Abstract
Cch1, a putative voltage-gated calcium ion channel, was investigated for its role in ascus development in Gibberella zeae. Gene replacement mutants of CCH1 were generated and found to have asci which did not forcibly discharge spores, although morphologically ascus and ascospore development in the majority of asci appeared normal. Additionally, mycelial growth was significantly slower, and sexual development was slightly delayed in the mutant; mutant mycelia showed a distinctive fluffy morphology, and no cirrhi were produced. Wheat infected with Δcch1 mutants developed symptoms comparable to wheat infected with the wild type; however, the mutants showed a reduced ability to protect the infected stalk from colonization by saprobic fungi. Transcriptional analysis of gene expression in mutants using the Affymetrix Fusarium microarray showed 2,449 genes with significant, twofold or greater, changes in transcript abundance across a developmental series. This work extends the role of CCH1 to forcible spore discharge in G. zeae and suggests that this channel has subtle effects on growth and development.
Forcible discharge of ascospores from asci is a mechanism common to the majority of fungi in the phylum Ascomycota. The mechanism is vital for initiating the disease cycle in many plant-pathogenic species, including Gibberella zeae (asexual stage Fusarium graminearum), the causal agent of fusarium head blight of wheat. Analysis of the epiplasmic fluid surrounding the spores within the ascus indicates that sugars (Ascobolus immersus) (14) or ions (G. zeae) (56) accumulate and act as osmolytes for generation of the turgor pressure necessary to fire the spores. In the case of G. zeae, the pressure drives an impressive acceleration—870,000 × g—the highest yet recorded in a biological system (56). While the physiological basis of the mechanism of discharge is beginning to be elucidated, the genetic basis remains largely unexplored.
Shifts in calcium levels across both internal and plasma membranes regulate many cellular processes (2). Calcium fluxes are generated by internal release or by influx through calcium channels. Although calcium signaling is not well understood in fungi, the components are conserved in fungal genomes (59), and several parts of the signaling machinery have been studied, including calcineurin (43, 10, 4, 48, 13), calmodulin (37), Ca2+/calmodulin-dependent kinases (11, 12, 35, 31, 58, 52), and phospholipase C (19).
CCH1 encodes a calcium ion channel localized in yeast to the plasma membrane (39). Cch1 has been described as a voltage-gated Ca2+ channel due to its highly conserved transmembrane voltage-sensing S4 domains related to the mammalian L-type voltage-gated calcium channel (15, 39). However, a recent comparison of the conserved charged residues (arginine and lysine) in the S4 region indicates that the full complement of residues is not present in the fungal proteins, and thus fungal Cch1 may be less sensitive to voltage than its mammalian orthologs (38). In Saccharomyces cerevisiae, Cch1 has been proposed to interact with the stretch-activated channel Mid1 (32). Both are implicated in a phenotype described as “mating pheromone-induced death”: on exposure to α-factor, deletion mutants of either gene in a MATa background die shortly after forming shmoos (26, 15). Based on phenotype and double-mutant studies, Locke and colleagues (39) suggest that Cch1 and Mid1 may function together in yeast, and coimmunoprecipitation studies have supported their association in vivo. However, Kanazaki and colleagues (32) have shown that Mid1 is capable of forming an independent, functional channel in Chinese hamster ovary cells (32), and Liu et al. (38) present evidence that Mid1 and Cch1 have independent roles under certain culture conditions.
G. zeae is a major crop pathogen, infecting wheat, barley, maize, and other cereal crops worldwide and causing extensive losses. In addition to reduced yield, the infected crop can be rendered unusable by the production of mycotoxins (9, 56), notably zearalenone (estrogenic) and trichothecenes (antifeedant and emetic). While the fungus possesses an asexual stage (F. graminearum) capable of producing abundant conidia, the primary inoculum of the disease is believed to be ascospores (49), which are forcibly discharged from perithecia forming on crop debris. The process of sexual development both in culture and in planta has been well described (54, 23). Recently, a detailed transcriptional analysis during the developmental stages of the perithecium was completed (24) on the Fusarium GeneChip (22).
As forcible ascospore discharge can be considered a possible target for control measures, we have been focusing on the physiological and genetic aspects of this phenomenon. In G. zeae, pharmacological evidence suggests that calcium plays a crucial role in forcible spore discharge (57). We have begun to investigate the role of calcium flux and signaling by the targeted disruption of the G. zeae homolog of S. cerevisiae CCH1. Our results suggest that Cch1 is critical to the discharge process.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The strain of G. zeae used for this study was a Michigan field isolate, PH-1 (FGSC 9075; NRRL 31084), and mutants derived from this wild-type strain. The genome of PH-1 has been sequenced (http://www.broad.mit.edu/annotation/genome/fusarium_graminearum/Home.html), and considerable annotation has been completed (http://mips.gsf.de/genre/proj/fusarium). All strains were maintained as mycelia and conidia (106 conidia ml−1) in 30% glycerol at −80°C and on sterile soil at −20°C. Conidia were generated for stocks and for quantification in carboxymethylcellulose (Sigma Chemical Co., St. Louis, MO) following standard techniques (7).
Construction of Δcch1 mutants.
The ortholog of S. cerevisiae CCH1 was identified in the G. zeae genome on the basis of BLAST sequence homology as fg01364 (mips.gsf.de/genre/proj/fusarium). Cultures were grown for 48 h in liquid yeast extract-sucrose medium (2% yeast extract-6% sucrose, pH 5.8), and mycelia were collected in Miracloth (Calbiochem-Novabiochem Corp., San Diego, CA) and lyophilized. DNA was extracted using CTAB (hexadecyl-trimethylammonium bromide) according to the method of Kerényi et al. (33). Primers (Table 1) were designed to 438-bp (primers 1 and 2) and 623-bp (primers 4 and 5) regions upstream and downstream, respectively, of the predicted transcriptional start and stop sites; the primers nearest the coding sequence had additional 25-base tails complementary to the hygromycin B phosphotransferase gene (hph) of plasmid pCB1004 (8) (Table 1). Primers 10 and 11 were used to amplify hph. The upstream and downstream flanking regions of CCH1 were combined with hph in a further round of PCR, using primers 1 and 5, and this construct was transformed into G. zeae. CCH1 was replaced by double-crossover integration of the gene replacement construct into the homologous portion of the genome. Protoplast-based transformation was performed as described by Gaffoor et al. (17). Hygromycin-resistant transformants were selected on V8 agar containing 150 μg ml−1 hygromycin B (Calbiochem-Novabiochem) and subsequently transferred to V8 agar containing 450 μg ml−1 hygromycin B (17). Conidia from hygromycin-resistant colonies were transferred to 2% water agar, and single conidial isolates were obtained for further analysis. Conidial isolates were grown for 3 to 4 days in carboxymethylcellulose broth, collected by centrifugation, and stored as above. Several mutants were confirmed by PCR and Southern analysis and used for further characterization. Transformants Δcch1-T12 and Δcch1-T14 were representative of the mutant phenotype and genotype and were used for further study, along with ectopic transformant Δcch1-T11.
TABLE 1.
