Abstract
Entomopathogenic fungi such as Metarhizium anisopliae infect insects by direct penetration of the cuticle, after which the fungus adapts to the high osmotic pressure of the hemolymph and multiplies. Here we characterize the M. anisopliae Mos1 gene and demonstrate that it encodes the osmosensor required for this process. MOS1 contains transmembrane regions and a C-terminal Src homology 3 domain similar to those of yeast osmotic adaptor proteins, and homologs of MOS1 are widely distributed in the fungal kingdom. Reverse transcription-PCR demonstrated that Mos1 is up-regulated in insect hemolymph as well as artificial media with high osmotic pressure. Transformants containing an antisense vector directed to the Mos1 mRNA depleted transcript levels by 80%. This produced selective alterations in regulation of genes involved in hyphal body formation, cell membrane stiffness, and generation of intracellular turgor pressure, suggesting that these processes are mediated by MOS1. Consistent with a role in stress responses, transcript depletion of Mos1 increased sensitivity to osmotic and oxidative stresses and to compounds that interfere with cell wall biosynthesis. It also disrupted developmental processes, including formation of appressoria and hyphal bodies. Insect bioassays confirmed that Mos1 knockdown significantly reduces virulence. Overall, our data show that M. anisopliae MOS1 mediates cellular responses to high osmotic pressure and subsequent adaptations to colonize host hemolymph.
Unlike bacteria and viruses that need to be ingested to cause disease, entomopathogenic fungi such as Metarhizium anisopliae kill insects by direct penetration of the cuticle followed by multiplication in the hemocoel (4). During infection processes the fungus therefore has to adapt to several distinct environments, including the hydrophobic wax-rich epicuticle, the protein-chitin procuticle, and the cellular hypodermis, before reaching the solute-rich hemolymph (4). Each step requires differential expression of hundreds of genes, including those for cuticle-degrading enzymes, toxin production, stress responses, and cell wall reorganization (5, 18). Adaptation to the hemolymph includes facing a major challenge from the insect immune system and M. anisopliae responds by secreting a collagen-like immune evasion protein, MCL1 (20). The hemolymph is also characterized by a high osmotic pressure (OP), in the range of 300 to 500 mOsmol/liter (3), that itself could constitute a stress. Studies with Saccharomyces cerevisiae and Candida albicans have shown that yeasts mediate osmotic stress responses through the high-osmolarity glycerol pathway and that there are at least two branches, with the SHO1 and SLN1 transmembrane proteins acting as their respective upstream sensors to trigger the mitogen-activated protein (MAP) kinase cascade (10, 12, 17, 23). Although they are widely referred to as osmosensing “receptors,” SHO1 and SLN1 are not likely to serve as receptors in the usual sense because they respond to a wide range of chemically distinct substances and stimuli. The C. albicans SHO1 osmosensor in particular is linked with oxidative stress and with alterations in cell wall biosynthesis and morphogenesis, so that treating Sho1 null mutants with H2O2, Congo red, or calcofluor white causes defects in growth (12). It is possible that these osmosensing proteins are activated through a physical stimulus that accompanies cell shrinkage or membrane remodeling (6).
In this study, we report the identification of an M. anisopliae homolog to the yeast osmosensor SHO1 that is up-regulated in insect hemolymph and artificial high-osmotic-pressure conditions. The gene was designated Mos1 for Metarhizium osmosensor-like protein. To study its function, we used an antisense RNA interference method to suppress transcript levels of Mos1 and demonstrate that the gene links osmotic stress responses with appressorium and hyphal body differentiation, cell wall biosynthesis and virulence.
MATERIALS AND METHODS
Gene cloning and phylogenetic analysis.
An EST study revealed a transcript (CN808248) that was highly expressed by M. anisopliae strain ARSEF 2575 grown in insect hemolymph (18). A BLAST search revealed similarity (E = 1 × 10−26) with an osmosensor protein SHO1 from the yeast Saccharomyces cerevisiae. To study the potential function of Mos1 the full cDNA sequence was acquired using primer walking. The deduced amino acid sequence was used for Blastp searches against GenBank and Broad Institute fungal genome databases. The homologs (defined as being at least E ≤ 1 × 10−5) from 23 fungal species were collected for a phylogenetic analysis of the evolutionary history of MOS1. The amino acid sequences were aligned with Clustal X and a neighbor-joining tree was generated with 1,000 bootstrap replicates using the program MEGA v3.1 (9).
Mos1 gene RNA interference.
