Abstract
The induction of heat shock genes (HSPs) is thought to be primarily regulated by heat shock transcription factors (HSFs), which bind target sequences on HSP promoters, called heat shock elements (HSEs). In this study, we investigated the 5′ untranslated regions of the Tetrahymena thermophila HSP70-1 gene, and we found, in addition to the canonical and divergent HSEs, multiple sets of GATA elements that have not been reported previously in protozoa. By means of in vivo analysis of a green fluorescent protein reporter transgene driven by the HSP70-1 promoter, we demonstrate that HSEs do not represent the minimal regulatory elements for heat shock induction, since the HSP70-1 is tightly regulated by both HSE and GATA elements. Electrophoretic mobility shift assay also showed that HSFs are constitutively bound to the HSEs, whereas GATA elements are engaged only after heat shock. This is the first demonstration by in vivo analysis of functional HSE and GATA elements in protozoa. Furthermore, we provide evidence of a functional link between HSE and GATA elements in the activation of the heat shock response.
The heat shock response is a universal property organisms use to cope with injuries caused by elevated temperatures, infections, exposure to toxic elements, and other environmental stresses. It has been established as a paradigm for inducible gene expression, leading researchers to unravel fundamental questions related to the molecular mechanisms of gene switches. The heat shock response involves a rapid and massive synthesis of a set of the so-called heat shock proteins (HSPs), among which those with a molecular mass of ∼70 KDa (HSP70s) play key roles (18).
The stress induction of the HSP70s is generally regulated at the transcriptional level by the binding heat shock transcription factors (HSFs) to the heat shock elements (HSEs) present in the 5′ flanking regions of heat shock genes. HSEs consist of contiguous inverted repeats of the sequence motif 5′-nGAAn3-′. The functional element usually contains a minimum of three repeated motifs with a maximum insertion of 5 bp between each motif (29). Nevertheless, functional HSEs may also digress from the consensus sequences. Among the deviations, the so-called gapped HSEs are frequently observed, containing a number of bases inserted between the repetitions. The Saccharomyces cerevisiae MDJ1 promoter is an example of a functional gapped HSE since it carries an insertion of 11 bp between the first and the second repetition [nTTCn-(11 bp)-nGAAn-(5 bp)-nGAAn]. This promoter also contains two functional contiguous nGAAn direct repeats (38). Recently, several examples of functional HSEs with direct repeats of nTTCn or nGAAn interrupted by 5-bp insertions were characterized in the promoter regions of several S. cerevisiae genes (34, 41).
Ciliated protozoa are unicellular eukaryotic organisms that are exposed to the natural environment for their entire life cycle. As a result, they serve as ideal organisms for studying molecular responses to the many stressful environmental conditions that are known to activate the heath shock response. Among ciliates, Tetrahymena thermophila is a very suitable model organism, given that its entire genome sequence is known (11) and given the ease with which it can be manipulated genetically (19, 31, 39). To date, the heat shock response in T. thermophila has been poorly analyzed at both the levels of gene and protein detection, while in T. pyriformis, the regulation of the HSP70 gene has been well analyzed at the transcriptional level (1, 2). However, no data are available regarding the organization of the HSP70 gene promoter and the responsive elements that regulate its induction in Tetrahymena.
In the present study, we analyzed induction of the T. thermophila HSP70-1 gene. We found that in addition to having a strictly regulated pattern of activation, it possesses a promoter region containing two conventional HSEs and an unexpected cluster of GATA motifs. GATA motifs have already been described in stress gene promoters, for example, in the Caenorhabditis elegans metallothionein genes mtl-1 and mtl-2 (28) and in the T. thermophila metallothionein gene MTT1 (10); however, these GATA motifs have never been shown to interact with the heat shock response machinery.
By means of in vivo mutational analysis and in vitro experiments, we show for the first time here that GATA motifs are directly involved in the heat shock response in T. thermophila. Moreover, we demonstrate that the association of the GATA motifs and one HSE is strictly required for the induction of HSP70-1 transcription in T. thermophila.
