Abstract
The nature of the cytoplasmic pathway of starch biosynthesis was investigated in the model glaucophyte Cyanophora paradoxa. The storage polysaccharide granules are shown to be composed of both amylose and amylopectin fractions, with a chain length distribution and crystalline organization similar to those of green algae and land plant starch. A preliminary characterization of the starch pathway demonstrates that Cyanophora paradoxa contains several UDP-glucose-utilizing soluble starch synthase activities related to those of the Rhodophyceae. In addition, Cyanophora paradoxa synthesizes amylose with a granule-bound starch synthase displaying a preference for UDP-glucose. A debranching enzyme of isoamylase specificity and multiple starch phosphorylases also are evidenced in the model glaucophyte. The picture emerging from our biochemical and molecular characterizations consists of the presence of a UDP-glucose-based pathway similar to that recently proposed for the red algae, the cryptophytes, and the alveolates. The correlative presence of isoamylase and starch among photosynthetic eukaryotes is discussed.
Glaucophyta are single-cell freshwater algae carrying peptidoglycan-containing plastids, called cyanelles, that strongly resemble their cyanobacterial ancestors. Together with the Rhodophyceae (red algae) and the Chloroplastida (green algae and land plants), they have been demonstrated to originate from a single endosymbiosis that gave birth to the Archaeplastida (including the plant kingdom) (1). This endosymbiosis involved a cyanobiont related to present-day cyanobacteria and a unicellular biflagellated heterotrophic eukaryotic host (43). In addition to acquiring photosynthesis, all three lines gained the ability to synthesize starch, a novel form of storage polysaccharide related to glycogen, the otherwise most widespread form of storage polysaccharide found in living cells.
Starch is composed of chains organized with the same chemical linkages as those of glycogen but defines a far more elaborate organization (for a review of starch structure, see reference 7). It consists of several distinct polysaccharide fractions. Amylose is composed of mostly linear α1,4-linked glucan chains. It is synthesized through the granule-bound starch synthase (GBSS), an elongation enzyme bound to and active within the starch granule (reviewed in reference 4). The latter is mostly composed of amylopectin, a moderately (5 to 6%) branched polymer that has an asymmetrical distribution of branches that allows the formation of chain clusters hooked together through a number of longer spacer chains. This organization enables the polymers to partially crystallize in the form of huge insoluble semicrystalline granules that come in various highly specific shapes and sizes within the plant kingdom. Unlike GBSS, the enzymes of amylopectin synthesis are active mostly within the soluble phase (reviewed in reference 5). The pathways of storage polysaccharide synthesis have been intensively studied in animals, fungi, and bacteria (for reviews, see references 20, 37, and 42). The bacterial and eukaryotic pathways of glycogen synthesis can be distinguished by a number of very important features. These include, but are not limited to, the nature of the glycosyl nucleotide used for polymerization, UDP-glucose or ADP-glucose in heterotrophic eukaryotes and bacteria, respectively.
Starch metabolism has been intensively studied in green plants and algae (reviewed in references 5, 23, 28, 46, and 54). While 6 to 12 genes generally are found in heterotrophic eukaryotes or bacteria for glycogen metabolism, green algae and land plants synthesize starch from ADP-glucose only (27, 53), with the help of 30 to 40 genes that are phylogenetically related to both cyanobacterial and heterotrophic eukaryote sequences (10, 36).
Red algae and glaucophytes can be distinguished from the green algae and land plants by the fact that they synthesize starch in the cytoplasm and not in their plastids.
In red algae and their secondary endosymbiotic starch-storing derivatives, such as cryptophytes and alveolates, both biochemical and bioinformatic data suggest the presence of a UDP-glucose-based pathway (34, 50). However, this pathway remains to be validated by genetic approaches.
In this paper we report the first detailed characterization of glaucophyte starch structure and metabolism. Supported by a combination of biochemical and molecular approaches, we propose that the model glaucophyte Cyanophora paradoxa synthesizes starch through a UDP-glucose pathway similar to those that recently were proposed for rhodophyceae, cryptophytes, and alveolates (dinoflagellates and apicomplexan parasites). The presence of a functional isoamylase complex in glaucophytes further strengthens the correlation established between this activity and the synthesis of starch in photosynthetic eukaryotes.
MATERIALS AND METHODS
Materials.
ADP-[U-14C]glucose, UDP-[U-14C]glucose, and [α-32P]dCTP were purchased from Amersham (Burkinghamshire, United Kingdom). ADP-glucose and UDP-glucose were obtained from Sigma. Sepharose CL-2B columns and Percoll were obtained from Amersham Pharmacia Biotech. The starch amyloglucosidase assay kit was obtained from Roche (Germany).
Cyanophora strains, growth, and media.
Cyanophora paradoxa strain 29.80 M was purchased at the EPSAG (University of Gottingen, Germany). The alga was grown in DY-V medium (R. A. Anderson and M. D. Keller, unpublished data) at 25°C under a 12-h day/12-h night cycle (30 μmol photons·m−1·s−1) and vigorous shaking.
Determination of starch levels, starch purification, and amylopectin structures.
A full account of amyloglucosidase assays, starch purification, and λmax (maximum wavelength of the iodine polysaccharide complex) measurements can be found in Delrue et al. (16). The separation of starch polysaccharides by gel permeation chromatography (GPC) on a Sepharose CL-2B column was performed as described in Wattebled et al. (51). The percentages of amylose and amylopectin were determined by pooling the large-mass amylopectin excluded from the column and the lower-mass amylose separately. The pooled amylopectin and amylose fractions each were assayed with the help of the amyloglucosidase assay. The chain length distributions of glaucophyte and chloroplastida amylopectins were obtained by debranching them with isoamylase and separating the chains through high-performance anion exchange with pulsed amperometric detection, as detailed in Fontaine et al. (19).
Crude extract preparation and compartmentalization studies.
Cells (3 liters at 2 × 106 cells ml−1) were harvested by centrifugation (4,000 × g for 15 min). The pellet was resuspended in 10 ml of 0.25 M Tris-HCl, pH 8.0, and disrupted by sonication. The lysate was centrifuged for 10 min at 10,000 × g. This supernatant was taken as the crude extract. For compartmentalization studies, a similar cell pellet was lysed by an osmotic shock procedure. This procedure consisted of resuspending the pellet in 2 ml of 1 M Tris-HCl, pH 8.0, for 4 min and subsequently adding 8 ml of 0.05 M Tris-HCl, pH 8.0, to this suspension for another 6 min. After centrifugation at 750 × g for 5 min, the supernatant was spun at 10,000 × g for 10 min. This defined our cytosolic extract. A second osmotic shock was performed on the pellet centrifuged at 750 × g as described above to lyse the few remaining cells. This cyanelle pellet then was resuspended in 10 ml of 0.25 M Tris-HCl, pH 8.0, and subjected to sonication as described above.
Enzyme purification and assays.
GBSS was assayed as described previously (16). In vitro synthesis of amylose was performed by using a previously detailed method (49).
Starch synthase and phosphorylase purifications.
The crude cytosolic extract for starch synthase and phosphorylase purifications was prepared as described above. The supernatant (17 ml) was loaded on an anion-exchange chromatography (AEC) column (MonoQ HR 5/5; 1-ml column volume) preequilibrated in buffer A (25 mM Tris-acetate, pH 7.5, 10 mM dithiothreitol [DTT]). The proteins were eluted at 1 ml·min−1 with a 50-ml linear gradient of 0 to 0.5 M NaCl in buffer A containing 1 M NaCl. Starch synthase and phosphorylase activities were monitored by zymogram analyses. Briefly, proteins from each fraction (0.5 ml) were separated by nondenaturing polyacrylamide gel electrophoresis (PAGE) containing 0.15% of rabbit glycogen (Sigma). After electrophoresis, gels were incubated overnight at room temperature either in starch synthase buffer [25 mM HEPES, pH 7.5, 30 mM (NH4)2SO4, 20 mM MgCl2, 5 mM β-mercaptoethanol, 1.2 mM UDP-glucose] or phosphorylase buffer (10 mM glucose-1-phosphate [G-1-P], 25 mM Tris-acetate, 10 mM DTT). The activities then were visualized as dark bands after the gels were stained with iodine solution. The fraction containing the starch synthase and phosphorylase activities were pooled and concentrated at 0.1 ml by using centrifugal filter devices (Amicon-ultra; Millipore). Starch synthase and phosphorylase activities were further purified by gel filtration chromatography (Superose 6 prepacked column; 1 cm by 30 cm; Amersham Biosciences). The samples (0.1 ml) were loaded onto a Superose 6 column preequilibrated in buffer A with a flow rate of 0.3 ml·min−1 using a fast-protein liquid chromatography system. Starch synthase and phosphorylase activities were monitored by zymogram analyses. The fractions (free of hydrolytic activities) were pooled and used for enzymatic characterization.