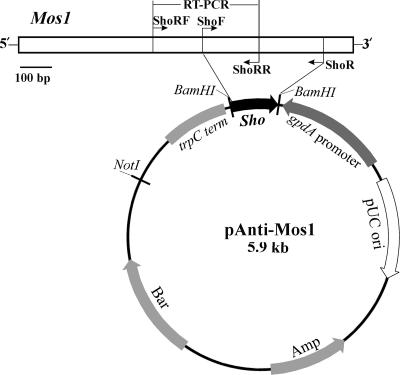
To study the function of Mos1, an RNA interference vector was constructed. The region (669 to 1132 bp) amplified by ShoF (CGGGATCCCCAAACATACCACGTTGCTG) and ShoR (CGGGATCCGCTTTGGCCTGATATGGGTA) was inserted into the BamHI site of pBARGPE1 (20) in the reverse direction to the GpdA promoter (Fig. 1). The orientation and sequence of the insert were verified by PCR and sequencing. The vector was linearized with ScaI, and 10 μg was used for protoplast transformation as described (20). The selected transformants were verified by PCR and maintained for five generations on potato dextrose agar (PDA) (Difco) before being used for further analyses.
FIG. 1.
Schematic map for vector construction. The sequences for primers are listed in Materials and Methods.
RT-PCR analysis.
To investigate the efficiency of RNA interference-mediated transcript depletion, wild-type and antisense-Mos1 strains were grown in Sabouraud dextrose broth (SDB) (Difco) for 36 h and the mycelia were collected and washed with sterile water. The mycelium (0.2 g [wet weight]) was then incubated (25°C for 8 h) for 1, 2, 4, or 8 h in 10 ml of Manduca sexta hemolymph or SDB amended with either 0.7 M KCl, 40 mM H2O2, 50 μg/ml calcofluor white, or 500 μg/ml Congo red. Gene expression by the wild-type and antisense-Mos1 strains was also compared in SDB at 37°C. RNA extraction and cDNA conversion (1 μg RNA was used) were performed as previously described (20). cDNA samples were diluted 20 times, and reverse transcription-PCR (RT-PCR) was conducted using the primer pairs ShoRF (ACTGCCTGACAAGGACAACC) and ShoRR (CATTGCTTGGTTATGCATCG) for the 555- to 899-bp region of Mos1 (Fig. 1). The small-subunit ribosomal gene (18S rRNA) was used for internal calibration (20). PCR products were separated on 1.5% agarose gels, and the density of each band was quantified using the Alpha Innotech gel documentation system. To allow comparison with antisense-Mos1 strains, we also used RT-PCR to look at the regulation of five additional genes involved with hemolymph or stress responses. These include the immune protective protein gene Mcl1, encoding a cell wall protein (20); the adhesin gene Mad1, encoding a cell wall protein involved with hyphal body differentiation (21); the perilipin-like gene Mpl1, encoding a protein that mediates intracellular OP by regulating release of glycerol from lipid droplets (21); a C4-methyl sterol oxidase gene (So) responsible for membrane permeability; and the stress response heat shock protein gene Hsp70 (18).
Growth and differentiation assays.
The native wild-type strain and its antisense-Mos1 transformants were inoculated onto PDA (Difco) or PDA supplemented with 0.7 or 1.5 M KCl and incubated at 25°C or 37°C for different time periods to observe growth differences. Germination of wild-type and antisense-Mos1 spores was also compared in SDB and in SDB amended with 0.7 M, 1.0 M, or 1.5 M KCl. Production of appressoria by the wild-type and antisense-Mos1 strains was induced by inoculating spores onto locust hind wings (19). To test the effects of compounds that interfere with cell wall synthesis or cause oxidative stress (12), the wild type and antisense-Mos1 transformants were inoculated on PDA amended with calcofluor white (25 or 50 μg/ml), Congo red (250 or 500 μg/ml), or H2O2 (20 or 40 mM). Liquid biomass assays were conducted by inoculating 0.2 g (wet weight) of mycelia into 10 ml of SDB or SDB amended with 1.5 M KCl, 50 μg/ml calcofluor white, 500 μg/ml Congo red, or 40 mM H2O2, and incubating at 25°C and 200 rpm for 3 days. The growth of fungal cultures in SDB was also assayed at 37°C. The mycelia were collected and dried at 70°C to constant weight before weighing. There were three replicates for each treatment, and three independent antisense-Mos1 transformants were tested to verify reproducibility.
Insect bioassays.