MATERIALS AND METHODS
Strains and culture conditions.
T. thermophila cells of strains Cu 428.2 (Mpr/Mpr [6-methylpurine sensitive, VII]) and Cu 427 (ChxI-I/ChxI- [cycloheximide sensitive, VI]) were grown in SPP medium (1% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% EDTA ferric sodium salt) at 30°C with moderate shaking. To prevent bacterial and fungal growth, the medium was enriched with penicillin G (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.025 μg/ml).
DNA extraction and amplification.
T. thermophila genomic DNA was extracted from Cu 428.2 cells by using a Quantum Prep Aquapure genomic DNA kit (Bio-Rad).
The primers used to amplify the first T. thermophila HSP70-1 gene fragment were designed based on the amino acid sequence of the Drosophila melanogaster HSP70 gene (AAN1354). In particular, the sense primer MBQG (5′-AAIGATCAAGGIAAIAGAACIACICC-3′; where I stands for inosine) was designed from the sequence N28DQGNRTTP36 of D. melanogaster HSP70, and the antisense oligonucleotide IIRE (5′-GCIGCIGCIGTTGGITCGTTIATIAT-3′) was designed from the sequence I169INEPTAAA177. The entire T. thermophila HSP70-1 open reading frame and related 5′ and 3′ flanking sequences were obtained by using the technique of anchored PCR with the Universal Vectorette System (Sigma).
Heat shock treatment and Northern blot analysis.
Cu 428 cells were cultured aerobically at 30°C in a 1-liter flask to a density of 2.5 × 105 cells/ml. Heat shock was performed at 34, 38, and 40°C. Prior to heat shock, 20-ml aliquots of SPP medium were heated at the temperatures indicated above. Samples of 7 × 106 cells were collected by centrifugation and suspended with the preheated medium. The shock duration was 30 min, after which total RNA was extracted by using an RNAspin minikit (GE Healthcare). For Northern blot analysis, 20-μg samples of total RNA were transferred onto Hybond-N membrane and hybridized with α-32P-labeled DNA.
To detect the HSP70-1 mRNA, a probe of 300 nucleotides (nt), corresponding to the sequence of the last 100 amino acids of the protein, was used. The DNA probe was amplified from the genomic DNA and labeled by the incorporation of [α-32P]dATP according to the random priming technique (25). Differences in sample loading were detected by rehybridization of the membrane to a 300-nt T. thermophila 17S rRNA probe labeled with [α-32P]dATP.
Construction of reporter plasmids.
The circular plasmid pD5H8, kindly provided by Geoffrey Kapler (Department of Biochemistry and Biophysics, Texas A&M University), was used to produce the transformation vectors used here. pD5H8 is a micronuclear ribosomal DNA (rDNA)-based plasmid that, upon insertion into conjugant cells, is rearranged in the newly formed macronuclei (42, 43, 44). The rDNA plasmid insert carries a mutation that confers paromomycin resistance to the exconjugant cells. Virtually total replacement of the endogenous rDNA is achieved upon prolonged selection of the transformant cell lines with paromomycin (12).
The wild-type plasmid, pWT, was generated by inserting the coding sequence of the green fluorescent protein (GFP) gene between the 5′ and 3′ untranslated regions (UTRs) of the T. thermophila HSP70-1 gene.