Primers used in this study
| Primer no. | Primer name | Sequence (5′-3′) | Contiga | Coordinatesa |
|---|---|---|---|---|
| 1 | Cch1 U5 | GATATCCAGTCGCGTCAC | 1.69 | 11544-11527 |
| 2 | Cch1 U3 | ACTTATTCAGGCGTAGCAACCAGGCGTCCAACGAGGTCCAATGTC | 1.69 | 11107-11124 |
| 3 | Cch1 U5 nested | ATGTTCGAAGCGTGTTTAGTGTGTTT | 1.69 | 11524-11499 |
| 4 | Cch1 D5 | TCAGCATCTTTTACTTTCACCAGCGTTGGAAGAGAAGATAGGTAC | 1.69 | 4504-4487 |
| 5 | Cch1 D3 | GTGTCTTTATCTAGCTTCAATAG | 1.69 | 3882-3904 |
| 6 | Cch1 D3 nested | ATCCTCCTGGGCGCCATCGCACCG | 1.69 | 3938-3961 |
| 7 | Cch1 internal F | CGTCAGTCTATCCCGCTGCAAG | 1.69 | 11036-11105 |
| 8 | Cch1 internal R | GGTTAGGCAGAGGTGGTCTTGG | 1.69 | 10166-10187 |
| 9 | Cch1 internal F rc | CTTGCAGCGGGATAGACTGACG | 1.69 | 11105-11036 |
| 10 | HC-CN1004Fb | AACGCTGGTGAAAGTAAAAGATGCTGAA | ||
| 11 | HC-CN1004Rb | ACGCCTGGTTGCTACGCCTGAATAAGT | ||
| 12 | Hyg internal 3′ | TTCGATCAGAAACTTCTCGACAGACG | ||
| 13 | Hyg internal 5′ | CGGCCGTCTGGACCGATGGCTGTGTA |
Contigs and coordinates are given for the G. zeae genome and do not apply to hygromycin primers 10 to 13, which amplify pCB1004.
Primers are courtesy of Jennifer Biezske.
When the expected amplicon was <2,000 bp, PCRs used Invitrogen Taq (Invitrogen, Carlsbad, CA), and the reaction protocol followed Tank and Sang (53). For amplicons of ≥2,000 bp (primer pair 1 and 5; assembling the deletion construct and amplifying the complementation template), Expand Long-Template Polymerase was used, following manufacturer's instructions (Roche Applied Science, Indianapolis, IN).
For Southern analysis, genomic DNA was cut to completion with BamHI, which was predicted to cut CCH1 twice, yielding one fragment of 3,507 bp that would hybridize to the CCH1 internal probe (amplified by primers 7 and 8). BamHI was not predicted to restrict the gene deletion construct carrying the hph gene. Restriction fragments were resolved by electrophoresis in 0.8% agarose in 1× Tris-acetate-EDTA (47) and transferred onto a Nytran Supercharge nylon membrane (Schleicher and Schuell Bioscience, Keene, NH). The membranes were probed twice with [32P]CTP-labeled probes, once with the CCH1 internal probe, and once with the hph probe. The residual probe was removed by immersion in boiling 0.1% sodium dodecyl sulfate in water between hybridizations (18). Primers 7 and 8 (within CCH1) and primers 4 and 9 (upstream of and within CCH1, respectively) were used to evaluate putative transformants for the absence of CCH1. The primer pair 3 and 13 and the pair 12 and 6 were used to evaluate putative transformants for the presence of the hygromycin resistance cassette in connection with the CCH1 flanking regions. Primer pair 7 and 8 and primer pair 10 and 11 were used as probes in Southern analysis, and pair 1 and 5 was used on PH-1 to amplify the entire CCH1 gene plus flanking regions for use in the complementation.
Complementation of the cch1 mutation.
Protoplasts from mutant Δcch1-T14 were transformed with the complete CCH1 genomic region including the upstream and downstream flanking regions (total size, 7,662 bp) (see “Construction of Δcch1 mutants” above) in the neomycin-resistant plasmid pYN06 (as described by Huo et al. [25]). Transformants were selected by their ability to grow on regeneration medium containing 200 μg ml−1 G418. Transformants were screened for the loss of the fluffy phenotype in the vegetative mycelia on carrot agar, and single spore isolates were obtained and stored as above. Genetic characterization of complemented mutants was done initially by PCR and subsequently by Southern analysis.
Characterization of mutant phenotypes.
Ten-microliter conidial stocks were center inoculated (for characterization of vegetative growth) or spread across the surface (for sexual development and GeneChip studies) of carrot agar (36) in a petri plate (60-mm diameter) and incubated at room temperature (22 to 24°C) until the mycelia reached the edge of the plate (4 to 6 days). For growth rates, radial growth from center-inoculated cultures (five replicates) of each mutant and the wild type were measured at 24-h intervals from the time of inoculation until the colony reached the edge of the plate. The first day's growth was discounted. G. zeae is homothallic, and sexual development was induced by gently removing the surface mycelium and applying 1 ml of 2.5% Tween 60 to the surface (6). To help visualize developmental stages, samples were collected daily thereafter from the surface of the agar. Samples were stained with acridine orange, squash mounted in water on a microscope slide, and examined at magnifications of ×200 and ×500 on a Zeiss standard epifluorescence microscope, as previously described (23). Wild-type PH-1 was used for comparison.
A previous study delineated six stages of sexual development corresponding to vegetative hyphae, dikaryotic hyphae, perithecium initiation, paraphysis development, ascus development, and ascospore formation. These developmental stages were designated as 0 h (vegetative hyphae and time of induction of sexual development) through 144 h (mature perithecia with discharging ascospores, with spores of some asci still coming to maturity). Although the cch1 mutants are developmentally delayed, we have retained the hourly designations of the wild type here for consistency. When the cultures reached the 144-h stage, a cork borer was used to remove a circular agar plug covered with perithecia. The plug was cut into two semicircles which were placed on a microscope slide in a moist chamber, with perithecia-covered surfaces perpendicular to the slide. The slides were examined after 15, 24, and 48 h for spore discharge. To account for potentially slower growth and development of the mutants, samples were also taken at 168 h postinduction and assayed for spore discharge.
Calcium supplementation.
The cch1 mutants were supplemented with calcium to determine if the addition of calcium would compensate for the mutation. Calcium supplementation was done as follows: CaCl2 was added to solidified carrot agar (i) simultaneously with inoculation or (ii) simultaneously with the Tween 60 at the time of induction of sexual development or (iii) at the initiation of the discharge assay, when the CaCl2 was allowed to permeate the agar plug. In all cases, CaCl2 at concentrations of 10, 100, 500, and 1000 mM was added to the agar, resulting in final concentrations of 0.77, 7.69, 38.46, and 76.92 mM, respectively.
In Cryptococcus neoformans, a Δcch1 mutant, was unable to grow when concentrations of extracellular calcium were very low (38). Accordingly, the wild type and Δcch1 mutants were grown on Bilay's medium (5) supplemented with a 1 mM concentration of the calcium chelator BAPTA (1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′,-tetraacetic acid). A 60-mm petri plate was center inoculated with 10 μl of conidial stock, and growth was observed over 1 week.
Pathogenicity assays.
To determine the effect of the Δcch1 mutation on pathogenicity of wheat, a method modified from Jin and Zhang (30) was used. Spring wheat (Triticum aestivum L. cv. “Bobwhite”) was grown in the greenhouse to anthesis and inoculated with 10 μl of conidial stock of either the wild type (control) or the Δcch1-T12 or the Δcch1-T14 mutant. Ten florets (one floret per head and one head per plant) were inoculated as previously described (23). Following inoculation, the plants were placed in a misting chamber for 96 h for the infection to develop; upon removal from the mist chamber, the wheat was observed for the development of symptoms. Colonization of stems by saprobic fungi occurred naturally in the greenhouse; these fungi were not inoculated onto the stems. All inoculated plants were housed together, with controls placed among the inoculated plants. The wheat matured in the greenhouse until fully senesced (approximately 28 days after infection), after which the stems were cut above the node closest to the soil line, and both stems and heads were allowed to air dry at 25°C for a minimum of 7 days in the laboratory. Two 7-cm segments, starting immediately below the head, were taken from each dried stem and placed in glass petri plates (100-mm diameter) containing 6 g (dry weight) of sterile vermiculite moistened with 5 ml of sterile distilled water. The stem fragments were incubated under continuous cool white light (F34T12/CW; Phillips United States, New York, NY) for 7 days at room temperature; then perithecia were quantified under a dissecting microscope (StereoZoom 7; Leica Microsystems GmbH, Wetzlar, Germany). Finally, each stem fragment was suspended in 9 ml of 0.2% Tween 20 and agitated briefly to remove spores; spores were collected from the resulting suspension by centrifugation. Spores were resuspended in a known volume of water, quantified using a hemocytometer, and identified. Spore ratios were compared using the chi-square test on a two-by-two contingency table with 1 degree of freedom.