To investigate the effect of depleting Mos1 transcripts on virulence, insect bioassays were conducted against newly emerged fifth-instar larvae of Manduca sexta (16). Conidia of the wild-type and antisense-Mos1 strains were applied either topically by immersion of larvae in an aqueous suspension containing 1 ×107 conidia/ml for 20 seconds or by injecting the second proleg with 10 μl of an aqueous suspension containing 5 × 106 spores per ml. Insects were maintained at 25°C and >90% relative humidity for 48 h and subsequently at 60 to 70% relative humidity. Each treatment had three replicates with 10 insects each, and the experiments were repeated twice. Mortality was recorded every 12 h. The median lethal time was calculated with cumulative mortality data. Additional insects were injected and bled 48 h later to observe the extent of hyphal body differentiation within the insect hemocoel.
Nucleotide sequence accession number.
The Mos1 mRNA sequence has been submitted to GenBank under accession number EU106866.
RESULTS
Protein structure and phylogenetic analysis.
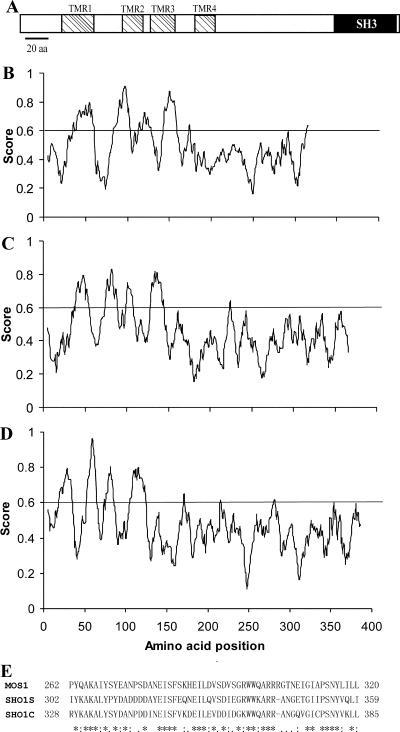
Analyses of the predicted MOS1 protein indicates that it is composed of 320 amino acid residues (34.6 kDa) with a predicted pI of 6.08. MOS1 shows homologies to S. cerevisiae osmosensor SHO1 (accession no. P40073; 34.1% identity) and C. albicans SHO1 (CAC81238; 31.5% identity). Consistent with the structures of S. cerevisiae and C. albicans osmosensors (11, 22), MOS1 does not have a signal peptide for secretion but contains four transmembrane regions and a C-terminal Src homology 3 (SH3) domain (262 to 317 residues) (Fig. 2A). Despite protein size differences, the hydrophobicity profiles characteristic of transmembrane regions were similar in MOS1 and yeast osmosensors (Fig. 2B to D), with the most highly conserved region being within the SH3 domain (Fig. 2E).
FIG. 2.
Protein characteristics. (A) Schematic structure of MOS1 protein. (B to D) Hydropathicity analysis of MOS1 (B), Saccharomyces cerevisiae osmosensor SHO1 (C), and Candida albicans osmosensor (D), scaled by the algorithm of Kyte and Doolittle. TMR, transmembrane region. (E) Alignment of the conserved SH3 domain between MOS1 and the S. cerevisiae (SHO1S) and C. albicans (SHO1C) osmosensors. *, consensus residues.
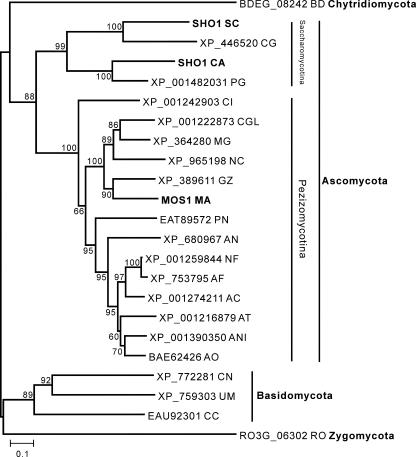
Blastp searches against available fungal genomes show that homologs of MOS1 are widely present in ascomycetes (MOS1 is most similar to that of Gibberella zeae [accession no. XP_389611; E = 1 × 10−105]), basidomycetes (Cryptococcus neoformans, Ustilago maydis, and Coprinopsis cinerea), zygomycetes (Rhizopus oryzae), and a chytrid (Batrachochytrium dendrobatidis). Interestingly, Blastp or tBlastn searches did not hit anything (E value set at the default of 1 × 10−3) in the latest genome release of the ascomycete Botrytis cinerea. A homolog is also absent in the microsporidian Encephalitozoon cuniculi, a species often used to root fungal phylogeny trees (7). A phylogenetic tree was constructed based on available sequences, and this was highly supported by bootstrap values (Fig. 3). The divergence of MOS1 and its homologs is congruent with current models of fungal speciation (7). Thus, clusters contain species of chytrids, ascomycetes (with two subgroups of Saccharomycotina and Pezizomycotina), basidomycetes, and zygomycetes (Fig. 3).