A 920-bp fragment including the HSP70-1 5′ UTR plus 21 nt encoding the first 7 amino acids of the protein was amplified by PCR using the primers PROM-FW and PROM-RV (Fig. 1). These primers introduced NotI and BglII restriction sites at the 5′ and 3′ ends of the 5′ UTR fragment, respectively. A 400-bp fragment corresponding to the HSP70-1 3′ UTR was produced using the primers TER-FW and TER-RV, which introduced XhoI and NotI restriction sites at the 5′ and 3′ ends, respectively. Similarly, BglII and XhoI restriction sites were generated at the 5′ and 3′ ends of the GFP coding sequence using the primers GFP-FW and GFP-RV. The 5′ UTR, the GFP gene, and 3′ UTR fragments were digested with the appropriate enzymes and then covalently coupled using T4 DNA ligase (Gibco) to form the HSP70-1 5′ UTR-GFP-HSP70-1 3′ UTR recombinant DNA. The ligation product was amplified from the ligation reaction, digested with NotI, and subsequently inserted into the pD5H8 vector at its unique NotI cloning site. Recombinant plasmids were transformed into Escherichia coli cells (strain DH5α). The plasmids were purified from bacterial lysates using a Qiagen media plasmid kit. Using inverse PCR strategies, we constructed four variants of pWT that included deletions of the HSE I, HSE II, and GATA elements (pΔHSE I, pΔHSE II, and pΔGATA I) or contained sequence mutations in the GATA elements (pΔGATA II).
FIG. 1.
Partial nucleotide sequence of the 5′ UTR of the HSP70-1 gene. The sequences comprising the putative HSE I and HSE II are boxed, and the individual sequence motifs are in boldface. The cluster of GATA elements is shaded. The initiating ATG is shown in boldface italics and is followed by the codons of the first seven amino acids of the protein. In the plasmids used in this study, these seven codons were fused in frame with the initiating ATG of the GFP, by means of a BglII site. The synthetic primers used to amplify this sequence from the genomic DNA are underlined, and the restriction sites NotI and BglII, at the 5′ and 3′ ends, respectively, are in boldface.
Three pairs of specific primers were designed to eliminate the HSE I (ΔH I-FW and ΔH I-RV), HSE II (ΔH II-FW andΔH II-RV), and GATA (ΔGI-FW and ΔGI-RV) sequences, respectively. A fourth pair of primers was designed to introduce point mutations in the GATA sequences (ΔGII-FW and ΔGII-RV). These primers were used to generate mutated plasmids from pWT using the Pfu polymerase, which shows proofreading activity but lacks the 5′-3′ exonuclease activity. Each of the four PCR products was digested with the enzyme DpnI to generate linear fragments of the template. After purification from agarose gels, each linear plasmid was treated with T4 DNA ligase to produce circular plasmids, which were then transformed individually into E. coli DH5α cells. Plasmids were purified from bacterial lysates, and each preparation was sequenced to verify the absence of amplification errors within the GFP coding sequence and the 5′ and 3′ UTRs.
Electroporation.
Electroporation was performed according to the method of Gaertig and Gorovsky (13). Equal numbers of cells of the Cu 428.2 and Cu 427 mating types were suspended in 10 mM Tris-HCl (pH 7.5) at final concentrations of 3 × 105 cell/ml, and the combined cells were starved for 20 h at 30°C with fast shaking to prevent mating. At 8 to 11 h before electroporation, the shaker was turned off, and starved cells were allowed to conjugate. After 3 h, the pairing efficiency was verified by direct counting of the cells, and nuclear events were monitored hourly by DAPI (4′,6′-diamidino-2-phenylindole) staining (37).
At the stage defined as “macronuclear development I” (26), cells were washed and resuspended in 1 ml of electroporation buffer (10 mM HEPES [pH 7.5]) to a final concentration of 1.5 × 107 cells/ml. A total of 240 μl of the cell suspension and 20 μg of plasmid DNA (resuspended in 10 μl of HEPES) were mixed, immediately transferred to a 0.4-cm Gene Pulser cuvette (Bio-Rad), and pulsed (0.44 kV with a 25-μF capacitor at 200 Ω) in a capacitor-discharge-based Cell Porator (Bio-Rad). After electroporation, cells were suspended in 5 ml of SPP plus antibiotics (penicillin G and streptomycin sulfate at final concentrations of 250 μg/ml each), followed by incubation at room temperature for 24 h before addition of the selective drug (paromomycin sulfate).