Microarray analysis.
The mycelium harvested at the time of induction, i.e., when the mycelium reached the edge of the petri dish, was considered the 0 h vegetative mycelium (4 days for the wild type and 5 days for the mutant). Surface mycelia or developing perithecia were similarly harvested from the induced cultures, at the stage in which ascus initials were present in immature perithecia (96 h) and at the time of ascus and ascospore maturity (multiseptate ascospores present in well-developed asci; 144 h). Three replicates were harvested for each developmental stage. Collection and analysis of developmental stages from the wild type have been previously described (24). All harvested samples were lyophilized and frozen at −80°C until RNA extraction.
RNA was extracted from lyophilized samples using the Trizol reagent (Invitrogen). A CTAB-chloroform step was incorporated into all RNA preparations due to high levels of polysaccharides in the 96-h and 144-h stages, as described in Hallen et al. (24). The samples were purified using an RNeasy Mini Kit (Qiagen) following the manufacturer's instructions. Purified RNA was processed using the Affymetrix One-Cycle Target Labeling procedure, following the Affymetrix manual (1) and hybridized to the Fusariuma520094 GeneChip (Affymetrix). Hybridization, washing and chip reading buffers, and procedures followed Affymetrix guidelines (1). The hybridization signals were scanned with a GeneChip GCS 3000 scanner (Affymetrix) and the cell intensity (CEL) files were obtained from GCOS 2.1 software (Affymetrix). CEL files are available at PLEXdb (http://www.plexdb.org/), accession numbers FG5 (wild type) and FG6 (mutant). CEL files were normalized in the Bioconductor package of R, version 2.3.0rc (20, 45) using RMA, an expression measure that accounts for background correction, quantile normalization, and variation between arrays (27, 28).
Comparisons between Δcch1 and the wild type for each developmental stage were conducted using the Limma package in Bioconductor (51). The list of differentially expressed genes was ranked based on the moderated t statistic introduced by Smyth (50). Statistical significance was empirically determined for each comparison by selecting the cutoff P value lower than the smallest P value found in any of the Affymetrix control probe sets, as recommended by Smyth (51). Genes for which significant differential transcript abundance was detected were functionally characterized using FunCat (46).
RESULTS
The putative G. zeae homolog of the yeast CCH1 was replaced with the coding sequence of the hph gene imparting hygromycin resistance. Sixteen hygromycin-resistant colonies were obtained from a single transformation experiment with the CCH1 deletion construct. Of the 16, six showed abnormal vegetative growth on carrot agar. All 16 were analyzed by PCR, and 5 of the abnormal colonies, designated Δcch1-T10, -T12, -T13, -T14, and -T15, were shown to be consistent with hph insertion, replacing CCH1. Of the transformants demonstrating wild-type growth (ectopic insertions of the hph gene), one isolate, designated Δcch1-T11, was used in further studies as a control. The status of transformants Δcch1-T10 through Δcch1-T14 was confirmed by Southern analysis with an internal fragment of CCH1 and with hph (Fig. 1).
FIG. 1.
Southern analysis of Δcch1 mutants and complements compared to the wild type using a probe internal to CCH1 (A) and a probe specific to hph1 (B). Lanes from left to right: 1-kb ladder, wild type (WT), transformant Δcch1-T10, Δcch1-T11, Δcch1-T12, Δcch1-T13, Δcch1-T14, and complements cch1comp6 and cch1comp5. There is an empty lane between the two complements. Transformant Δcch1-T11 is an ectopic insertion of the replacement construct, including hph. Ten micrograms was loaded per lane. Genomic DNA was digested with BamHI. The CCH1 internal probe hybridizes to a BamHI fragment of 2,891 bp in the wild type. The hph1 probe hybridizes to a BamHI fragment of 3,465 bp in Δcch1 replacement mutants. Numbers on the left indicate size in kb.
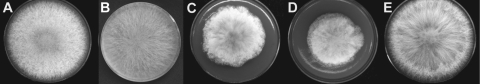
Mutants of G. zeae lacking a functional CCH1 displayed abnormal, delayed vegetative growth on carrot agar. Colony morphology was distinct, with a dense, fluffy growth not observed in the wild type (Fig. 2). Radial colony growth of the wild type was 17.8 mm 24 h−1 until the edge of the petri plate was reached, compared with 10.3 for Δcch1-T12 and 10.6 for Δcch1-T14 (standard deviations of 1.4, 0.5, and 0.3, respectively). The ectopic transformant Δcch1-T11 exhibited radial colony growth of 21.2 mm 24 h−1 (standard deviation, 2.6), and the complemented strain Δcch1comp6 grew 20.7 mm 24 h−1 (standard deviation, 0.7).
FIG. 2.
Vegetative growth on carrot agar 96 h after inoculation. (A) Wild type. (B) Ectopic transformant Δcch1-T11. (C and D) Mutants Δcch1-T12 and Δcch1-T14. Note the dense, fluffy growth. (E) Δcch1comp6. Complementation of the Δcch1 deletion mutant restores the wild-type phenotype.
Following induction of sexual development, the Δcch1 mutants continued to exhibit slower development, with a delay of approximately 24 h compared with the wild type to full maturity. While there were obvious differences in gross colony morphology, these were not quantifiable at the microscopic level when we examined hyphal tip growth. Normal morphological development of the perithecia was present in the mutants. Asci viewed under the microscope were morphologically normal, and ascospores were viable (86 out of 97 [88%] ascospores germinated, compared with 130 out of 140 [92%] for the wild type; results not significantly different, with the chi-square test). Spore discharge was not detected either at the time of mutant perithecium maturity (168 h postinduction; multiseptate ascospores present in perithecium) or 24 h after attaining maturity (192 h postinduction) (Fig. 3). Examination of lids of petri dishes containing mutant cultures up to 3 weeks after maturation failed to reveal any released ascospores, whereas the lids of wild-type cultures exhibited copious amounts of spores that had been discharged from the perithecia. Notably, 38% of the Δcch1 mutant asci contained at least one abnormal spore (Fig. 4). Abnormal ascospores have also been occasionally observed in the wild type. Abnormal spores were much reduced in size and were not included in the germination assays.
FIG. 3.
Forcible ascospore discharge in the wild type (A), ectopic transformant Δcch1-T11 (B), mutant Δcch1-T12 (C), complement Δcch1comp6 (D), and mutant Δcch1-T12 supplemented with 38.46 mM CaCl2 (E). Photographs were taken 24 h after the assay was initiated. Other mutants shared a phenotype similar that of to Δcch1-T12.
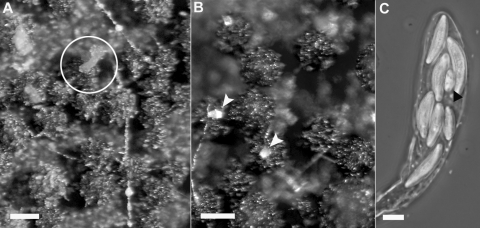
FIG. 4.