FIG. 3.
Phylogenetic relationship of MOS1 with fungal homologs. Fungal species: BD, Batrachochytrium dendrobatidis; SC, Saccharomyces cerevisiae; CG, Candida glabrata; CA, Candida albicans; PG, Pichia guilliermondii; CI, Coccidioides immitis; CGL, Chaetomium globosum; MG, Magnaporthe grisea; NC, Neurospora crassa; GZ, Gibberella zeae; MA, Metarhizium anisopliae; PN, Phaeosphaeria nodorum; AN, Aspergillus nidulans; NF, Neosartorya fischeri; AC, Aspergillus clavatus; AF, Aspergillus fumigatus; AT, Aspergillus terreus; ANI, Aspergillus niger; AO, Aspergillus oryzae; CN, Cryptococcus neoformans var. neoformans; UM, Ustilago maydis; CC, Coprinopsis cinerea; RO, Rhizopus oryzae.
Gene expression and RNA interference.
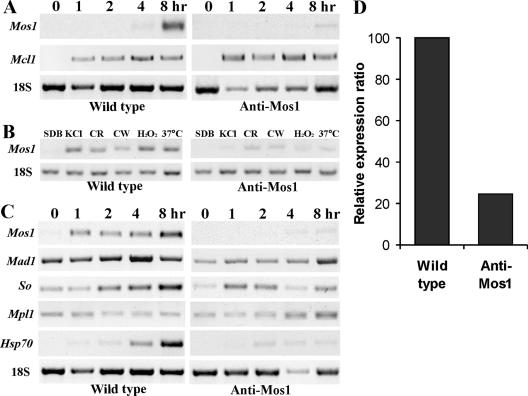
During growth of the wild type in SDB, Mos1 transcripts were detectable by RT-PCR only at the 1-μg/μl RNA level (data not shown). Expression was sharply up-regulated after 4 hours in Manduca hemolymph or 1 hour in SDB amended with 0.7 M KCl, with the highest transcript levels at the final time point tested (8 h) (Fig. 4A to C). In addition to transcriptional induction by osmotic stress, the wild type also showed up-regulation of Mos1 when subjected to heat (37°C) or oxidative stress (H2O2), or when challenged with compounds that interfere with cell wall synthesis (calcofluor white or Congo red) (Fig. 4B). The Mos1 antisense transformant was depleted in Mos1 transcripts under all stress conditions (Fig. 4B) and transcript levels were reduced >4-fold after 8 h in insect hemolymph or SDB plus KCl medium (Fig. 4D).
FIG. 4.
Gene expressions and Mos1 RNA interference. (A) RT-PCR analysis of Mos1 gene expression by wild-type and antisense-Mos1 strains in Manduca sexta hemolymph. (B) Mos1 gene expression by mycelia incubated in SDB amended with 0.7 M KCl, 500 μg/ml Congo red (CR), 50 μg/ml calcofluor white (CW), or 40 mM H2O2 at 25°C or in SDB at 37°C. (C). RT-PCR analysis of differential gene expression by mycelium in SDB amended with 0.7 M KCl. Mycelial inoculum from 36-h SDB cultures was transferred into insect hemolymph or SDB plus KCl medium for 0, 1, 2, 4, and 8 h. (D) Relative expression of Mos1 by wild type (calibrated as 100%) and antisense-Mos1 strains. The genes for Mcl1, Mad1, So, Mpl1, Hsp70, and 18S are described in Materials and Methods.
We also analyzed the effects of depleting Mos1 on expression of other Metarhizium genes known to be involved with immune or stress responses. Knocking down Mos1 transcripts did not affect transcription of the hemolymph-specific gene Mcl1 (Fig. 4A) but altered expression of the Mad1, So, Mpl1, and Hsp70 genes compared to the wild type (Fig. 4C). The Mad1 gene in the antisense-Mos1 transformant was down-regulated >2-fold between 0 and 4 h in the SDB plus KCl medium, and transcript levels reached wild-type levels only at 8 h postinoculation. The up-regulation of So and Hsp70 that occurred in the wild type did not occur in the transformant. Conversely, after 4 h of incubation in SDB plus KCl, Mpl1, encoding a protein that covers lipid droplets, was down-regulated ∼2-fold by the wild type but up-regulated >2-fold by the antisense-Mos1 transformant (Fig. 4C).