The starch synthase activities were measured in the synthesis direction. Standard assays were performed for 15 min at 35°C with 20 μl of enzyme preparation and 80 μl of a mixture containing HEPES-KOH (50 mM; pH 7.5), (NH4)2SO4 (0.1 M), β-mercaptoethanol (5 mM), MgCl2 (5 mM), bovine serum albumin (BSA) (0.5 mg·ml−1), glycogen or amylopectin (10 mg·ml−1), UDP-Glc (2 mM), and UDP-[U-14C]Glc (2 μM). At the end of the incubation, glycogen or amylopectin was precipitated with 1 ml methanol-KCl (45), incubated at −20°C for 10 min, and collected by centrifugation (3,000 × g, 5 min, 4°C). The pellet was resuspended in 0.2 ml distilled water; the precipitation, centrifugation, and resuspension procedure was repeated twice more before the pellets were resuspended in 0.2 ml distilled water and mixed with 2.5 ml scintillation liquid. The radioactivity incorporated was determined by liquid scintillation counting.
The phosphorylase activity was measured in the degradation direction. The phosphorolysis of polysaccharides was assayed in a final volume of 300 μl by incubating purified phosphorylase (0.4 mg) in 25 mM Tris-acetate, pH 7.5, 10 mM Pi (orthophosphate), and 10 mg·ml−1 of rabbit liver glycogen or amylopectin for 1 h at 30°C. The reaction was stopped by being boiled for 5 min at 95°C. The production of G-1-P was assayed by adding 300 μl of 50 mM Tris-HCl, 120 mM MgCl2, 0.05 mM glucose-1,6-diphosphate, and 0.5 mM NADP buffer. The production of NADPH,H+ was monitored spectrophotometrically at 365 nm after the addition of 4 U of phosphoglucomutase (Sigma Chemical Co., St. Louis, MO) and 2 U of glucose-6-phosphate dehydrogenase (Sigma). The phosphorylase activity is expressed as nanomoles of G-1-P produced per minute per milligram of protein. Apparent Km values were determined by using GraphPad Prism 5 software.
The starch hydrolase activities were purified by loading 42 ml of supernatant on an AEC column (HiTrapQ FF [5 ml]) preequilibrated in buffer A (25 mM Tris-acetate, pH 7.5, 10 mM DTT). The proteins were eluted at 5 ml·min−1 with a 15-ml linear gradient of 0 to 0.5 M NaCl in buffer A containing 1 M NaCl. Starch metabolism enzymes were monitored by zymogram analyses. Proteins from each fraction (2.5 ml) were separated by nondenaturing PAGE. After electrophoresis, proteins were transferred overnight by electroblotting to a polyacrylamide gel containing 0.3% (wt/vol) polysaccharide (9). Starch synthase activities were monitored after being electrotransferred (2 h, 30 V, 300 mA) to glycogen-containing gels. The gels were incubated overnight at room temperature in the incubation buffer described above containing either 1.2 mM ADP-Glc or 1.2 mM UDP-Glc. The electroblots were stained with iodine solution. Because blue-staining isoamylase-like activities migrated very slowly in this zymogram system, in a fashion similar to that of isoamylase in green algae and plants, we chose to purify this enzyme by GPC. Isoamylase was purified by gel filtration chromatography (Superose 6 prepacked column; 1 cm by 30 cm; Amersham Biosciences). The isoamylase-containing fractions (2 ml by 2.5 ml) that were free of all other starch hydrolases were pooled and concentrated to 0.5 ml by using centrifugal filter devices (Amicon-ultra; Millipore). The sample (0.1 ml) was loaded onto a Superose 6 column preequilibrated in buffer A with a flow rate of 0.3 ml·min−1 using a fast-protein liquid chromatography system. The isoamylase activity was monitored according to the procedure of Miller (30) based on the chemical interaction between the 3,5 dinitrosalicyclic acid (DNS) and sugar reducing ends. For each fraction (0.6 ml), 50 μl of isoamylase activity was mixed with 50 μl of 1% (wt/vol) potato amylopectin dispersed in buffer A. The reaction was stopped by adding 100 μl of DNS. The mixtures were heated for 10 min at 100°C. The optical density was measured at 570 nm. The glucose was used as a standard for the reducing end value. The isoamylase activity is expressed as the equivalent of 1 μg of glucose reducing end produced min−1 mg−1 of protein. The isoamylase-containing fractions were checked for the absence of other starch hydrolytic enzymes by zymogram analysis. The fractions containing the purified isoamylase were pooled and used to perform further characterizations. Debranching enzyme activity toward amylopectin, glycogen, beta-limit dextrin (BLD), and pullulan was monitored by measuring the production of reducing ends over a time course. Fifty microliters of isoamylase activity (20 μg of reducing ends produced/min) was incubated at 37°C in 950 μl of 1% (wt/vol) polysaccharide dispersed in buffer A. The production of reducing ends was monitored over a time course of 0 to 3 h. Aliquots of 100 μl were removed after each half hour and mixed with 100 μl of DNS as described above. The total quantity of reducing ends released in the same experimental conditions for each polysaccharide was determined using commercial isomylase (Pseudomonas amylodermosa isoamylase). The percentage of debranched polysaccharide was calculated by the formula (microgram of glucose equivalent of reducing ends measured/microgram of glucose equivalent of reducing ends released after complete debranching of the polysaccharide) × 100, thereby reducing all errors due to the different reactions of glucose and MOS with respect to DNS. The relative debranching activity toward AP and BLD was determined with respect to Gly, which was chosen arbitrarily as the reference value (set as 1) after taking into consideration the differences in the percentage of branching points for each polysaccharide for amylopectin (4.17%), BLD (9.25%), and Gly (7.12%) obtained from Manners (29).
NMR analysis.
Fifty microliters of the purified isoamylase activity (20 μg of reducing end produced min−1) was mixed with a 950-μl solution containing 5 mg of rabbit liver glycogen dispersed in 25 mM Tris-acetate, pH 7, 10 mM DTT overnight at 37°C. The reaction was boiled for 5 min at 99°C. Proteins were removed by loading them onto a Dowex 50:2 (1 cm by 6 cm) column immediately coupled to a Dowex 1:2 (1 cm by 6 cm) column equilibrated with water as described previously. The lyophilized samples then were subjected to proton nuclear magnetic resonance (NMR) as described previously (8, 31). The spectra were calibrated on δ-CH3 of dimethyl sulfoxide (2.6 parts/million).
Apparent Km determination of GBSSI.
The protocols used for the apparent Km determination of granule-bound starch synthase I (GBSSI) were described by Ral et al. (39).
cDNA screening and cloning for starch synthase, starch phosphorylase, and GBSS of Cyanophora paradoxa.
The Cyanophora paradoxa λ ZAP cDNA library was provided by W. Löffelhardt. The cDNA probes were designed using expressed sequence tag (EST) sequences obtained from the TbestDB project web site (http://tbestdb.bcm.umontreal.ca/searches/login.php). The protocol used for cDNA library screening is described in Jakowitsch et al. (21) with the following modifications. DNA probe labeling and detection were performed with the DIG luminescent detection kit (Roche Molecular Biochemicals) using the protocol provided by the manufacturer.
Nucleotide sequence accession numbers.
Sequences determined in the course of this work have been deposited in GenBank under accession numbers EU165054 to EU165056.
RESULTS
Cyanophora paradoxa accumulates semicrystalline high-amylose storage polysaccharide granules that are similar to plant starch.