Cirrhus formation and ascospore morphology. Cirrhi were present in the wild type (A; circled) and absent in the mutant Δcch1 (B). Occasional clusters of conidia (arrowheads) were seen on the surface of the mutant perithecia. A moderate number of cch1 mutant asci harbored abnormal spores (C; arrowhead). For panels A and B an Axioskop 2 Plus microscope (Carl Zeiss, Inc., Hallbergmoos, Germany) equipped with differential interference contrast optics was used. An AxioCam HRC (Hallbergmoos) photomicrographic system attached to the microscope was used to capture images. The photograph in panel C was taken on a Nikon Labophot 2 microscope using phase optics and a Kodak DCS Pro camera (Rochester, NY). Scale bars, 180 μm (A and B) and 5 μm (C).
Wild-type perithecia will commonly exude spores which have not been discharged as the colonies age and dry. These exuded spore masses, called cirrhi, are prominent within several days of maturation in the wild type (Fig. 4A). No cirrhi were observed at any time as the mutant cultures aged (Fig. 4B). The presence of a single cluster of conidia was frequently observed on the surface of the perithecia (Fig. 4B). Strikingly, older cultures of the wild type normally become covered with conidial clusters (reduced sporodochia), but the Δcch1 mutants never accumulated more than these single clusters. There was no difference in conidia production between the wild type and the mutants in standard conidia-inducing medium made with carboxymethylcellulose.
A complementation construct was prepared by subcloning a 7.6-kb fragment surrounding and including the CCH1 coding region into pYN06 and using that construct to complement the Δcch1-T14 mutant. Two G418-resistant colonies, Δcch1comp5 and Δcch1comp6, were obtained from one transformation experiment, and both harbored the wild-type CCH1. The wild-type vegetative phenotype and active spore discharge were restored with the complementation (Fig. 2 and 3), which was confirmed initially by PCR analysis (results not shown) and subsequently by Southern analysis (Fig. 1).
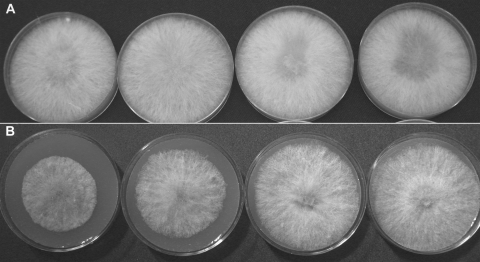
To test whether additional calcium could complement the Δcch1 mutation, as had been shown in S. cerevisiae (15), transformants were supplemented with calcium (CaCl2) during several different cultural and developmental stages. Carrot agar was amended to a final concentration of 0.77, 7.69, 38.46, or 76.92 mM CaCl2 at each of three time points: at the time of initial inoculation, concurrent with induction of sexual development, and at the initiation of the ascospore discharge assay. To ensure that higher than average calcium levels did not exert a negative effect, wild-type PH-1 was also supplemented. No phenotypic differences in either wild-type or mutant cultures were noted at the two lowest concentrations of CaCl2. At the higher levels, the wild-type vegetative phenotype was partially restored in the mutants (Fig. 5). In addition, spore discharge was restored in the mutants at the two highest levels of CaCl2 (shown for 38.46 mM CaCl2 in Fig. 3), although at the highest level (76.92 mM) spore discharge began to diminish in the wild type, probably due to a reduction in the turgor in the ascus (57). To test, by contrast, the phenotype of the wild type and mutants under calcium stress, the calcium chelator BAPTA was added to solid Bilay's medium, which was then center inoculated with conidia of the wild type and Δcch1 mutants. Growth was poor for all cultures; however, wild-type PH-1 was capable of extending hyphae beyond the point of inoculation, while the Δcch1 cultures never grew beyond the point of inoculation (Fig. 6), although some aerial hyphal growth was visible.
FIG. 5.
The effect of supplemental calcium (CaCl2) on vegetative growth. (A) Wild-type PH-1. (B) Mutant Δcch1-T12 (other Δcch1 mutants showed the same effect). From left to right, the CaCl2 concentrations are 0.77, 7.69, 38.46, and 76.92 mM.
FIG. 6.
The effects of the calcium chelator BAPTA on growth of the wild type (A), ectopic transformant Δcch1-T11 (B), and mutant Δcch1-T12 (C) at 9 days postinoculation. Mutant Δcch1-T14 (not shown) gave a similar phenotype to Δcch1-T12.
Examination of wheat infected with Δcch1 and with the wild type revealed no differences in symptom severity, with all inoculated plants (10 plants each for the wild type and the Δcch1 mutants) exhibiting well-developed symptoms (senescence of heads and stems extending below the top node) by 14 days postinoculation. Mycelia were readily observed within the pith of the stems when the plants were harvested at 28 days postinoculation. Perithecia were formed in 5 days when dry wheat stem sections were incubated in moist vermiculite under light. Perithecia were produced abundantly by both the wild type and the mutants in approximately equal numbers (average, 114 perithecia per 7 cm stem fragment). Stems colonized by the Δcch1 mutants and by the wild type revealed both obvious colonization and conidiation by both G. zeae and other nonpathogenic fungi from the greenhouse (largely Penicillium and Aspergillus spp.). In both wild-type- and mutant-infected stems, G. zeae conidia were in the minority compared with those of the other fungi; however, the number of G. zeae conidia was smaller, and the number of other conidia was greater in the stems colonized by the mutants. Thus, there were 8.75 × 103 G. zeae conidia per stem in the Δcch1 mutants, compared with 5.04 × 105 conidia from other organisms. In the wild-type-infected stems, there were 1.32 × 104 G. zeae conidia per stem and 2.08 × 105 conidia from other organisms (results were averaged over seven stems). The ratios were 1/59 (G. zeae conidia/other conidia) in wheat stems infected by a cch1 mutant and 1/16 in stems infected by the wild type. These values differed significantly at a P of <0.001 according to the chi-square distribution.
As calcium fluxes are known to affect gene transcription, gene expression was compared between the wild type and Δcch1-T14. RNA was extracted at three time points, representing vegetative growth, immature perithecia (containing paraphyses, croziers, and immature asci), and mature perithecia with multiseptate ascospores. These time points in Δcch1 were developmentally comparable to the wild type 0 h, 96 h, and 144 h, respectively (24). In total, 2,449 probe sets showed a statistically significant change in abundance of twofold or greater between mutant and the wild type for all three time points inclusive. Specifically, 651 transcripts were differentially regulated in vegetative growth, 1,428 at 96 h and 515 at 144 h (see Table S1 in the supplemental material for a complete list of differentially expressed transcripts). There is comparatively little overlap between time points, with only 111 transcripts showing differential accumulation versus the wild type at more than one time point. Table 2 shows those significantly different from the wild type at both 0 and 96 h. CCH1 (probe set fgd69-40_at) shows a decrease in transcript abundance of 6.18- to 8.62-fold in the Δcch1 mutants compared with the wild type, indicating that hybridization on the GeneChip to the CCH1 probe set is essentially absent in the mutants but present in the wild type, as expected for gene replacement mutants.
TABLE 2.