Biological characterization.
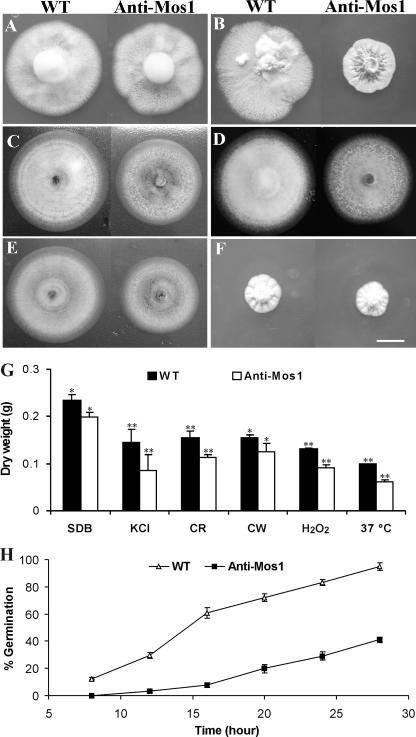
Growth of the wild type (4.72 ± 0.10 cm in colony diameter) and the antisense-Mos1 transformant (4.67 ± 0.08 cm in diameter) was not significantly different (t = 6.31; P = 0.25) at 9 days postinoculation on PDA (Fig. 5A). Both the wild-type and antisense-Mos1 strains demonstrated similar low growth rates (ca. 3.5 cm in diameter after 30 days) on PDA amended with 0.7 M KCl, but inhibition of antisense-Mos1 transformants was significantly greater than that of the wild type with 1.5 M KCl (t = 5.15; P = 7.08 × 10−3) (Fig. 5B). In addition, as described for Candida Sho1 deletion mutants (11), growth of antisense-Mos1 transformants was significantly repressed on PDA medium amended with calcofluor white at 25 μg/ml (t = 16.56; P = 2.67 × 10−5) (Fig. 5C) or 50 μg/ml (t = 14.31; P = 2.96 × 10−5) and by Congo red at 250 μg/ml (t = 8.41; P = 5.45 × 10−4) (Fig. 5D) or 500 μg/ml (t = 6.21; P = 0.025). Growth of antisense-Mos1 transformants was also repressed by addition of 20 mM H2O2 (t = 16.56; P = 3.89 × 10−5) (Fig. 5E) or 40 mM H2O2 (t = 6.31; P = 3.96 × 10−4) to induce oxidative stress. Both the wild type and antisense-Mos1 strains grew very slowly during 9 days at 37°C, but the difference between them was not significant (P = 6.31; t = 0.25) (Fig. 5F). Liquid biomass assays demonstrated similar effects as plate assays for growth repression by different compounds or stressful growth conditions (Fig. 5G). However, using dry weight measurements, we detected significant differences (α < 0.05) in growth in unamended SDB between the wild type and the antisense-Mos1 transformant.
FIG. 5.
Culture growth and spore germination assays. (A and B) Growth of wild-type (WT) and antisense-Mos1 strains after 9 days on PDA (A) or after 30 days on PDA amended with 1.5 M KCl (B). (C to E) Growth after 9 days at 25°C in the presence of 25 μg/ml calcofluor white (C), 250 μg/ml Congo red (D), or 20 mM H2O2 (E). (F) Growth after 9 days on PDA at 37°C. Bar, 1 cm. (G) Liquid biomass assays in SDB or SDB amended with 1.5 M KCl, 250 μg/ml Congo red (CR), 25 μg/ml calcofluor white (CW), or 20 mM H2O 2 after incubation for 3 days at either 25°C or 37°C. *, significant difference at an α value of <0.05; **, significant difference at an α value of <0.01. (H) Percentages of spore germination by the wild-type and antisense-Mos1 strains in SDB amended with 0.7 M KCl for different times as indicated. Error bars indicate standard deviations.
Spore germination rates of antisense-Mos1 spores were significantly (>2-fold) lower than those of wild-type spores both in SDB and in SDB amended with KCl (Fig. 5H). Spores germinating on locust wings demonstrated phenotypic differences between the wild-type and antisense-Mos1 strains. More than 90% of wild-type and antisense-Mos1 conidia germinated on the cuticle, but the wild type grew significantly longer germ tubes (14.2 ± 1.8 μm; t = 8.2, P = 0.0002) than did antisense-Mos1 spores (9.5 ± 1.1 μm) before they differentiated appressoria (Fig. 6A and B). Consistent with our previous observations (19), the wild type produced bar-shaped single-celled hyphal bodies during growth in hemolymph (Fig. 6C). The antisense-Mos1 transformants produced a high percentage (72.3% ± 5.8%) of multicellular and branched hyphal chains (Fig. 6D), as previously observed with adhesin gene Mad1 null mutants (21). Similar results were obtained from three independent antisense-Mos1 transformants (data not shown).