For all transformation experiments, three different cell lines were selected by increasing the quantity of the paromomycin sulfate, beginning at 100 μg/ml and ending at 1,000 μg/ml. To increase the replacement of the endogenous rDNA by phenotypic assortment, the selected cell lines were grown for 2 weeks in selective medium containing 1 mg of paromomycin/ml. The presence of the synthetic rDNA-GFP constructs in the exconjugant cells was verified by PCR analysis. The PCR products were sequenced to check that point mutations or other rearrangements did not occur during transfection.
GFP synthesis was established by in vivo microscopic observation of the cells throughout the 1-h heat shock treatment. At 2 min prior to observation, the cells were treated with dibucaine hydrochloride at a final concentration of 0.3 mM to reduce the swimming speed without affecting viability.
Synthesis of cDNA.
Total RNA was extracted from 5 ml of wild-type or recombinant Tetrahymena cell cultures that were heat shocked at 38°C until GFP fluorescence was observed. The RNA spin mini RNA isolation kit (GE Healthcare) was used. DNase I treatment of total RNA was performed directly on the silica membrane. RNA quality was checked by electrophoresis on a 1.2% formaldehyde agarose gel. Before retrotranscription, RNA samples were amplified by PCR using 17S rDNA specific primers to exclude the presence of DNA contamination. First-strand cDNA was produced from 300-ng samples of total RNA using 200 ng of random hexamer primers and Moloney murine leukemia virus reverse transcriptase (Fermentas).
Nuclear extracts.
The protocol used to prepare nuclear extracts was derived from the large-scale procedure of Dignani et al. (9). Briefly, 3 × 107 cells were harvested from a 100-ml culture by centrifugation at 3,500 rpm for 3 min, and the pellet was resuspended in 400 ml of lysis buffer containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 250 mM sucrose, 0.5% Triton X-100, a protease cocktail (1 mM phenylmethylsulfonyl fluoride, 0.2 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], 0.1 mM EDTA, 13 mM bestatin, 0.14 mM E-64, 0.1 mM leupeptin, 3 nM aprotinin), and phosphatase inhibitors (10 mM sodium β-glycerophosphatase and 0.5 mM dithiothreitol [DTT]). Cell lysate, was centrifuged at 15,000 × g for 10 min at 4°C, and the supernatant fraction was discarded. The presence of intact nuclei in the pellet was confirmed by propidium iodide staining. The pellet was resuspended in 300 μl of cold extraction buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA), and the suspension was incubated on ice for 20 min. Cellular debris was removed by centrifugation for 5 min at 4°C, and the resulting supernatant fraction was divided into aliquots of 20 μl and stored at −80°C. The protein concentration was estimated using the Bradford method (5).
EMSA analysis.
Transcription factor binding to HSE (or GATA) elements was assayed by incubating samples of nuclear extract (10 μg of protein) and of 32P-labeled DNA target (10 pg) in 10 mM Tris-HCl (pH 7.5) containing 50 mM NaCl, 2.5 mM MgCl2, 4% glycerol, 0.5 mM DTT, and 1 μg of poly(dI-dC) (21). Reactions were diluted to a final volume of 20 μl and incubated for 20 min at room temperature. The reactions were stopped by addition of 4 μl of 5× gel loading buffer (300 mM Tris-HCl [pH 7.5], 50% glycerol, 0.05% bromophenol blue), and the samples were electrophoresed on 5% nondenaturing polyacrylamide gels at 120 V for 1.5 h in a buffer containing 50 mM Tris-HCl (pH 8.5), 0.38 M glycine, and 2 mM EDTA. Gels were dried and exposed to film overnight at −80°C. Binding competition assays were performed by addition of 50- and 100-fold molar excesses of wild-type or mutated DNA targets (unlabeled) or with labeled mutant targets substituted for wild-type targets.
DNA binding targets.