Transcripts showing statistically significant twofold or greater differential accumulation between mutant and wild type at multiple time points
| Type of change in transcript abundance and transcript identifiera | Transcript abundance in the mutant relative to wild type at the indicated time pointb
|
Annotation | |||||
|---|---|---|---|---|---|---|---|
| 0 h | P0 h | 96 h | P96 h | 144 h | P144 h | ||
| Twofold or greater increase between wild type and mutant at all time points: fgd246-110_at | 4.252137 | 1.70E-07 | 2.961257 | 0.0003721 | 4.35791 | 5.88E-07 | Hypothetical protein |
| Twofold or greater decrease between wild type and mutant at all time points | |||||||
| fg06105_s_at | −1.667428 | 3.26E-05 | −1.544056 | 0.00103707 | −1.81548 | 4.83E-06 | Probable trimethyllysine dioxygenase |
| fgd361-40_s_at | −2.310062 | 7.23E-06 | −2.286179 | 0.00069665 | −1.875791 | 5.85E-06 | Conserved hypothetical protein |
| Increase at 0 h and 96 h* | |||||||
| fgd318-850_at | 4.205249 | 5.17E-06 | 1.885949 | 0.00306174 | −0.775595 | 0.308734 | Conserved hypothetical protein |
| fgd185-60_at | 3.210368 | 1.31E-06 | 2.112676 | 0.00275112 | −0.015636 | 0.975858 | Conserved hypothetical protein |
| fgd197-120_at | 2.983299 | 2.00E-05 | 3.231011 | 0.00374232 | −0.789604 | 0.072781 | Hypothetical protein |
| fgd21-20_at | 2.585006 | 1.33E-05 | 3.126629 | 0.000714 | −0.521372 | 0.301871 | Hypothetical protein |
| fgd158-490_at | 2.584465 | 3.26E-09 | 3.758745 | 0.00031145 | −0.09127 | 0.742611 | Probable pectin lyase precursor |
| fgd233-360_s_at | 2.518178 | 2.38E-05 | 2.804031 | 0.00354073 | −0.346337 | 0.20056 | Hypothetical protein |
| fg09187_s_at | 2.208471 | 2.86E-05 | 1.88661 | 0.00203256 | −0.636026 | 0.107265 | Related to VerA protein |
| fgd160-740_at | 2.162682 | 4.44E-08 | 2.443102 | 7.39E-05 | 0.166141 | 0.547707 | Conserved hypothetical protein |
| fg07734_at | 1.818964 | 2.82E-05 | 3.329558 | 0.00023151 | 1.547734 | 0.004251 | Related to global transactivator |
| Increase at 0 h and decrease at 96 h* | |||||||
| fgd292-290_x_at | 4.260392 | 4.53E-10 | −1.659034 | 0.00190987 | 0.348952 | 0.033907 | Putative protein (EST hit) |
| fgd458-640_at | 3.146793 | 3.02E-07 | −1.276051 | 0.00170183 | −0.482921 | 0.101694 | Related to putative tartrate transporter |
| fgd4-40_at | 3.124855 | 7.92E-06 | −1.618002 | 0.00441397 | −0.065222 | 0.833894 | TOX1; related to KP4 killer toxin |
| fg12249_at | 2.651301 | 9.82E-06 | −1.947163 | 0.00309896 | 0.067724 | 0.859242 | Probable amino acid transport protein GAP1 |
| fg07325_s_at | 2.186069 | 3.70E-05 | −1.067731 | 0.00077608 | −1.441705 | 0.003597 | Related to ATP-binding cassette transporter protein YOR1 |
| fgd320-650_at | 1.260561 | 3.12E-07 | −1.959269 | 0.00170962 | −0.004734 | 0.979366 | Hypothetical protein |
| Decrease at 0 h and increase at 96 h* | |||||||
| fgd199-110_s_at | −1.122248 | 3.28E-05 | 2.398922 | 0.00446521 | 0.085527 | 0.791343 | Hypothetical protein |
| fgd112-310_at | −1.911777 | 8.49E-06 | 1.598495 | 0.00482901 | 2.132243 | 0.000159 | Related to ARG8, acetylornithine aminotransferase |
| fgd266-180_at | −1.930477 | 2.96E-08 | 1.938448 | 0.00304904 | −0.034996 | 0.915222 | Conserved hypothetical protein |
| fgd425-330_at | −2.12576 | 1.26E-05 | 2.153474 | 0.0028155 | −1.021158 | 0.028486 | Related to ANON-37CS protein |
| fgd77-70_at | −2.285935 | 5.13E-06 | 3.955246 | 0.00321478 | −0.319272 | 0.191412 | Probable regulatory subunit of protein phosphatase-1 |
| fgd148-1240_at | −2.324533 | 2.33E-05 | 2.198154 | 0.00390104 | −0.61492 | 0.135032 | Conserved hypothetical protein |
| fg04626_s_at | −2.640947 | 3.24E-05 | 3.575254 | 0.00025086 | −0.026996 | 0.935111 | Hypothetical protein |
| Decrease at 0 h and 96 h* | |||||||
| fg01182_s_at | −1.213758 | 6.88E-06 | −1.160318 | 0.00016456 | −1.044243 | 0.000145 | Conserved hypothetical protein |
| fg06960_s_at | −1.36357 | 4.90E-06 | −1.061768 | 0.00012174 | −1.312393 | 0.00465 | Related to Ku70-binding protein |
| fgd56-90_at | −1.384284 | 2.98E-06 | −2.039921 | 0.00023909 | 0.357408 | 0.189783 | Probable multiubiquitin chain binding protein (MBP1) |
| fgd450-20_at | −1.398908 | 1.96E-05 | −1.399369 | 0.00086921 | −0.564188 | 0.330237 | Probable phosphatidylinositol/phosphatidylcholine transfer protein SEC14 |
| fgd104-290_at | −1.432652 | 6.95E-06 | −1.847161 | 0.0029308 | −2.325124 | 0.000334 | Conserved hypothetical protein |
| fgd422-230_at | −1.54976 | 3.70E-06 | −1.312596 | 0.00028198 | −1.461646 | 0.034723 | Probable coatomer epsilon subunit |
| fg00331_s_at | −1.584002 | 3.51E-05 | −2.637301 | 0.00105877 | 0.20298 | 0.541548 | putative protein (EST hit) |
| fgd35-590_at | −1.626283 | 4.53E-06 | −2.011254 | 0.00309951 | −1.186748 | 0.001245 | Probable septum formation maf |
| fg00430_s_at | −1.753789 | 2.95E-05 | −1.739395 | 0.00224068 | −1.538549 | 0.000397 | Probable oligosaccharyltransferase |
| fg08308_s_at | −1.83793 | 3.73E-06 | −1.467145 | 0.00483457 | 0.147557 | 0.543416 | ABC2; related to multidrug resistance protein |
| fgd383-190_at | −1.997796 | 2.90E-07 | −1.792443 | 0.00110624 | −0.905406 | 0.005111 | Conserved hypothetical protein |
| fgd24-30_at | −2.028044 | 1.84E-05 | −1.579857 | 0.00059991 | −1.650269 | 0.000555 | Conserved hypothetical protein |
| fgd248-140_at | −2.053918 | 2.44E-05 | −1.838453 | 0.00458005 | −1.713851 | 0.001132 | Probable aldo-keto reductase YPR1 |
| fgd104-420_at | −2.108565 | 7.59E-06 | −1.449971 | 0.00035639 | −0.236224 | 0.159022 | Probable branched-chain alpha-ketoacid dehydrogenase kinase; mitochondrial precursor |
| fgd168-1760_s_at | −2.108666 | 5.61E-09 | −1.108928 | 0.00270452 | -1.463851 | 0.054713 | Related to type 2C protein phosphatase |
| fgd335-320_at | −2.145885 | 1.17E-06 | −1.382001 | 0.00043239 | 2.014346 | 0.010176 | Probable glycine decarboxylase P subunit |
| fgd367-120_at | −2.170614 | 6.10E-08 | −1.815004 | 0.00365397 | 1.029645 | 0.009561 | Related to n-carbamoyl-l-amino acid hydrolase |
| fgd59-140_at | −2.553609 | 3.19E-06 | −1.51113 | 0.00069322 | −0.825075 | 0.009643 | Probable GTPase-activating protein of the rho/rac family (LRG1 protein) |
| fgd212-830_at | −2.58426 | 5.02E-07 | −1.522224 | 0.0020098 | −1.344456 | 0.098771 | Probable 3-hydroxyisobutyrate dehydrogenase |
| fgd170-50_s_at | −2.753081 | 1.54E-06 | −1.113897 | 0.00290648 | −0.848131 | 0.000775 | Probable potassium transporter TRK-1 |
| fgd192-1180_at | −3.780183 | 2.01E-08 | −1.404931 | 0.00321436 | 0.313153 | 0.495277 | Related to serine-type carboxypeptidase F precursor |
| fgd90-10_at | −4.045339 | 2.81E-05 | −1.489611 | 0.00080104 | −3.030282 | 0.001579 | Probable DIP5-glutamate and aspartate permease; able to mediate transport of other amino acids |
Significant changes in abundance in the mutant are indicated by an asterisk.