FIG. 6.
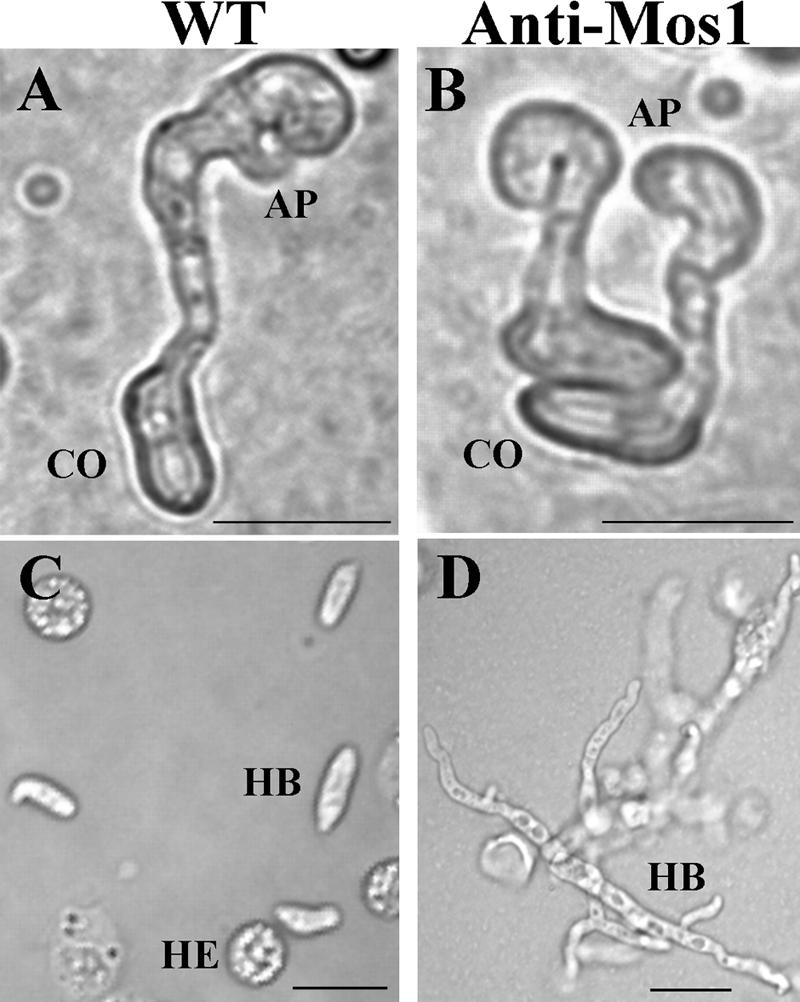
Cell differentiation. (A) Appressorium differentiation on locust hind wing by wild-type (WT) and antisense-Mos1 (B) strains at 18 h postinoculation. (C) Hyphal body differentiation by WT and (D) antisense-Mos1 strains in Manduca hemocoel at 48 h postinjection. CO, conidia; AP, appressorium; HB, hyphal body; HE, hemocytes of insect. Bar, 10 μm.
Mos1 affects fungal virulence.
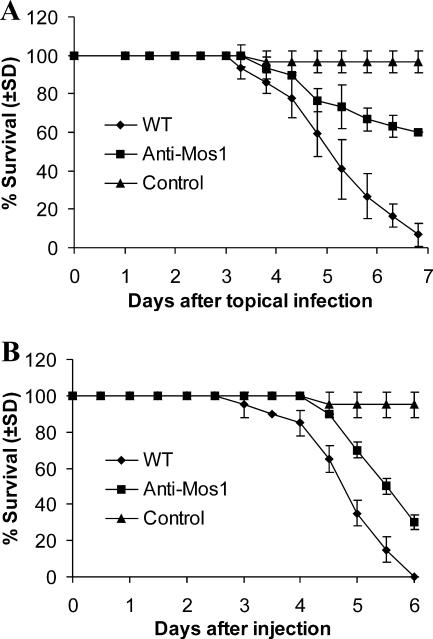
Virulence tests using fifth instar larvae of M. sexta revealed a significant reduction in mortality and speed of kill by the antisense-Mos1 transformants compared to the wild type. Following topical infection, mortality rates for the antisense-Mos1 transformant failed to reach 50% before insects pupated (Fig. 7A). Antisense-Mos1 transformants caused 70% mortality during injection assays, but the median time (5.8 ± 0.6 days) was significantly longer (t = 4.0; P = 0.03) than that for the wild type (4.6 ± 0.3 days) (Fig. 7B).