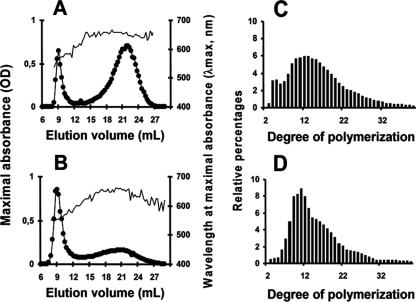
Starch was purified from late-log-phase Cyanophora paradoxa cells by sedimentation and density gradient purification. The dispersed polysaccharide was subjected to the separation of polysaccharide fractions through GPC. Results displayed in Fig. 1A demonstrate that this material contained both a high- and low-mass polysaccharide fraction similar to amylopectin and amylose, respectively, from the unicellular green alga Chlamydomonas reinhardtii (Fig. 1B). The amylose/amylopectin ratio was calculated by harvesting their respective fractions after GPC and measuring the two pooled fractions with the amyloglucosidase assay. The amylose content of the glaucophyte was systematically high to very high, varying from 30% of the total polysaccharide in nitrogen-supplied medium in alternating day and night cultures to 60% in continuous light. At variance with the results obtained with green algae, nitrogen starvation had little impact on the relative amylose content. Chlamydomonas reinhardtii wild-type strain 137C accumulates from 2% to at most 10% amylose when supplied with nitrogen and from 10 to 30% under nitrogen starvation. Amylopectin from Cyanophora paradoxa was collected after GPC and subjected to debranching through the action of an isoamylase. The chain length distribution of the debranched chains then was examined by high-performance anion exchange with pulsed amperometric detection. Results displayed in Fig. 1C show that Cyanophora paradoxa contains a starch-type bimodal distribution of glucans that is selectively enriched in very short chains compared to those of Chlamydomonas reinhardtii (Fig. 1D). Starch granules diffract and give two types of X-ray diffraction patterns corresponding to two different assembly geometries that have been named A type and B type, respectively (7). The starch granule typically is considered semicrystalline. Indeed, only sections of the amylopectin molecule assemble into crystals. Amylose usually is thought to be mostly amorphous. Nevertheless, a high amylose content has a tendency to yield a B type of diffraction pattern for the remaining amylopectin. The Cyanophora paradoxa starch granules thus were further analyzed using X-ray diffraction. The samples used for the analysis produced clear B-type diffractograms with a crystallinity percentage around 30% (Fig. 2A). The morphology of the starch granules was further investigated by scanning electron microscopy (Fig. 2B). Classical starch granules were evidenced with occasional cavities or morphology alterations that could be due either to in vivo polysaccharide degradation or to the growth of granules in close contact with other cellular structures. However, when present, the cavities did not match the size and frequency recently observed in the cryptophyte floridean starch granules (18). The size of the Cyanophora granules varied between 0.5 and 2 to 3 μm in diameter, figures comparable to those of the starch found in green algae and plant leaves.
FIG. 1.
Starch structure analysis by size-exclusion chromatography and chain length distribution analysis of amylopectin. Constituent fractions of starch granules were separated using GPC (see Materials and Methods). Amylopectin is a massive molecule that is excluded from the gel; linear amylose is eluted later. Glucans eluted in each fraction were detected by their interactions with iodine. (A and B) The x axis represents the column elution volume corresponding to each fraction. The left y axis measures the maximum absorbance (•) of the iodine-polysaccharide complex, while the right y axis represents the wavelength at which this maximum absorbance is measured (λmax; thin unbroken line). (C and D) Branched glucans of the amylopectin fraction purified from the column then are analyzed by isoamylase-mediated debranching and the determination of the chain length distribution. Chain lengths (degrees of polymerization) are indicated on the x axis. The percentage of each class of glucans is presented on the y axis. Results of two exclusion chromatography and chain length distributions are presented for Cyanophora starch (A and C) and Chlamydomonas reinhardtii starch (B and D), respectively. OD, optical density.
FIG. 2.
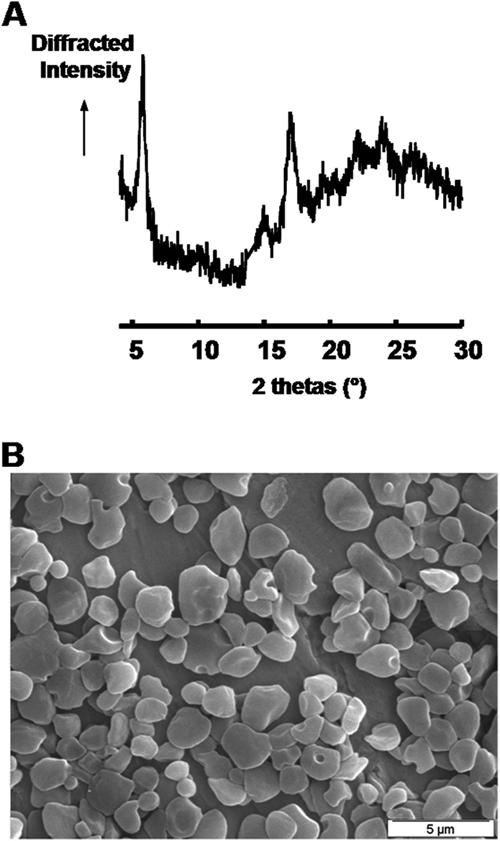
X-ray analysis and scanning electron microscopy of the starch granules of Cyanophora paradoxa. (A) Diffraction peaks at 20 values (Bragg angles) of 9.9°, 11.2°, 15°, 17°, 18.1°, and 23.3° characterize the A-type starches, whereas diffraction peaks at 26 values of 5.6°, 15°, 17°, 22°, and 24° typify B-type starches. Thus, this wide-angle X-ray scattering diagram is characteristic of a pure B-type starch with a degree of crystallinity of 31% ± 3%. (B) Electron micrographs of purified starches from Cyanophora paradoxa (scale bar, 5 μm).
Based on the results of all of these studies, we conclude that Cyanophora paradoxa accumulates a typical semicrystalline high-amylose starch.
Glaucophyte granule-bound starch synthase activity can use both ADP-glucose and UDP-glucose.
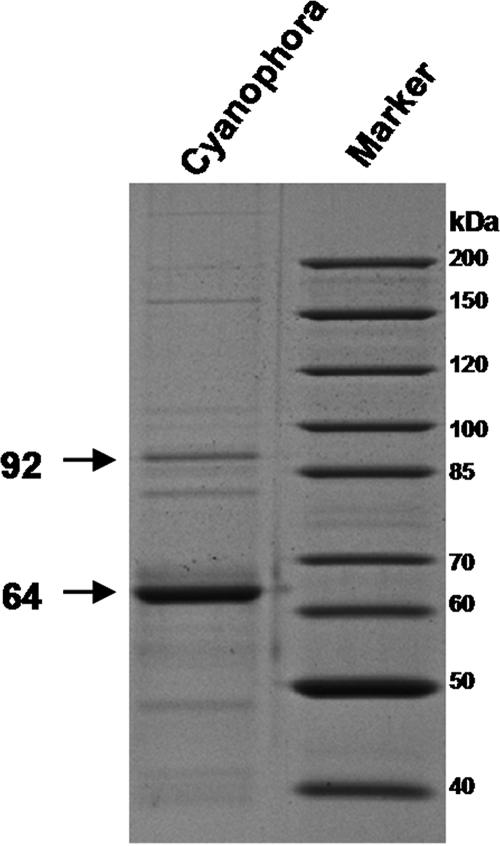
Amylose generally is associated with the presence of granule-bound starch synthase, the enzyme that, in plants, is responsible for the processive synthesis of such long glucans (for a review, see reference 4). We thus measured the granule-associated starch synthase activity from freshly purified starch granules. The Cyanophora paradoxa GBSS was able to use both ADP-glucose and UDP-glucose (apparent Km values of 8 mM ADP-Glc and 15 mM UDP-Glc) but displayed a sixfold higher Vmax with UDP-glucose (0.23 ± 0.04 nmol ADP-Glc incorporated/min/mg of starch and 1.48 ± 0.5 nmol UDP-Glc incorporated/min/mg of starch). We further characterized the structure of the polysaccharide produced with both ADP-glucose and UDP-glucose. Results displayed in Fig. 3 demonstrate that the behavior of the starch-bound activity of the two glycosyl nucleotides was identical and was analogous to that evidenced for cryptophyte (18), green alga (49), and plant (17) GBSSI. Indeed, GBSSI is known in the absence of added malto-oligosaccharides to polymerize glucose residues to the outer chains of amylopectin. These chains subsequently are cleaved off amylopectin to generate mature amylose (Fig. 3A and B). On the other hand, in the presence of malto-oligosaccharides that act as competitive primers with amylopectin, the enzyme is observed to directly synthesize amylose-like molecules, and very little incorporation occurs in the amylopectin (Fig. 3C and D). In addition to this typical GBSS type of activity, we found a 64-kDa major granule-associated protein (Fig. 4) that displayed significant homology to plant GBSSI sequences. The protein was hydrolyzed with trypsin, and peptides were sequenced. This enabled us to correlate the protein with the full GBSSI-like sequence that we have cloned from our Cyanophora paradoxa cDNA bank (GenBank accession no. EU165054).