Log 2 relative change. Negative numbers and positive numbers represent a decrease and increase, respectively, in mutant transcript abundance compared with wild type. P values are identified by time point (e.g., P value for change in transcript abundance at 0 h is Po h). The cutoffs for statistical significance vary between treatments.
A functional categorization of genes showing a statistically significant decrease in transcript accumulation of twofold or greater in the Δcch1 mutant compared to wild type is shown in Table 3 (for the list of individual genes and complete breakdown into FunCat categories see Table S2 in the supplemental material). Of particular interest are those categories for which there is an enrichment of genes showing a decrease in transcript abundance in the mutant compared to the genome as a whole. This enrichment is significant at P values of less than 0.05 (29). This analysis shows a significant enrichment of genes involved in metabolism (FunCat category 01) in the pool of down-regulated Δcch1 genes. FunCats 02 and 14 (energy and protein fate, respectively) also contained significant proportions of genes with lowered transcript levels at 0 and 96 h compared to the wild type, while FunCats 30 and 40 (cellular communication/signal transduction mechanism and cell fate, respectively) contained significant proportions of genes with lowered abundance at 96 and 144 h for the same comparison. Genes involved in cell cycle and DNA processing (FunCat 10) were significantly reduced in transcript levels at all time points in the mutant.
TABLE 3.
Functional categories of genes showing a statistically significant decrease in transcript accumulation of twofold or greater in the Δcch1 mutant compared to wild typea
| FunCat code | Category name (description) | % of genome | Relative decrease in transcript of the mutant and statistical significance at the indicated time point
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0 h
|
96 h
|
144 h
|
||||||
| No. of genes (%) | P | No. of genes (%) | P | No. of genes (%) | P | |||
| 01 | Metabolism | 13.7 | 95 (24.2) | 9.43E-09 | 172 (20.7) | 6.54E-09 | 37 (9.86) | 1 |
| 02 | Energy | 2.96 | 22 (5.62) | 0.003189 | 44 (5.3) | 0.000129 | 4 (1.06) | 1 |
| 10 | Cell cycle and DNA processing | 4.36 | 32 (8.18) | 0.000491 | 79 (9.52) | 3.05E-11 | 32 (8.53) | 0.000235 |
| 11 | Transcription | 4.77 | 20 (5.11) | 0.408944 | 55 (6.63) | 0.008169 | 31 (8.26) | 0.002126 |
| 12 | Protein synthesis | 2.84 | 4 (1.02) | 1 | 26 (3.13) | 0.32809 | 9 (2.4) | 1 |
| 14 | Protein fate (folding, modification, destination) | 6.44 | 38 (9.71) | 0.007318 | 124 (14.9) | 1.79E-19 | 31 (8.26) | 0.091332 |
| 16 | Protein with binding function or cofactor requirement (structural or catalytic) | 7.36 | 39 (9.97) | 0.032521 | 101 (12.1) | 2.53E-07 | 25 (6.66) | 1 |
| 18 | Regulation of metabolism and protein function | 1.14 | 10 (2.55) | 0.014227 | 27 (3.25) | 6.93E-07 | 10 (2.66) | 0.010869 |
| 20 | Cellular transport, transport facilities and transport routes | 7.5 | 40 (10.2) | 0.027795 | 107 (12.9) | 1.35E-08 | 27 (7.2) | 1 |
| 30 | Cellular communication/signal transduction mechanism | 1.86 | 9 (2.3) | 0.307383 | 29 (3.49) | 0.000818 | 17 (4.53) | 0.000686 |
| 32 | Cell rescue, defense and virulence | 4.73 | 23 (5.88) | 0.16665 | 61 (7.35) | 0.000378 | 16 (4.26) | 1 |
| 34 | Interaction with the environment | 3.29 | 15 (3.83) | 0.308379 | 42 (5.06) | 0.003558 | 19 (5.06) | 0.042005 |
| 36 | Systemic interaction with the environment | 0.18 | 3 (0.76) | 0.035313 | 4 (0.48) | 0.065461 | 1 (0.26) | 0.508159 |
| 40 | Cell fate | 1.49 | 9 (2.3) | 0.131456 | 33 (3.98) | 2.15E-07 | 14 (3.73) | 0.001518 |
| 41 | Development (systemic) | 0.3 | 2 (0.51) | 0.34069 | 5 (0.6) | 0.110039 | 1 (0.26) | 1 |
| 42 | Biogenesis of cellular components | 4.34 | 20 (5.11) | 0.257292 | 74 (8.92) | 2.12E-09 | 18 (4.8) | 0.366487 |
| 43 | Cell type differentiation | 2.5 | 16 (4.09) | 0.037512 | 47 (5.66) | 1.15E-07 | 15 (4) | 0.050677 |
| 99 | Unclassified proteins | 69.3 | 215 (54.9) | 1 | 413 (49.8) | 1 | 259 (69) | 0.568122 |
As some genes can be classified in more than one FunCat category, percentages sum to >100.
DISCUSSION
Arguably, the most unique cell in the life cycle of the ascomycetous fungi is the ascus. Asci elongate in response to increased turgor pressure and eventually rupture at the tip to fire spores into the air (54). In the perithecium-forming fungi, the asci fire singly in succession—approximately 45 s apart in G. zeae under optimal conditions (56)—which suggests the presence of a regulatory mechanism that coordinates discharge. One possibility for a regulatory mechanism is a calcium signaling cascade. In this study, we have identified one component of the calcium signaling machinery, CCH1, which affects discharge in G. zeae when genetically disrupted.
Cch1 has been implicated as part of the high-affinity calcium-acquiring machinery (42). The role of Cch1 in G. zeae in facilitating calcium influx at low concentrations was aptly demonstrated by the lack of growth of the mutants in the presence of the calcium chelator BAPTA in contrast to the wild type and ectopic Δcch1 mutant, which both exhibited growth (Fig. 6). These results support the role of Cch1 in obtaining calcium in low-calcium environments, as has been suggested in studies of C. neoformans (38) and yeast (39, 42). However, the fact that discharge was restored at high concentrations of Ca2+ indicates that another channel is present—possibly a distinct low-affinity Ca2+ influx system, as postulated by Muller and colleagues (42)—and able to substitute for Cch1.