FIG. 7.
Survival of Manduca sexta larvae after infection with wild-type or antisense-Mos1 conidia. (A) Survival of Manduca larvae following topical application with suspensions of 1 × 107 conidia/ml of wild-type or antisense-Mos1 strains. Control insects were dipped in water. (B) Survival of Manduca larvae following injection into the second proleg with 10 μl of suspensions of 5 × 106 conidia/ml. Control insects were injected with 10 μl water. Error bars indicate standard deviations.
DISCUSSION
Adaptation is essential for fungi to survive under nonoptimal osmotic conditions, and studies on yeasts have identified several of the elements and pathways involved (14, 15). In this study, we characterized an M. anisopliae gene (encoding MOS1) that contains the transmembrane regions and C-terminal SH3 domain of the yeast SHO1 osmosensors. Phylogenetic analysis indicates that MOS1 homologs are widely distributed among fungi and show an evolutionary history that follows fungal speciation.
Expression of Mos1 is up-regulated in insect hemolymph or in artificial media with high osmolarity. Presumably, low-level expression in the absence of hyperosmotic stress is to provide sufficient MOS1 for surveillance of osmotic conditions. Up-regulation may provide increased sensitivity and signaling capacity in hyperosmotic growth conditions. In yeast, HOG1 inhibits SHO1 as part of a negative feedback loop, leading to diminished cellular responses to an osmotic-stress stimulus (6). Up-regulation of Mos1 could function to bypass this system during prolonged osmotic stress. Gene silencing by RNA interference is being increasingly used for fungal functional genomic studies (11). To explore the functions of Mos1, we used an expression vector that generates antisense RNA to the transcript encoding MOS1. Compared to those of the wild type, the spores of the antisense Mos1 transformants showed greater delays in germination with increasing concentrations of KCl, demonstrating a reduced ability to respond to osmotic stress. The ability of the transformant to still grow, albeit very slowly, on media with high osmolarity could be due to residual Mos1 mRNA or to the alternative osmotic signaling branches known to exist in yeast (12, 17).
Similar to the case for C. albicans Sho1 null mutants (12), Mos1 knockdown transformants were more sensitive to oxidative stress and perturbation of cell wall biosynthesis than the wild type. The mechanisms by which osmosensors interact with fungal cell wall development and oxidative stress responses are not clear. In yeast, SHO1 acts as a scaffold protein in the MAP kinase pathway during adaptation to high OP (12, 24). This could explain the pleiotropic effects produced by repressing Mos1, as fungal MAP kinase pathways are involved in a wide range of biological functions, including development, oxidative stress responses, and virulence (1, 8, 13). Knockdown of Mos1 did not affect expression of the immune coat protein gene Mcl1, which is specifically expressed in hemolymph (20). Mcl1 may therefore use inducers that are more precisely diagnostic of hemolymph than OP. However, knockdown of Mos1 represses expression of a subset of genes that are coordinately up-regulated during growth in hemolymph as well as some other media. OP may therefore act as a pathogenicity-related signal for M. anisopliae. The down-regulated genes included Mad1, an adhesin involved in spore germination and hyphal body differentiation (21). This could at least in part explain the delay in germination and the formation of multicellular branched hyphal bodies by antisense-Mos1 transformants that mimic the effects of disrupting Mad1. Yeast C4-methyl sterol oxidase (encoded by So) is an important component in biosynthesis of ergosterol, the compound responsible for cell membrane permeability and stiffness (2). The homolog of Metarhizium So was among the most common transcripts during growth in insect hemolymph (18). Up-regulation of So by the wild type implies that membrane permeability decreases and stiffness increases under osmotic stress, providing a barrier to solutes and cellular deformation. Conversely, down-regulation of So in antisense-Mos1 transformants suggests a diminished ability to control membrane permeability and stiffness. The perilipin homolog MPL1 covers lipid droplets, masking them from enzymolysis. Levels of MPL1 control turgor pressure and appressorium differentiation by regulating breakdown of triacylglycerols and consequent production of the osmoticant glycerol (22). Mpl1 is down-regulated by the wild type at high OP, which should allow an accumulation of glycerol in the cell, increasing intracellular OP. Conversely, Mpl1 was up-regulated after Mos1 knockdown, indicating a diminished ability to increase intracellular OP in response to osmotic stress. Heat shock proteins are known to chaperone biological responses under nonoptimal conditions, including those provided by the “shocks” of heat, cold, nutrient deprivation, and oxidative stress, etc. The knockdown mutants showed a dramatic down-regulation of Hsp70 indicative of a general decrease in osmotic and oxidative stress responses.