FIG. 3.
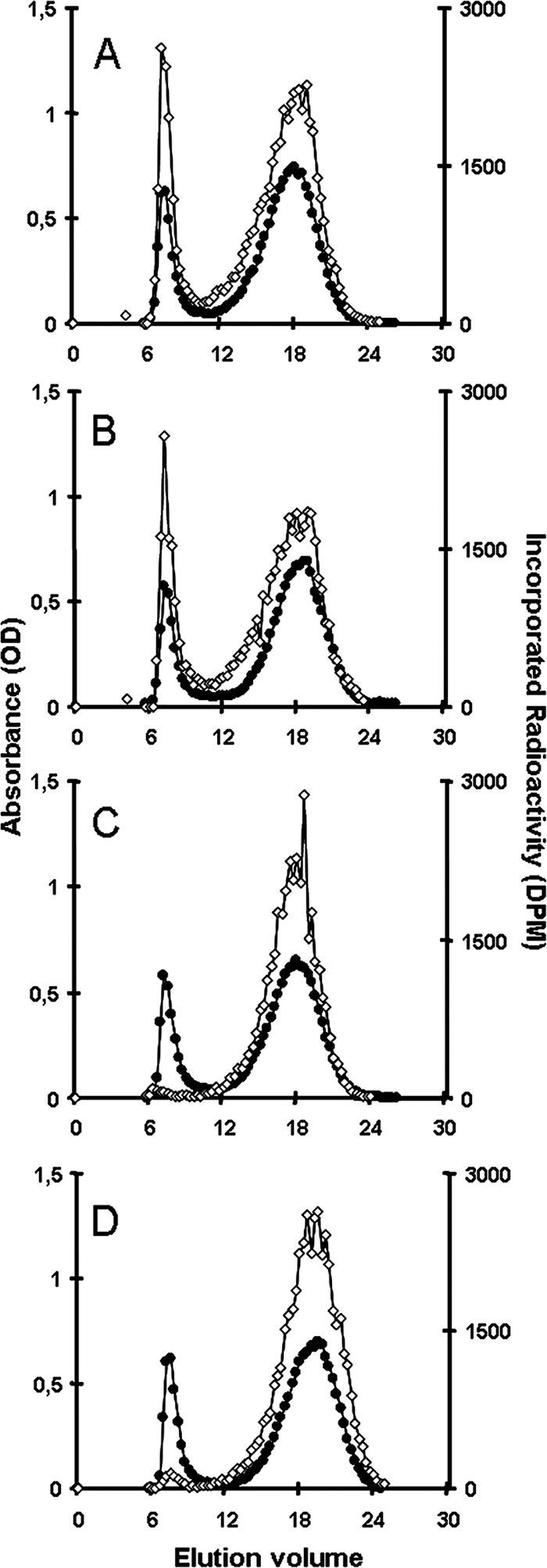
Fractionation of starch granules isolated from Cyanophora and subjected to in vitro synthesis of amylose. After incubation with radiolabeled ADP-glucose (A) or UDP-glucose (B), starch granules were subjected to CL-2B size-exclusion chromatography in order to detect the fractions in which radiolabeled glucose residues were incorporated. The isotopic dilution for ADP-glucose was three times lower than that for UDP-glucose. The same experiment with ADP-glucose (C) or UDP-glucose (D) was done with the addition of 50 mM maltotriose. The x axis indicates the elution volume, the left axis shows the iodine-polysaccharide complex absorbance of each fraction eluted from the column (•), and the right axis shows the radioactivity of each fraction (⋄). Without the addition of maltotriose (DP3), radioactive material was incorporated in both the excluded amylopectin fraction and the amylose fraction. The addition of DP3 restricts incorporation in the amylose fraction only. This behavior is a specific property of GBSS-like starch synthases. OD, optical density.
FIG. 4.
Visualization of the proteins associated with Cyanophora paradoxa starch granules. Granule-bound proteins were extracted by boiling them in sodium dodecyl sulfate (SDS) and were separated by SDS-PAGE (18). A molecular mass marker also is displayed. The major band at around 64 kDa was hydrolyzed with trypsin, and the resulting peptides were eluted from the gel and identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) and mass spectrometry-mass spectrometry (MS-MS) sequencing. The peptides YDQYFDAWDTSVR, VTFLLHNLLYQGR, DLPVNALATR, VFYETKGKDR, and SVPTTPLLAFVGR correspond to a GBSS and were found in the sequence isolated by cDNA screening. The band at around 92 kDa was hydrolyzed with trypsin, and the resulting peptides were eluted from the gel and identified by MALDI-TOF and MS-MS sequencing. The peptide MQAVQQRY corresponds to a starch phosphorylase and was found in recent Cyanophora paradoxa EST resources (GenBank accession no. EC661027) (35).
Cyanophora paradoxa contains a major GT5 family UDP-glucose-utilizing soluble starch synthase.
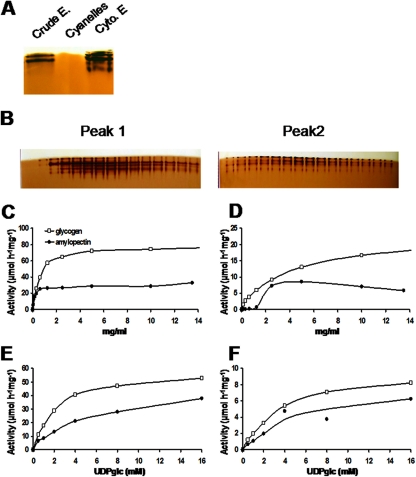
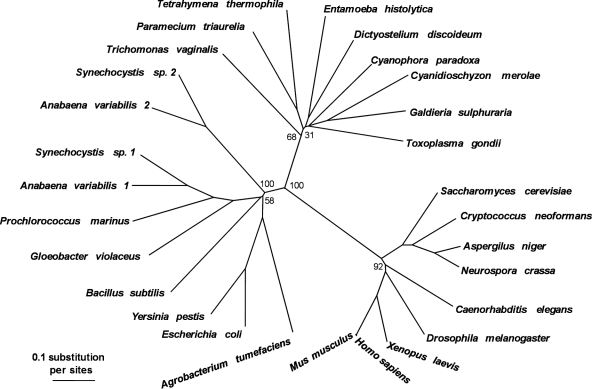
Because the kinetic characterization of GBSSI did not support a very dramatic preference for UDP-glucose over ADP-glucose, we investigated the properties of the soluble starch synthases. Our glycogen-primed zymogram gels demonstrated the presence of several UDP-glucose-utilizing soluble starch synthase activity bands with close mobility. These activities were localized in the cytoplasmic compartment (Fig. 5A). No ADP-glucose-utilizing elongation enzymes were evidenced by these procedures (results not shown). We chose to partly purify the enzyme activities and investigate their kinetic properties. Results displayed in Fig. 5B demonstrate that two types of soluble starch synthase activities could be purified free of contaminating starch hydrolases by AEC followed by GPC. Peaks 1 and 2 amounted to 87 and 13%, respectively, of the total assayable starch synthase activity, with an optimal temperature of 35 and 25°C, respectively, and an optimal pH of 7.5 and 8, respectively (data not shown). These activities display a marked preference for UDP-glucose and cannot use ADP-glucose at significant rates (<2% of the activity measured in the presence of UDP-glucose) (data not shown). Interestingly, the second activity (peak 2) displayed clear sigmoidal kinetics in the presence of increasing concentrations of amylopectin (Fig. 5D), while peak 1 showed a classical Michaelis-Menten type of kinetics in the presence of glycogen or amylopectin, as did peak 2 in the presence of glycogen (Fig. 5C and D). The behavior of peak 2 recalls that of the starch synthase that recently was purified from Gracilaria tenuistipitata (34). For peak 1, increasing concentrations of UDP-glucose in the presence of either glycogen or amylopectin enabled us to measure apparent Km values of 2.2 ± 0.2 mM in the presence of 10 mg·ml−1 glycogen and 5.2 ± 0.9 mM in the presence of 10 mg·ml−1 amylopectin (Fig. 5E). Peak 2 displayed Km values of 4 ± 0.3 mM in the presence of 10 mg·ml−1 glycogen and 4.6 ± 2.6 mM in the presence of 10 mg·ml−1 amylopectin (Fig. 5F). We then searched for the presence of ESTs encoding soluble starch synthase and found a small sequence, which was used to select out longer cDNAs. We obtained an incomplete 1,669-bp cDNA containing most of the core sequence corresponding to soluble starch synthases of the GT5 family according to the CAZY classification, but it still lacked the variable N-terminal extension evidenced both in rhodophyceae and chloroplastida (GenBank accession no. EU165055). Phylogenetic trees built with this core sequence clearly support the grouping of this cDNA sequence with those evidenced in rhodophyceae and alveolates (Fig. 6).