Double-stranded DNA targets of ∼80 bp were labeled at their 3′ ends with [α-32P]dATP. Wild-type GATA and HSE I target sequences were first amplified from the pWT plasmid by using synthetic primers that introduced EcoRI restriction sites at both the 5′ and 3′ ends (HSE I-FW, HSE I-RV, GATA-FW, and GATA-RV). Mutated HSE I target used in competition assays was obtained by using a two-step PCR amplification. In the first amplification the primer HSE Imut-FW was used in combination with the primer HSE I-RV to introduce the desired point mutations. The fragments obtained were used as a template for the second round of amplification with the primers HSE I-FW and HSE I-RV in order to add the EcoRI site at both ends. The mutated GATA was amplified directly from the pΔGATA II plasmid by using the primers GATA-FW and GATA-RV to introduce the EcoRI site at both ends. After purification from an agarose gel, the fragments were digested with EcoRI and then precipitated with 2.5 M ammonium acetate and 2 volumes of absolute ethanol to eliminate the small fragments generated by the cleavage.
Targets (500 ng) were labeled with [α-32P]dATP using the Klenow fragment, and unincorporated nucleotides were eliminated by precipitation.
Primers.
The synthetic primers and their positions, according to Fig. 1, were as follows (restriction sites sequences are underlined): PROM-FW (5′-TCTTGCGGCCGCTATTTTCAAAAAATTTTAATTA-3′; from nucleotide [nt] −910 to nt −879); PROM-RV (5′-CCTACAGATCTAGTTTTATCGTTTTTACC-3′; from nt +31 to nt +1); ΔH I-FW (5′-ATTAGAGAGTTAAAAATAAAAAATGAA-3′; from nt −349 to nt −322); ΔH I-RV (5′-AAATCGAATTATTATAAATGATAAATTC-3′; from nt −366 to nt −393); ΔH II-FW (5′-CGATTTAGAAGAGTTCCAGAATATT-3′; from nt −361 to nt −346); ΔH II-RV (5′-AATAAATGGAATTAATACCCACAATC-3′; from nt −400 to nt −426); ΔGI-FW (5′-ATGCTTTTTACTTAATTTGTAGGAGAGGTTA-3′; from nt −271 to nt −240); ΔGI-RV (5′-TATTAATTCATTTTTTATTTTTAACTCTC-3′; from nt −317 to nt −345); ΔGII-FW (5′-TAATTGTATGATCGAAACATAAATATTAAGATG-3′; from nt −301 to nt −269); ΔGII-RV (5′-ATCTATGTATTTGGCTATTAATTCATTTTT-3′; from nt −302 to nt −331); HSE I-FW (5′-ATTAGAATTCATCATTTATAATAATTCGATTTAC-3′; from nt −398 to nt −364); HSE Imut-FW (5′-CATTTATAATAATTCGATTTAGATGAGTACCACAAT-3′; from nt −385 to nt −350); HSE I-RV (5′-TATCTATCTATGTATCTATCTATGAATTC-3′; from nt −297 to nt −325); GATA-FW (5′-TATTAGAGAGTTGAATTCAAAAAATGAATT-3′; from nt −350 to nt −321); and GATA-RV (5′-CCTCTCCATCGAATTCAGTAAAAAGCA-3′; from nt −244 to nt −270).
RESULTS
Characterization of the T. thermophila HSP70 coding and regulatory regions.
Using heterologous degenerate primers and anchored PCR, we cloned and sequenced the T. thermophila HSP70-1 coding region, which has been deposited in GenBank under accession number AY028663. Analysis of the deduced amino acid sequence revealed that the protein harbors the three canonical HSP70 domains: the ATPase domain of 45 kDa, the substrate-binding domain of 15 kDa, and a 10-kDa variable domain. It also contains the three motifs IDLGTTYSC, LVFDLGGGTFDVSLL, and IVLVGGSSRIPKVIQ, typically recognized as signatures of the HSP70 family (3). Two major sequence features distinguish HSP70-1 from most other eukaryotic HSP70s: a lack of the Gly/Pro-rich segment at the C-terminal domain and the substitution of an Asp for a Glu in the terminal tetrapeptide (DEVD versus the more common EEVD). In fact, each of the five members of the T. thermophila HSP70 family possesses the same substitution.