In an effort to simulate perithecium formation in crop residue in the field, we have established a protocol to induce development of perithecia on colonized wheat stems, a process which also allows conidiation. An unexpected outcome of this experiment was the observation that the Δcch1 mutant allowed more conidiation from contaminating fungi than did the wild type. This suggests that, while the Δcch1 mutant is competent in infecting and colonizing wheat, it may not be as capable as the wild type in defending its resources against competitors in the crop residue. One possible explanation may be the reduced expression of the polyketide synthase gene responsible for aurofusarin production (fg12040) (17, 34, 41). fg12040 showed a 16-fold reduction in transcript abundance in the Δcch1 mutant compared to the wild type during vegetative growth; this is of interest as rubrofusarin, a precursor to aurofusarin and also produced by the fg12040 protein, is known to possess antimicrobial properties (16, 21). The antimicrobial properties of aurofusarin have not been determined. More rigorous testing of this hypothesis using a strain deficient in aurofusarin production is under way. Although the growth of the Δcch1 mutant is slower in culture, the progress of disease symptoms is identical to that of the wild type, suggesting that its development is not slowed in the plant. Regardless, the stem assay uses fully colonized stems, so the slow growth of the Δcch1 mutant compared with wild-type G. zeae should not affect the outcome of this assay.
In wild-type G. zeae, CCH1 transcripts are detected at moderate levels throughout growth and development and do not appear to be exclusive to sexual development (24) (PlexDB accession number FG5). Results from the Fusarium GeneChip analysis of the Δcch1 mutant showed surprisingly little overlap in differentially accumulated transcripts between time points examined. This suggests that the role of Cch1 in G. zeae is a dynamic one, changing to suit different needs during organismal development and differentiation. In support of Cch1's role in sexual development, 20 genes implicated in meiosis (FunCat 10.03.02), out of 77 meiosis genes predicted in the G. zeae genome, exhibited a significant decrease in transcript abundance in the mutant during at least one of the three time points examined. Transcripts for genes involved in the mitotic cell cycle showed a significant decrease in all time points, as did transcripts for DNA recombination and repair genes. The highest category of genes showing reduced mRNA accumulation in the mutant at 0 h (during mycelial growth) was metabolism. The decrease in these classes of genes may explain the delay in mycelial growth and development. It is surprising, with the large number of genes whose expression is affected, that the in vitro effects of the mutation are so subtle. The mutant lags in development by approximately 24 h following induction. Mycelial growth (preinduction) is also noticeably slower. Thus, differences between gene expression in the vegetative state might include differences in mycelial age. However, developmental stages reflect formation of discrete cell types (perithecium initials, ascogenous hyphae, paraphyses, and ascospores), and therefore our method of sampling based on developmental stage should ensure an equitable comparison.
For the study of sexual development and differentiation, 96 h is arguably the most interesting of the time points we examined: the perithecium has reached its full size and pigmentation, and asci are forming. Ascus structure will be completed over the next 24 h, and the spores will mature over the next 48 h (55, 44). A decline in meiosis-related transcript abundance was observed in each of the time points in the mutant; however, meiosis takes place during ascus development from about 96 to 120 h (24). It is not surprising, therefore, that 96 h showed the most meiosis genes down-regulated (12; also see Table S2 in the supplemental material). Also at 96 h, more genes are differentially regulated in the Δcch1 mutant than in the wild type, and this difference is greater than that at 0 h and 144 h (Table 3; also see Table S2 in the supplemental material). Some of these changes in gene expression may be reflected in the increased number of spores with aberrant morphologies in the asci of the mutant.
Understanding the mechanism by which Cch1 affects discharge will reveal important aspects of ascus function. In mammalian cells, the L-type ion channels are triggered to open by membrane depolarization. Previous work has indicated that the fungal channels are less sensitive to voltage fluctuations and may have a different gating mechanism (38). Interestingly, we previously reported the effect of the L-type ion channel inhibitor, verapamil, resulting in nearly complete arrest of ascospore discharge (57). Mycelial growth of treated cultures was also reduced by nearly 50%. Thus, the application of verapamil to the cultures resulted in a phenotype that closely mimics that of the Δcch1 mutation, and the target of verapamil is, therefore, probably Cch1. Because verapamil is effective on perithecia whose asci are mature and whose nuclei are already enclosed in the spores, it seems likely that the effect of Cch1 on discharging asci is directly on the cell components to effect spore propulsion rather than by influencing transcription. This hypothesis is further supported by the calcium addition experiment reported here, in which the addition of exogenous calcium elicits discharge, indicating that the structural and physiological components of the discharge apparatus are intact. Calcium signaling is known to regulate cytoskeletal rearrangements (for example, Mace et al. [40]) and other physiological functions (3) in mammals. There are no gross cytoskeletal defects in the mutant, or the shape of the asci would be affected. However, more subtle defects in cytoskeleton function may be preventing ascus function. We will explore mechanisms both upstream and downstream of Cch1 in future experiments.
Supplementary Material
Acknowledgments
We thank John Guenther and Shawna Desrosiers for development of the assay to produce perithecia on colonized straw. Kurt Stepnitz and Ralph Common performed the photography for Fig. 3 and 4C, respectively. Affymetrix GeneChip hybridization was performed by Annette Thelen at the Michigan State University Research Technology Support Facility.
This research was supported by a grant from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (grant no. 2004-35604-14327 to F.T. and collaborators H. Corby Kistler and Jin-Rong Xu) and the Michigan Agricultural Experiment Station.
Footnotes
Published ahead of print on 14 December 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Affymetrix. 2004. GeneChip expression analysis. Affymetrix, Santa Clara, CA.
- 2.Berridge, M. J., P. Lipp, and M. D. Bootman. 2000. The versatility and universality of calcium signaling. Nat. Rev. Mol. Cell Biol. 111-21. [DOI] [PubMed] [Google Scholar]
- 3.Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signaling: dynamics, homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 4517-529. [DOI] [PubMed] [Google Scholar]
- 4.Blankenship, J. R., F. L. Wormley, M. K. Boyce, W. A. Schell, S. G. Filler, J. R. Perfect, and J. Heitman. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, C. 1971. The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, England.
- 6.Bowden, R. L., and J. F. Leslie. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89182-188. [DOI] [PubMed] [Google Scholar]
- 7.Cappellini, R. A., and J. L. Peterson. 1965. Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57962-966. [Google Scholar]
- 8.Carroll, A. M., J. A. Sweigard, and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 4122. [Google Scholar]
- 9.Council for Agricultural Science and Technology. 2003. Mycotoxins: risks in plant, animal, and human systems. Task force report no. 139. Council for Agricultural Science and Technology, Ames, IA.