FIG. 5.
Characterization of starch synthase activities. (A) One hundred micrograms of total protein from crude extract (Crude E.), purified cyanelles, and cytosolic extracts (Cyto. E) was loaded on a native PAGE containing 0.15% glycogen. After migration, the gel was incubated with 1.2 mM UDP-glucose. Starch synthase activities were visualized as brownish bands after iodine staining only in the crude extract and the cytosolic extract. (B) Cytosolic proteins were further fractionated on an AEC column (MonoQ HR5/5) with a gradient of 1 M NaCl. Zymogram analyses performed on each fraction (10 μl) revealed the presence of two peaks of starch synthase activities. Fractions 15 to 29 (peak 1), which were very clearly separated from fractions 36 to 50 (peak 2), were subjected to an additional purification step by gel filtration chromatography in order to determine kinetic parameters (see Materials and Methods). Twenty microliters of purified starch synthases from peak 1 (specific activity, 61 μmol·h−1·mg−1 of protein) (C and E) and peak 2 (specific activity, 10 μmol·h−1·mg−1 of protein) (D and F) were incubated with 0, 0.08, 0.15, 0.31, 0.62, 1.25, 2.5, 5, 10, and 13.5 mg·ml−1 of amylopectin and glycogen in the presence of 16 mM UDP-Glc and 2 μM UDP-[U-14C]Glc (C and D) or were incubated with 0, 0.5, 1, 2, 4, 8, or 16 mM UDP-Glc and 2 μM UDP-[U-14C]Glc in the presence of 10 mg·ml−1 of amylopectin or glycogen (E and F). [U-14C]Glc incorporated onto polysaccharides was measured after the precipitation and washing steps. The results are expressed as micromoles of glucose incorporated per hour per milligram of protein. Purification factors were 41.5 and 8.1 for peaks 1 and 2, respectively.
FIG. 6.
Neighbor-joining distance tree inferred for glycogen synthases and starch synthases. Amino acid sequences were aligned using ClustalW (47), and alignment gaps were manually removed. Neighbor-joining trees were calculated for 10,000 bootstrap replicates using Neighbor (Phylip package; http://evolution.genetics.washington.edu/phylip.html). Trees were edited using Treeview (Roderic D. M. Page, University of Glasgow, Scotland, United Kingdom). Numbers at nodes represent bootstrap values. A more detailed unrooted maximum-likelihood tree can be found elsewhere (Deschamps et al., submitted).
Cyanophora paradoxa contains an isoamylase type of debranching enzyme and multiple starch phosphorylases.
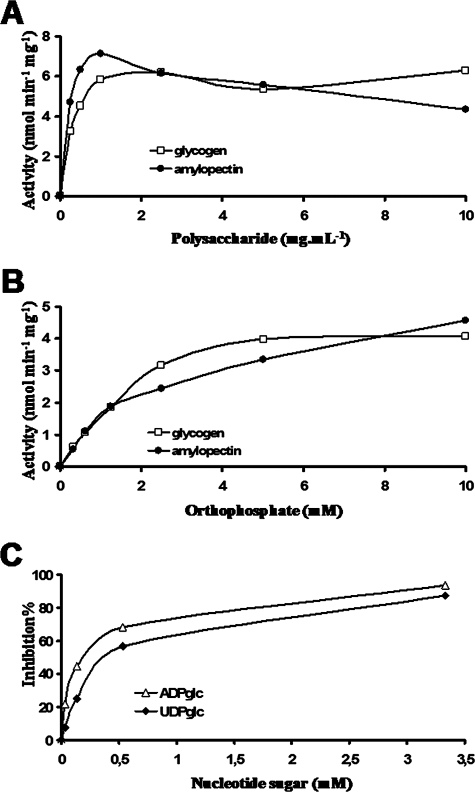
Starch phosphorylase zymograms display activity bands that barely entered the polysaccharide-containing gels. We tracked this phosphorylase activity during the semipurification and assayed the enzyme(s) in semipure fractions lacking starch synthase or hydrolase activities. The kinetics displayed in Fig. 7A demonstrate the presence of classical starch phosphorylase activities with high affinity toward both glycogen (apparent Km, 0.5 ± 0.08 mg·ml−1) and amylopectin (apparent Km, 0.65 ± 0.05 mg·ml−1). However, the enzyme clearly was inhibited by high amounts of amylopectin substrate. Figure 7B displays a similar experiment performed with increasing concentrations of orthophosphate in the presence of either 10 mg·ml−1 glycogen (apparent Km, 2 ± 0.8 mM Pi) or 10 mg·ml−1 amylopectin (apparent Km, 3 ± 0.7 mM Pi). Interestingly, as displayed in Fig. 7C, this activity was inhibited by both UDP-glucose (Ki, 0.35 ± 0.1 mM) and ADP-glucose (Ki, 0.1 ± 0.05 mM) in a fashion similar to that evidenced in green algae (2, 14, 32). As was described for other phosphorylases, the inhibition was competitive with respect to orthophosphate. The finding of a short EST that possibly could encode starch phosphorylase gave us a probe to select out the full-length cDNA and therefore enabled us to get the full enzyme sequence (GenBank accession no. EU165056). Another EST recently was selected from the growing genomic resources (GenBank accession no. EC661027) (35). This EST displayed a clearly distinct sequence, suggesting the presence of several different phosphorylase subunits or enzymes. The mass of several peptides generated by the analysis of a 92-kDa starch granule-associated protein (Fig. 4) matched that predicted by the EST encoding this second phosphorylase.
FIG. 7.
Characterization of phosphorylase activity. Phosphorylase was assayed in the degradation direction by using 0, 0.25, 0.5, 1, 2.5, 5, and 10 mg·ml−1 of glycogen or amylopectin in the presence of 10 mM orthophosphate (A) and by using 0, 0.312, 0.625, 1.25, 2.5, 5, and 10 mM orthophosphate in the presence of 10 mg·ml−1 of amylopectin or glycogen (B). The amount of G-1-P produced was determined by monitoring the production of NADPH,H+ at 365 nm after the addition of phosphoglucomutase and glucose-6-phosphate dehydrogenase. The phosphorylase activity is expressed as nanomoles of G-1-P produced per minute per milligram of protein. The apparent Km values for polysaccharides were determined at 0.5 ± 0.08 mg·ml−1 and 0.65 ± 0.05 mg·ml−1 for glycogen and amylopectin, respectively. The apparent Km values for Pi were determined at 2 ± 0.8 mM and 3 ± 0.7 mM in the presence of 10 mg·ml−1 of glycogen and amylopectin, respectively. (C) The inhibition of phosphorylase activity regarding nucleotide sugars was analyzed using 0, 0.033, 0.153, 0.533, and 3.3 mM of either UDP-Glc or ADP-Glc in the presence of 10 mg·ml−1 glycogen at 2 mM Pi. Both sugar nucleotides inhibit the phosphorylase activity (Ki for UDP-Glu, 0.35 ± 0.1 mM; Ki for ADP-Glu, 0.1 ± 0.05 mM).
We then probed for the existence of starch hydrolases and found the usual set of zymogram bands on starch-containing gels from crude extracts (Fig. 8A) and after semipurification (Fig. 8B). In addition, we looked for the presence of these enzymes within cyanelles and found no traces of such activities. Finally, among the starch hydrolases, we probed for the existence of isoamylase, a starch debranching enzyme, through standard zymogram procedures and enzyme purification. Mutant work performed with green algae and vascular plants strongly suggested that isoamylase is involved in trimming the misplaced chains in a hydrophilic precursor of amylopectin to generate an insoluble semicrystalline form of the polysaccharide (3). This suggestion now is supported by a wealth of molecular and biochemical data obtained solely from green plants and algae (12, 13, 15, 22, 31, 33, 38, 52).