Because T. thermophila macronuclear chromosomes contain short intergenic regions, we analyzed 1,500 nt of the 5′ UTR of the gene for the presence of putative regulatory elements. As shown in Fig. 1, we identified two HSEs: the first lying between the nucleotides at positions −365 and −350 (depicted as HSE-I and matching perfectly the consensus sequence) and the second lying between the nucleotides at positions −402 and −370 (depicted as HSE-II). The latter is a gapped-like HSE because it contains an insertion of 15 nt between the second and the third repetitions. Other GAA and TTC motifs are present in the 5′ UTR downstream of the HSE I element, but their wide separation and their proximity to the ATG start codon make it unlikely that they constitute functional HSEs.
In addition to the HSEs, which are common in stress inducible genes, we identified a cluster of GATA motifs, positioned between the nucleotides at positions −317 and −271, that consist of five repetitions of the consensus sequence W GATA R organized as tandem repeats. Because GATA motifs have never been directly correlated with the heat shock response, we investigated, by means of mutagenesis and in vivo analyses, whether they are required for the expression of a reporter gene either in association with, or independent of, the presence of one or both of the HSEs.
In vivo characterization of the T. thermophila HSP70-1 gene regulatory regions.
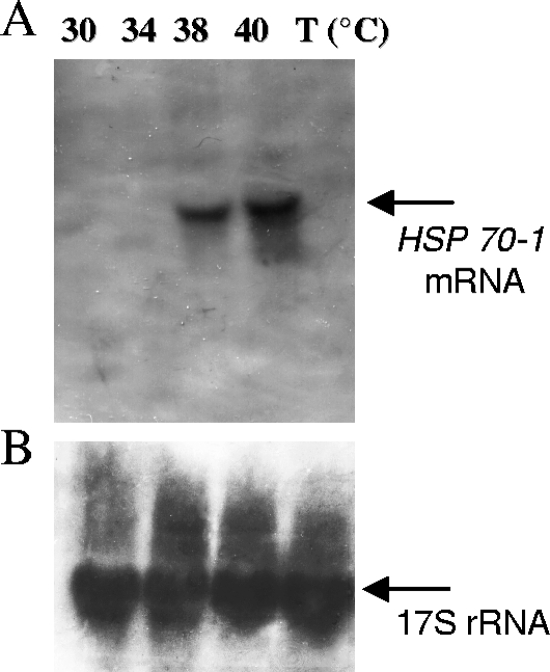
The heat shock response of T. thermophila was analyzed by Northern blotting to determine the trend of the expression of the HSP70-1 gene with increasing temperature. Subsequently, the same blots were stripped and reprobed for 17S rRNA to normalize for differential RNA recovery and/or gel loading. No HSP70-1 mRNA signal was visible at 30°C (optimal growing temperature) or 34°C, whereas a strong signal was visible at 38°C and increased by threefold at 40°C (Fig. 2).
FIG. 2.
Northern blot analysis of total RNA samples extracted from T. thermophila cells after heat shock treatments performed at the indicated temperatures. (A) Samples of 20 μg of total RNA were transferred onto Hybond N membrane and assayed with labeled probe specific for the HSP70-1 mRNA. (B) Differences in sample loading were detected by reprobing the membrane with a labeled probe of the same length and complementary to the T. thermophila 17S rRNA.
To evaluate the role of the HSE and GATA motifs on the regulation of transcription of this gene under conditions of heat shock, we produced a set of five plasmids and used them to transform T. thermophila cells by conjugant electroporation. The wild-type plasmid (pWT) contained nucleotides from positions +21 to −900 of the HSP70-1 gene fused in frame with the GFP coding region (Fig. 1). The second and third plasmids, pΔHSE-I and pΔHSE-II, differed from pWT, with the former lacking the HSE I element and the latter lacking the HSE II element. The fourth and fifth plasmids, pΔGATA-I and pΔGATA-II, either lacked the GATA motifs (pΔGATA-I) or contained mutated motifs (pΔGATA II). In the latter case, the GATA sequences were changed from GATAGATACATAGATAGATAGATAGATAGATAGATAAATAGATAAGATG to GTTAAATACATAGATTAATTGTATGATCGAAATCATAAATATTAATGATG.
