [DOI] [PubMed] [Google Scholar]
- 30.Monahan, B. J., M. C. Askin, M. J. Hynes, and M. A. Davis. 2006. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell 5226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monahan, B. J., J. A. Fraser, M. J. Hynes, and M. A. Davis. 2002. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 185-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15473-497. [Google Scholar]
- 33.Mylonakis, E., R. Moreno, J. B. El Khoury, A. Idnurm, J. Heitman, S. B. Calderwood, F. M. Ausubel, and A. Diener. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 733842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 714831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, K., G. M. Cox, A. P. Litvintseva, E. Mylonakis, S. D. Malliaris, D. K. Benjamin, S. S. Giles, T. G. Mitchell, A. Casadevall, J. R. Perfect, and J. Heitman. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 734922-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen, K., R. E. Marra, F. Hagen, T. Boekhout, T. G. Mitchell, G. M. Cox, and J. Heitman. 2005. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 171975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perfect, J. R., N. Ketabchi, G. M. Cox, C. W. Ingram, and C. L. Beiser. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 313305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 82974-2985. [DOI] [PubMed] [Google Scholar]
- 39.Smith, D. G., M. D. Garcia-Pedrajas, S. E. Gold, and M. H. Perlin. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50259-275. [DOI] [PubMed] [Google Scholar]
- 40.Sukroongreung, S., K. Kitiniyom, C. Nilakul, and S. Tantimavanich. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36419-424. [PubMed] [Google Scholar]
- 41.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1751405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treco, D. A., and V. Lundblad. 1993. Current protocols in molecular biology, vol. 2, unit 13.1, p. 2. John Wiley & Son, Inc., New York, NY. [Google Scholar]
- 43.Van Nuland, A., P. Vandormael, M. Donaton, M. Alenquer, A. Lourenço, E. Quintino, M. Versele, and J. M. Thevelein. 2006. Ammonium permease based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 591485-1505. [DOI] [PubMed] [Google Scholar]
- 44.Wang, P., C. B. Nichols, K. B. Lengeler, M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2002. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot. Cell 1257-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Sci. USA 937327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue, C., Y. Tada, X. Dong, and J. Heitman. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1263-273. [DOI] [PubMed] [Google Scholar]
- 47.Yue, C., L. M. Cavallo, J. A. Alspaugh, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 1999. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 1531601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, L., D. Kostrewa, S. Berneche, F. K. Winkler, and X. D. Li. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 10117090-17095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmer, B. L., H. O. Hempel, and N. L. Goodman. 1984. Pathogenicity of the basidiospores of Filobasidiella neoformans. Mycopathologia 85149-153. [DOI] [PubMed] [Google Scholar]