FIG. 8.
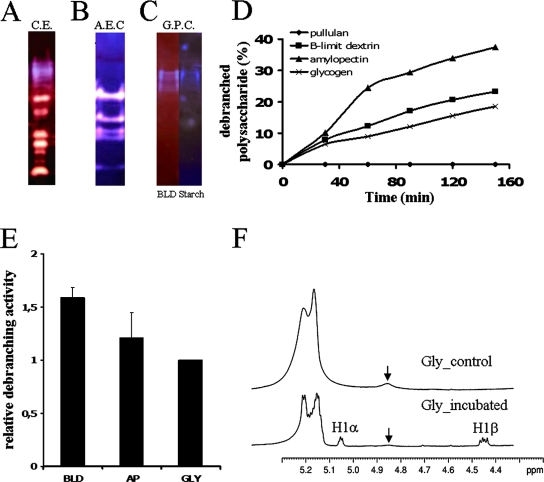
Characterization of the debranching enzyme activity in Cyanophora paradoxa. Starch-metabolizing enzymes from crude extracts (A) were fractionated by AEC (B) and by gel filtration (C). The activities from crude extracts, from fraction F16 of the anion exchange, and from fraction F10 of the gel filtration were separated by nondenaturing PAGE and visualized after their transfer to a BLD gel (A), a starch gel (B), or both a BLD and starch gel (C). After iodine staining, starch-containing and BLD-containing gels create a blue (A to C) and a purple (C) background, respectively. According to the specificity of the starch-metabolizing activities, the substrate in the gel becomes more branched (red-staining bands), completely hydrolyzed (white band), or completely debranched (blue band). The size of the blue bands was estimated to be around 390 kDa by GPC. (C) The debranching activity of isoamylase from fraction F10 of the gel filtration was incubated with amylopectin (▴), BLDs (▪), GLY (×), and pullulan (⧫). The activity was measured by monitoring the increase in the amount of reducing ends. The results are expressed as the percentage of debranched polysaccharides. (D) The relative debranching activity toward AP, BLD, and PUL was determined with respect to GLY (E), which was arbitrarily chosen as the reference (set as 1), after taking into consideration the differences in the percentages of branching points for each polysaccharide. (F) NMR analysis of rabbit liver glycogen polysaccharide incubated with or without isoamylase activity. 1H spectra were made in 80% (vol/vol) dimethylsulfoxide-20% (vol/vol) D2O at 70°C, as described in Mouille et al. (31).
We searched for the presence of isoamylase by tracking a slow-migrating set of blue-stained bands evidenced on amylopectin- or starch-containing zymogram gels after iodine staining of the incubated gels. A blue-stained band is indicative of a debranching-type hydrolase digesting the polysaccharide trapped within the gels. The blue stain is due to the remaining slowly diffusing longer debranched oligosaccharides. Because this is only an indication of the presence of a debranching enzyme, we subjected crude extracts (Fig. 8A) to purification by AEC (Fig. 8B) followed by GPC until the fraction suspected to contain debranching enzyme was devoid of all other types of starch hydrolases (amylases, branching enzyme, and D-enzyme) (Fig. 8C). The substrate specificity of the suspected debranching enzyme was measured with enzyme assays involving the production of reducing ends (Fig. 8D and E). Definitive proof that the purified fraction contained only debranching enzyme activity finally was obtained by proton NMR analysis (Fig. 8F). The signals at 5.3 to 5.1 ppm and 4.85 ppm correspond to the proton resonances from the carbon atoms engaged in an α-1,4 linkage and an α-1,6 linkage, respectively. The NMR analysis performed at 70°C instead of 90°C yields a two- to threefold underestimation of the number of branching points. After incubation with isoamylase activity, the proton resonances from the carbon atoms of the reducing end appear as two signals, at 5.05 and 4.45 ppm, corresponding to α (1Hα) and β (1Hβ) anomers, respectively. This is correlated with a dramatic decrease of the signal at 4.85 ppm, corresponding to the 1H from the carbon atoms engaged in an α-1,6 linkage. The results clearly indicated the presence in Cyanophora paradoxa of a high-mass (390 kDa) isoamylase complex (Fig. 8C), as measured by GPC on a Superose 6 column. The Cyanophora enzyme was able to debranch glycogen, amylopectin, and amylopectin β-limit dextrin with comparable efficiencies (Fig. 8D and E). It was, however, unable to debranch pullulan at significant rates.
DISCUSSION
Our characterization of Cyanophora paradoxa soluble starch synthases clearly shows that these activities are able to elongate glucans by the use of a UDP-glucose substrate. However, these enzymes are unable to use ADP-glucose at significant rates.
The sole existence of a UDP-glucose-based starch metabolism pathway was further ascertained by assaying for the presence of ADP-glucose pyrophosphorylase activity and protein (results not shown). We found no evidence for an enzyme activity responding to 3-phosphoglyceric acid activation in crude extracts. In addition, an antibody directed against the maize small subunit yielded a clear cross-reaction with the enzyme found in Chlamydomonas. This cross-reaction disappears in sta6 mutants of Chlamydomonas reinhardtii that lack the corresponding small subunit. We were unable to find any comparable cross-reaction with Cyanophora extracts, thereby suggesting the absence of the corresponding protein or the presence of modified enzyme despite the conservation of cross-reactions throughout the green lineage.
We cloned a sequence encoding a protein that bears strong resemblance to the soluble starch synthase sequences identified in cyanidiale red algae and in aveolates, including the glycogen-storing ciliates and the amylopectin-storing apicomplexans. Interestingly, alveolates also are currently thought to derive from red algae by a secondary endosymbiosis event (for a general review, see reference 6). The substrate specificity, which is the same as that described for the glycogen synthase of heterotrophic eukaryotes, argues in favor of a eukaryotic (host) origin of this enzyme gene. However, the phylogenetic tree produced in this work clearly distinguishes the glaucophyte, rhodophyceae, and alveolate enzymes from those of fungi and animals. Indeed, according to the CAZY classification, the latter belong to a family (family GT3) of glycosyltransferases distinct from that that includes bacterial glycogen synthases and green plant starch synthases (family GT5). The rhodophyceae and glaucophyte transferases described here both are GT5-type enzymes and therefore bear more resemblance to the bacterial glycogen synthases or the green plant starch synthases than to the fungal or animal enzyme. Interestingly, both Dictyostelium discoideum and Entamoeba histolytica and the very distantly related parabasalid Trichomonas vaginalis display a similar type of enzyme, arguing that these genes are of eukaryotic (host) origin (P. Deschamps, C. Colleoni, D. Moreira, Y. Nakamura, E. Suzuki, J. L. Putaux, A. Buléon, S. Haebel, G. Ritte, M. Steup, L. I. Falcón, W. Loffelhardt, J. Nirmal Raj, C. Plancke, C. d'Hulst, D. Dauvillée, and S. Ball, submitted for publication). Several distinct soluble starch synthases were evidenced by our purification studies. It is presently unknown if the activities reflect alternative regulated states of the same enzyme or if they are encoded by distinct proteins.
Another important finding reported in this work consists in the characterization of a glaucophyte GBSS responsible for amylose synthesis. This enzyme could use both ADP-glucose and UDP-glucose, but the specific activity measured with UDP-glucose was sixfold higher. The ambiguous selectivity of GBSSI toward ADP-glucose and UDP-glucose is a common trait for the GBSSI genes currently reported. Indeed, the maize enzyme initially was detected through its activity toward UDP-glucose (25), while ADP-glucose was recognized only later as the preferred substrate (40). Similarly, we have recently characterized a GBSSI from the cryptophyte Guillardia theta that showed only a slight preference for UDP-glucose (18). The reason for the low selectivity of the bound enzyme compared to that of the soluble starch synthases is discussed elsewhere (P. Deschamps et al., submitted).