Bioassay data indicate that proper Mos1 functioning is important for virulence against M. sexta. Repressing Mos1 causes dysfunctional appressorium differentiation on the cuticle surface. However, the Mos1 knockdown was also less virulent if the cuticle was bypassed by injection, demonstrating the importance of adaptive responses to hemolymph OP in maintaining virulence. Further studies will be required to better understand how MOS1 interacts with other proteins/pathways during adaptation to osmotic stress and regulation of OP-mediated developmental and behavioral functions.
Acknowledgments
This work was supported by the National Hi-Tech Research and Development Program of China (grant 2006AA10A212), by the Science and Technology Commission of Shanghai Municipality (grant 07PJ14101), and by an NSF grant (MCB-0542904).
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Aguirre, J., W. Hansberg, and R. Navarro. 2006. Fungal responses to reactive oxygen species. Med. Mycol. 44101-107. [DOI] [PubMed] [Google Scholar]
- 2.Bard, M., D. A. Bruner, C. A. Pierson, N. D. Lees, B. Biermann, L. Frye, C. Koegel, and R. Barbuch. 1996. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA 93186-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman, R. F. 1998. The insects: structure and function. Cambridge University Press, Cambridge, United Kingdom.
- 4.Charnley, A. K. 2003. Fungal pathogens of insects: cuticle degrading enzymes and toxins. Adv. Bot. Res. 40241-321. [Google Scholar]
- 5.Freimoser, F. M., G. Hu, and R. J. St. Leger. 2005. Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiology 151361-371. [DOI] [PubMed] [Google Scholar]
- 6.Hao, N., M. Behar, S. C. Parnell, M. P. Torres, C. H. Borchers, T. C. Elston, and H. G. Dohlman. 2007. A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr. Biol. 17659-667. [DOI] [PubMed] [Google Scholar]
- 7.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G. H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schussler, J. E. Longcore, K. O'Donnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443818-822. [DOI] [PubMed] [Google Scholar]
- 8.Klosterman, S. J., M. H. Perlin, M. Garcia-Pedrajas, S. F. Covert, and S. E. Gold. 2007. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv. Genet. 571-47. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 10.Monge, R. A., E. Roman, C. Nombela, and J. Pla. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152905-912. [DOI] [PubMed] [Google Scholar]
- 11.Nakayashiki, H. 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 5795950-5957. [DOI] [PubMed] [Google Scholar]
- 12.Roman, E., C. Nombela, and J. Pla. 2005. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 2510611-10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roman, E., D. M. Arana, C. Nombela, R. Alonso-Monge, and J. Pla. 2007. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15181-190. [DOI] [PubMed] [Google Scholar]
- 14.Saito, H., and K. Tatebayashi. 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. (Tokyo) 136267-272. [DOI] [PubMed] [Google Scholar]
- 15.Seet, B. T., and T. Pawson. 2004. MAPK signaling: Sho business. Curr. Biol. 14R708-R710. [DOI] [PubMed] [Google Scholar]
- 16.St. Leger, R. J., L. Joshi, M. J. Bidochka, and D. W. Roberts. 1996. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 936349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatebayashi, K., K. Tanaka, H. Y. Yang, K. Yamamoto, Y. Matsushita, T. Tomida, M. Imai, and H. Saito. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 263521-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, C., G. Hu, and R. J. St. Leger. 2005. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet. Biol. 42704-718. [DOI] [PubMed] [Google Scholar]
- 19.Wang, C., and R. J. St. Leger. 2005. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot. Cell 4937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, C., and R. J. St. Leger. 2006. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc. Natl. Acad. Sci. USA 1036647-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, C., and R. J. St. Leger. 2007. The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot. Cell 6808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, C., and R. J. St. Leger. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 28221110-21115. [DOI] [PubMed] [Google Scholar]
- 23.Westfall, P. J., and J. Thorner. 2006. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot. Cell 51215-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarrinpar, A., R. P. Bhattacharyya, M. P. Nittler, and W. A. Lim. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 14825-832. [DOI] [PubMed] [Google Scholar]