- 10.Cyert, M. S. 2001. Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu. Rev. Genet. 35647-672. [DOI] [PubMed] [Google Scholar]
- 11.Dayton, J. S., and A. R. Means. 1996. Ca2+/calmodulin-dependent kinase is essential for both growth and nuclear division in Aspergillus nidulans. Mol. Biol. Cell 71511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayton, J. S., M. Sumi, N. N. Nanthakumar, and A. R. Means. 1997. Expression of a constitutively active Ca2+/calmodulin-dependent kinase in Aspergillus nidulans spores prevents germination and entry into the cell cycle. J. Biol. Chem. 2723223-3230. [DOI] [PubMed] [Google Scholar]
- 13.Deng, L., R. Sugiura, M. Takeuchi, M. Suzuki, H. Ebina, T. Takami, A. Koike, S. Iba, and T. Kuno. 2006. Real-time monitoring of calcineurin activity in living cells: evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol. Biol. Cell 174790-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, M., J. Cox, D. J. Davis, A. Wagner, R. Taylor, A. J. Huerta, and N. P. Money. 2004. New information on the mechanism of forcible ascospore discharge from Ascobolus immersus. Fungal Genet. Biol. 41698-707. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, M., N. Schnell, J. Chattaway, P. Davies, G. Dixon, and D. Sanders. 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419259-262. [DOI] [PubMed] [Google Scholar]
- 16.Frandsen, R. J. N., N. J. Nielsen, N. Maolanon, J. C. Sørensen, S. Olsson, J. Nielsen, and H. Giese. 2006. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol. Microbiol. 611069-1080. [DOI] [PubMed] [Google Scholar]
- 17.Gaffoor, I., D. W. Brown, R. Plattner, R. H. Proctor, W. Qi, and F. Trail. 2005. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 41926-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaffoor, I., and F. Trail. 2006. Characterization of two polyketide synthase genes involved in zearalanone biosynthesis in Gibberella zeae. Appl. Environ. Microbiol. 721793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavric, O., D. Becker dos Santos, and A. Griffiths. 2007. Mutation and divergence of the phospholipase C gene in Neurospora crassa. Fungal Genet. Biol. 44242-249. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. H. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, J. G., H. J. Zhang, S. I. Pendland, B. D. Santarsiero, A. D. Mesecar, F. Cabieses, and N. R. Farnsworth. 2004. Antimycobacterial naphthopyrones from Senna oblique. J. Nat. Prod. 67225-227. [DOI] [PubMed] [Google Scholar]
- 22.Güldener, U., K.-Y. Seong, J. Boddu, S. Cho, F. Trail, J.-R. Xu, G. Adam, H.-W. Mewes, G. J. Muehlbauer, and H. C. Kistler. 2006. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet. Biol. 43316-345. [DOI] [PubMed] [Google Scholar]
- 23.Guenther, J. C., and F. Trail. 2005. The development and differentiation of Gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia 97229-237. [DOI] [PubMed] [Google Scholar]
- 24.Hallen, H. E., M. Huebner, S.-H. Shiu, U. Güldener, and F. Trail. 2007. Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet. Biol. 441146-1156. [DOI] [PubMed] [Google Scholar]
- 25.Hou, Z., C. Yuo, Y. Peng, T. Katan, H. C. Kistler, and J.-R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 151119-1127. [DOI] [PubMed] [Google Scholar]
- 26.Iida, H., H. Nakamura, T. Ono, M. S. Okumura, and Y. Anraku. 1994. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol. Cell. Biol. 148259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry, R. A., B. M. Bolstad, S. F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry, R. A., B. Hobbs, F. Collin, F. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 29.Jansen, R., and M. Gerstein. 2000. Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res. 281481-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, Y., and X. Zhang. 1998. Mass production of ascospores of Gibberella zeae. Phytopathology 88S44. (Abstract.) [Google Scholar]
- 31.Joseph, J. D., and A. R. Means. 2000. Identification and characterization of two Ca2+/CaM-dependent protein kinases required for normal nuclear division in Aspergillus nidulans. J. Biol. Chem. 27538230-38238. [DOI] [PubMed] [Google Scholar]
- 32.Kanzaki, M., M. Nagasawa, I. Kojima, C. Sato, K. Naruse, M. Sokabe, and H. Iida. 1999. Molecular identification of a eukaryotic, stretch-activated, non-selective cation channel. Science 285882-886. [DOI] [PubMed] [Google Scholar]
- 33.Kerényi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 654071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, J.-E., K.-H. Han, J. Jin, H. Kim, J.-C. Kim, S.-H. Yun, and Y.-W. Lee. 2005. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl. Environ. Microbiol. 711701-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, Y. K., D. X. Li, and P. E. Kolattukudy. 1998. Induction of Ca2+-calmodulin signaling by hard-surface contact primes Colletotrichum gloeosporioides conidia to germinate and form appressoria. J. Bacteriol. 1805144-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klittich, C. J. R., and J. F. Leslie. 1988. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuori). Genetics 118417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraus, P. R., C. B. Nichols, and J. Heitman. 2005. Calcium- and calcineurin-independent roles for calmodulin in Cryptococcus neoformans morphogenesis and high-temperature growth. Eukaryot. Cell 41079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, M., P. Du, G. Heinrich, G. M. Cox, and A. Gelli. 2006. Cch1 mediates calcium entry in Cryptococcus neoformans and is essential in low-calcium environments. Eukaryot. Cell 51788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locke, E. G., M. Bonilla, L. Liang, Y. Takita, and K. W. Cunningham. 2000. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Mol. Cell. Biol. 206686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mace, O. J., E. L. Morgan, J. A. Affleck, N. Lister, and G. L. Kellett. 2007. Calcium absorption by Cav1.3 induces terminal web myosin II phosphorylation and apical GLUT2 insertion in rat intestine. J. Physiol. 580605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malz, S., M. N. Grell, C. Thrane, F. J. Maier, P. Rosager, A. Felk, K. S. Albertsen, S. Salomon, L. Bohn, W. Schäfer, and H. Giese. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42420-433. [DOI] [PubMed] [Google Scholar]
- 42.Muller, E. M., E. G. Locke, and K. W. Cunningham. 2001. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 1591527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 162576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi, W., C. Kwon, and F. Trail. 2006. Microarray analysis of transcript accumulation during perithecium development in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Mol. Genet. Genomics 27687-100. [DOI] [PubMed] [Google Scholar]
- 45.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 46.Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani, M. Mokrejs, I. Tetko, U. Güldener, G. Mannhaupt, M. Münsterkötter, and H. W. Mewes. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 185539-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sanglard, D., F. Ischer, O. Marchetti, J. Entenza, and J. Bille. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48959-976. [DOI] [PubMed] [Google Scholar]
- 49.Shaner, S. 2003. Epidemiology of Fusarium head blight of small grain cereals in North America. In K. J. Leonard, and W. R. Bushnell (ed.), Fusarium head blight of wheat and barley. APS Press, St. Paul, MN.
- 50.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 33. [DOI] [PubMed] [Google Scholar]
- 51.Smyth, G. K. 2005. Limma: linear models for microarray data. In R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, NY.
- 52.Solomon, P. S., K. Rybak, R. D. Trengove, and R. P. Oliver. 2006. Investigating the role of calcium/calmodulin-dependent protein kinases in Stagonospora nodorum. Mol. Microbiol. 62367-381. [DOI] [PubMed] [Google Scholar]
- 53.Tank, D. C., and T. Sang. 2001. Phylogenetic utility of the glycerol-3-phosphate acyltransferase gene: evolution and implications in Paeonia (Paeoniaceae). Mol. Phylogenet. Evol. 19421-429. [DOI] [PubMed] [Google Scholar]
- 54.Trail, F. 2007. Fungal cannons: explosive spore discharge in the Ascomycota. FEMS Microbiol. Lett. 27612-18. [DOI] [PubMed] [Google Scholar]
- 55.Trail, F., and R. Common. 2000. Perithecial development by Gibberella zeae: a light microscopy study. Mycologia 92130-138. [Google Scholar]
- 56.Trail, F., I. Gaffoor, and S. Vogel. 2005. Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 42528-533. [DOI] [PubMed] [Google Scholar]
- 57.Trail, F., H. Xu, R. Loranger, and D. Gadoury. 2002. Physiological and environmental aspects of ascospore discharge in Gibberella zeae (anamorph Fusarium graminearum) Mycologia 94181-189. [PubMed] [Google Scholar]
- 58.Yang, Y. H., P. Cheng, G. Zhi, and Y. Liu. 2001. Identification of calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J. Biol. Chem. 27641064-41072. [DOI] [PubMed] [Google Scholar]
- 59.Zelter, A., M. Bencina, B. J. Bowman, O. Yarden, and N. D. Read. 2004. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea and Saccharomyces cerevisiae. Fungal Genet. Biol. 41827-841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.