Kinetic characterization of both soluble and granule-bound enzymes showed that both activities display comparable affinities toward their UDP-glucose substrate. This distinguishes glaucophytes from their chloroplastida relatives. In green algae and land plants, the affinity of the soluble enzyme for ADP-glucose is at least 4-fold to more than 10-fold higher than that of the bound activity (4). When the supply of ADP-glucose is limited, amylopectin synthesis is relatively favored over that of amylose. This explains why mutants partly defective for ADP-glucose synthesis display a significant relative enrichment of amylopectin over that of amylose (48). In Chlamydomonas, cultures supplied with normal levels of nutrients contain little amylose (<5%), while nutrient-starved cultures accumulate between 10 to 25% of this starch fraction (26). In Cyanophora paradoxa, the relative amount of amylose always remains at a very high level (between 30 to 60%), which is only rarely reached in some plant mutants. Moreover, this high level is not significantly affected by the supply of nutrients. These levels are three- to sixfold higher than those reported for green algae and plants. We believe this to be due solely to the comparable affinities of the granule-bound and soluble starch synthase toward UDP-glucose. Indeed, the amount of protein within the granule and the enzyme-specific activity are significantly inferior to those we measure for the Chlamydomonas enzyme and therefore cannot explain the relatively high amylose content.
The finding we report for GBSSI in glaucophytes echoes the description that was recently made for rhodophyceae (44). GBSSI thus is found in all three lineages that presently define the archaeplastida and very likely was present in the ancestor of all plants. This has important implications with respect to our understanding of the evolution of storage polysaccharide metabolism. It has been demonstrated previously that GBSSI requires the presence of a preformed semicrystalline starch granule to be normally active (11). This in turn suggests that the ancestor of all plants synthesized such a structure, probably in its cytoplasm. An ancient cytoplasmic location for the synthesis of starch is suggested by the finding of a similar compartmentalization of storage polysaccharide in rhodophyceae and glaucophytes. Although some nuclear gene phylogenies have not been able to resolve the divergence order of the three archaeplastida lineages, both more recent nuclear gene phylogenies as well as plastid phylogenies indicate that glaucophytes diverged first from the common ancestor (41, 43). In addition, a recent study concerning the emergence of the light-harvesting complex (LHC) proteins can be explained only with a similar divergence order (24). The hypothesis of a cytoplasmic location of starch synthesis in the common ancestor of all plants thus is more parsimonious in this respect.
It must be stressed that the phylogeny of all sequences obtained either in this work or by bioinformatics analysis of the growing Cyanophora paradoxa (Deschamps et al., submitted) genomic resources are in complete agreement with the monophyletic origin of plants. Of special relevance are the phosphorylase enzyme sequences, which, as in red and green algae, display a common eukaryotic (host) origin.
All positive findings reported in this work support the existence of a UDP-glucose-based pathway of starch synthesis in glaucophytes that is very similar to that that has been proposed for rhodophyceae. We cannot exclude, however, the existence of minor forms of soluble starch synthases able to use ADP-glucose that would be phylogenetically related to the chloroplastidial or cyanobacterial enzymes, especially when faced with the exquisite sensitivity of the Cyanophora paradoxa phosphorylase to inhibition by ADP-glucose. A definitive negative conclusion in this respect can be reached only when the full genome sequence of Cyanophora paradoxa is available. Finally, we report the finding of a multimeric isoamylase complex within the cytoplasm of Cyanophora paradoxa. Isoamylase has been suggested to be involved in the maturation and aggregation of amylopectin into semicrystalline granules (3). In Chlamydomonas, the organization of isoamylase into large heteromultimeric complexes per se has been demonstrated to be required for the function of this enzyme in amylopectin synthesis (12, 13). Our report of a large-mass isoamylase in the cytoplasm of Cyanophora paradoxa defines the first report of such an enzyme in a nongreen eukaryote. The presence of this activity, the genes of which are clearly of cyanobacterial phylogeny, correlates with the presence of starch in Cyanophora paradoxa. While the activity was identified through biochemical means only, the existence of genes in rhodophyceae that bear strong homologies to plant isoamylases strengthens the correlation between the presence of starch and that of isoamylase. In turn, this correlation agrees with the critical function suggested for isoamylase in the biogenesis and aggregation of the starch granule (3).
Acknowledgments
This research was funded by the French Ministry of Education, the Centre National de la Recherche Scientifique (CNRS), l'Institut National de la Recherche Agronomique, by the Region Nord Pas de Calais, the European Union, and by the Deutsche Forschungsgemeinschaft (SFB 429 TP B2 [M.S.] and SFB 429 TP B7 [G.R.]).
We thank Curtis Hannah for providing us with the antibody directed against the small subunit of ADP-glucose pyrophosphorylase. We thank Emmanuel Maes for performing the NMR analysis.
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. Mc-Court, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52399-451. [DOI] [PubMed] [Google Scholar]
- 2.Ball, S., T. Marianne, L. Dirick, M. Fresnoy, B. Delrue, and A. Decq. 1991. A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 18517-26. [DOI] [PubMed] [Google Scholar]
- 3.Ball, S., H. P. Guan, M. James, A. Myers, P. Keeling, G. Mouille, A. Buléon, P. Colonna, and J. Preiss. 1996. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell 86349-352. [DOI] [PubMed] [Google Scholar]
- 4.Ball, S. G., M. H. B. J. van de Wal, and R. G. F. Visser. 1998. Progress in understanding the biosynthesis of amylose. Trends Plant Sci. 3462-467. [Google Scholar]
- 5.Ball, S. G., and M. K. Morell. 2003. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54207-233. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, D., H. S. Yoon, and J. D. Hackett. 2004. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 2650-60. [DOI] [PubMed] [Google Scholar]
- 7.Buléon, A., P. Colonna, V. Planchot, and S. Ball. 1998. Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 2385-112. [DOI] [PubMed] [Google Scholar]
- 8.Colleoni, C., D. Dauvillée, G. Mouille, M. Morell, M. Samuel, M. C. Slomiany, L. Liénard, F. Wattebled, C. D'Hulst, and S. Ball. 1999. Biochemical characterization of the Chlamydomonas reinhardtii alpha-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 1201005-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colleoni, C., A. M. Myers, and M. G. James. 2003. One- and two-dimensional native PAGE activity gel analyses of maize endosperm proteins reveal functional interaction between specific starch metabolizing enzymes. J. Appl. Glycosci. 50207-212. [Google Scholar]
- 10.Coppin, A., J. S. Varre, L. Lienard, D. Dauvillée, Y. Guerardel, M. O. Soyer-Gobillard, A. Buléon, S. Ball, and S. Tomavo. 2005. Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J. Mol. Evol. 60257-267. [DOI] [PubMed] [Google Scholar]
- 11.Dauvillée, D., C. Colleoni, E. Shaw, G. Mouille, C. D'Hulst, M. Morell, M. S. Samuel, B. Bouchet, D. J. Gallant, A. Sinskey, and S. Ball. 1999. Novel, starch-like polysaccharides are synthesized by an unbound form of granule-bound starch synthase in glycogen-accumulating mutants of Chlamydomonas reinhardtii. Plant Physiol. 119321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauvillée, D., C. Colleoni, G. Mouille, A. Buléon, D. J. Gallant, B. Bouchet, M. K. Morell, C. D'Hulst, A. M. Myers, and S. Ball. 2001. Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 1251710-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauvillée, D., C. Colleoni, G. Mouille, M. K. Morell, C. D'Hulst, F. Wattebled, L. Liénard, D. Delvallé, J. P. Ral, A. M. Myers, and S. Ball. 2001. Biochemical characterization of wild type and mutant isoamylases of Chlamydomonas reinhardtii supports a function of the multimeric enzyme organization in amylopectin maturation. Plant Physiol. 1251723-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauvillée, D., V. Chochois, M. Steup, S. Haebel, N. Eckermann, G. Ritte, J. P. Ral, C. Colleoni, G. Hicks, F. Wattebled, P. Deschamps, C. d'Hulst, L. Lienard, L. Cournac, J. L. Putaux, D. Dupeyre, and S. Ball. 2006. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48274-285. [DOI] [PubMed] [Google Scholar]
- 15.Dauvillée, D., V. Mestre, C. Colleoni, M. C. Slomianny, G. Mouille, B. Delrue, C. d'Hulst, C. Bliard, J. M. Nuzillard, and S. Ball. 2000. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Sci. 157145-156. [DOI] [PubMed] [Google Scholar]
- 16.Delrue, B., T. Fontaine, F. Routier, A. Decq, J. M. Wieruszeski, N. Van den Koornhuyse, M. L. Maddelein, B. Fournet, and S. Ball. 1992. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J. Bacteriol. 1743612-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denyer, K., B. Clarke, C. Hylton, H. Tatge, and A. Smith. 1996. The elongation of amylose and amylopectin chains in isolated starch granules. Plant J. 101135-1143. [Google Scholar]
- 18.Deschamps, P., I. Haferkamp, D. Dauvillée, S. Haebel, M. Steup, A. Buléon, J. L. Putaux, C. Colleoni, C. d'Hulst, C. Plancke, S. Gould, U. Maier, H. E. Neuhaus, and S. Ball. 2006. Nature of the periplastidial pathway of starch synthesis in the cryptophyte Guillardia theta. Eukaryot. Cell 5954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontaine, T., C. D'Hulst, M. L. Maddelein, F. Routier, T. M. Pepin, A. Decq, J. M. Wieruszeski, B. Delrue, N. Van den Koornhuyse, J. P. Bossu, B. Fournet, and S. Ball. 1993. Toward an understanding of the biogenesis of the starch granule. Evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J. Biol. Chem. 26816223-16330. [PubMed] [Google Scholar]
- 20.François, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25125-145. [DOI] [PubMed] [Google Scholar]
- 21.Jakowitsch, J., M. G. Bayer, T. L. Maier, A. Lüttke, U. B. Gebhart, M. Brandtner, B. Hamilton, C. Neumann-Spallart, C. B. Michalowski, H. J. Bohnert, H. E. A. Schenk, and W. Löffelhardt. 1993. Sequence analysis of pre-ferredoxin-NADP+-reductase cDNA from Cyanophora paradoxa specifying a precursor for a nucleus-encoded cyanelle polypeptide. Plant Mol. Biol. 211023-1033. [DOI] [PubMed] [Google Scholar]
- 22.James, M. G., D. S. Robertson, and A. M. Myers. 1995. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7417-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James, M. G., K. Denyer, and A. M. Myers. 2003. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6215-222. [DOI] [PubMed] [Google Scholar]
- 24.Koziol, A. G., T. Borza, K. I. Ishida, P. Keeling, R. W. Lee, and D. G. Durnford. 2007. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 1431802-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leloir, L. F., M. A. de Fekete, and C. E. Cardini. 1961. Starch and oligosaccharide synthesis from uridine diphosphate glucose. J. Biol. Chem. 236636-641. [PubMed] [Google Scholar]
- 26.Libessart, N., M. L. Maddelein, N. Van Den Koornhuyse, A. Decq, B. Delrue, G. Mouille, C. D'Hulst, and S. G. Ball. 1995. Storage, photosynthesis and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas reinhardtii. Plant Cell 71117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, T. P., T. Caspar, C. Somerville, and J. Preiss. 1988. Isolation and characterisation of a starchless mutant of Arabidopsis thaliana (L.) Heynth lacking ADP-glucose pyrophosphorylase activity. Plant Physiol. 861131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Y., and T. Sharkey. 2006. The importance of maltose in transitory starch breakdown. Plant Cell Environ. 29353-366. [DOI] [PubMed] [Google Scholar]
- 29.Manners, D. J. 1991. Recent developments in our understanding of glycogen structure. Carbohydrate Polymers 1637-82. [Google Scholar]
- 30.Miller, G. L. 1959. Cardiac arrest. Miss. Doc. 37149-151. [PubMed] [Google Scholar]
- 31.Mouille, G., M. L. Maddelein, N. Libessart, P. Talaga, A. Decq, B. Delrue, and S. Ball. 1996. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 81353-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, Y., and M. Imamura. 1983. Characteristics of a glucan phosphorylase from Chlorella vulgaris. Phytochemistry 22835-840. [Google Scholar]
- 33.Nakamura, Y., A. Kubo, T. Shimamune, T. Matsuda, K. Harada, and H. Satoh. 1997. Correlation between activities of starch debranching enzymes and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 12143-153. [Google Scholar]
- 34.Nyvall, P., J. Pelloux, H. V. Davies, M. Pedersen, and R. Viola. 1999. Purification and characterisation of a novel starch synthase selective for uridine 5′-diphosphate glucose from the red alga Gracilaria tenuistipitata. Planta 209143-152. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, E. A., L. B. Koski, Y. Zhang, L. Yang, E. Wang, M. W. Gray, G. Burger, and B. F. Lang. 2007. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res. 35D445-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patron, N. J., and P. K. Keeling. 2005. Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J. Phycol. 411131-1141. [Google Scholar]
- 37.Preiss, J., and T. Romeo. 1989. Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv. Microb. Physiol. 30183-238. [DOI] [PubMed] [Google Scholar]
- 38.Rahman, A., K. S. Wong, J. L. Jane, A. M. Myers, and M. G. James. 1998. Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 117425-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ral, J. P., E. Derelle, C. Ferraz, F. Wattebled, B. Farinas, F. Corellou, A. Buléon, M. C. Slomiany, D. Delvalle, C. d'Hulst, S. Rombauts, H. Moreau, and S. Ball. 2004. Starch division and partitioning a mechanism for granule propagation and maintenance in the picophytoplanktonic green alga Ostreococcus tauri. Plant Physiol. 1363333-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recondo, E., and L. Leloir. 1961. Adenosine diphosphate glucose and starch biosynthesis. Biochem. Biophys. Res. Commun. 685-88. [DOI] [PubMed] [Google Scholar]
- 41.Reyes-Prieto, A., and D. Bhattacharya. 2007. Phylogeny of nuclear encoded plastid targeted proteins supports an early divergence of glaucophytes within Plantae. Mol. Biol. Evol. 242358-2361. [DOI] [PubMed] [Google Scholar]
- 42.Roach, P. J. 2002. Glycogen and its metabolism. Curr. Mol. Med. 2101-120. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-Ezpeleta, N., H. Brinkmann, S. C. Burey, B. Roure, G. Burger, W. Loffelhardt, H. J. Bohnert, H. Philippe, and B. F. Lang. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 151325-1330. [DOI] [PubMed] [Google Scholar]
- 44.Shimonaga, T., S. Fujiwara, M. Kaneko, A. Izumo, S. Nihei, P. B. Francisco, A. Satoh, N. Fujita, Y. Nakamura, and M. Tsuzuki. 2007. Variation in storage alpha-polyglucans of red algae: amylose and semi-amylopectin types in porphyridium and glycogen type in cyanidium. Mar. Biotechnol. 9192-202. [DOI] [PubMed] [Google Scholar]
- 45.Smith, A. M. 1990. Enzymes of starch synthesis, p. 93-102. In P. J. Lea (ed.), Methods in plant biochemistry, vol. 3. Academic Press, London, United Kingdom. [Google Scholar]
- 46.Tetlow, E. J., M. K. Morell, and M. J. Emes. 2004. Recent developments in understanding the regulation of starch metabolism in higher plants. J. Exp. Biol. 552131-2145. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van den Koornhuyse, N., N. Libessart, B. Delrue, C. Zabawinski, A. Decq, A. Iglesias, J. Preiss, and S. Ball. 1996. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J. Biol. Chem. 27116281-16288. [DOI] [PubMed] [Google Scholar]
- 49.Van de Wal, M., C. D'Hulst, J. P. Vincken, A. Buléon, R. Visser, and S. Ball. 1998. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J. Biol. Chem. 27322232-22240. [DOI] [PubMed] [Google Scholar]
- 50.Viola, R., P. Nyvall, and M. Pedersen. 2001. The unique features of starch metabolism in red algae. Proc. R. Soc. Lond. B 2681417-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wattebled, F., J. P. Ral, D. Dauvillée, A. Myers, M. James, R. Schlichting, C. Giersch, S. Ball, and C. D'Hulst. 2003. STA11, a Chlamydomonas reinhardtii locus required for normal starch granule biogenesis, encodes disproportionating enzyme. Further evidence for a function of α-1,4 glucanotransferases during starch granule biosynthesis in green algae. Plant Physiol. 132137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wattebled, F., Y. Dong, S. Dumez, D. Delvallé, V. Planchot, P. Berbezy, D. Vyas, P. Colonna, M. Chatterjee, S. Ball, and C. d'Hulst. 2005. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zabawinski, C., N. Van den Koornhuyse, C. D'Hulst, R. Slichting, A. Decq, C. Giersch, B. Delrue, J. M. Lacroix, J. Preiss, and S. Ball. 2001. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of an heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 1831069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeeman, S. C., S. M. Smith, and A. M. Smith. 2007. The diurnal metabolism of leaf starch. Biochem. J. 40113-28. [DOI] [PubMed] [Google Scholar]