A stretch of 400 bp of the 3′ UTR of HSP70-1 was also included in all of the plasmids and positioned after the stop codon of the GFP coding region.
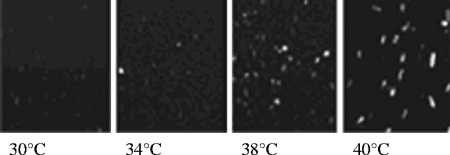
To test the GFP-reporter methodology, cells were first transfected with pWT-GFP, and transformants were selected with paromomycin sulfate as described in Materials and Methods. Three independently transformed cell lines were shocked at increasing temperatures (30, 34, 38, and 40°C), and the degree of fluorescence at each temperature was compared to the Northern blot results. The three cell lines showed identical trends of fluorescence, in good agreement with the trend of the HSP70-1 mRNA expression (an example is shown in Fig. 3). GFP fluorescence was not detected at 30°C, whereas the percentage of fluorescent cells increased from ∼5% at 34°C to ∼80% at 38°C. At 40°C, 100% of the cells were fluorescent. The transition from small numbers of fluorescent cells at 34°C to large numbers at 38°C is likely to reflect individual variability in the threshold temperature to activate the heat shock response, possibly due to asynchrony in the physiological stage of the cells. Although at 40°C the percentage of cells exhibiting fluorescence was the highest, when visualized by microscopy they appeared impaired, with low mobility, vacuolization, and cell death. At 38°C, in contrast, the cells were active and healthy. Therefore, 38°C was chosen as the optimal inducing temperature for the following in vivo promoter analyses.
FIG. 3.
Visualization of the fluorescence emission by T. thermophila cells transfected with the pWT (containing the wild-type 5′ flanking region of the T. thermophila HSP70-1). Samples of 20 ml of cell cultures were incubated at the indicated temperatures, and the GFP fluorescence emission was detected in vivo, throughout 1 h, by using an Olympus IX71 fluorescence microscope equipped with fluorescein isothiocyanate filters. Pictures were taken by the Olimpus U-CMAD3 digital camera at ×100 magnification. Two minutes prior to observation, the cells were treated with dibucaine hydrochloride at a final concentration of 0.3 mM to reduce the swimming speed.
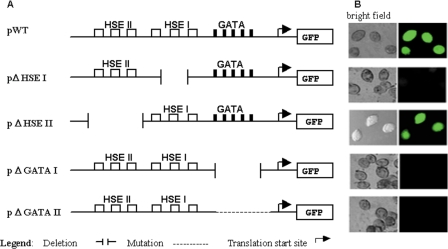
Having established the optimal conditions for heat shock, we next examined cells transfected with wild-type and mutant reporter constructs. Figure 4 shows that cells transfected with pΔHSE-I were unable to produce fluorescence, which indicates that the HSE I element is necessary for the induction of GFP expression. In contrast, cells transfected with pΔHSE-II showed the same degree of GFP expression as the pWT transformants. These first experiments indicated that HSE II (which is the gapped-like element) is ineffective in the induction of transcription of the reporter gene under heat shock conditions. A second implication of these results is that the widespread, separated GAA and TTC distributed motifs are incapable of serving as a functional HSE that can compensate for the deletion of HSE I.
FIG. 4.
Mutation analysis of the 5′ UTR of the T. thermophila HSP70-1 gene. (A) Schematic representation of the wild-type and mutated sequences used to produce the synthetic plasmids listed below: pWT, containing 900 bp of the wild-type T. thermophila HSP70-1 5′ UTR; pΔHSE I, derived from the pWT, lacking 16 nt containing the putative HSE I; pΔHSE II, derived from the pWT, lacking 28 nt containing the putative HSE II; pΔGATA I, derived from the pWT, lacking 45 nt containing the cluster of GATA motifs; pΔGATA II, derived from the pWT, maintaining the original sequence length and subjected to site-directed mutagenesis on the GATA sequences that were changed from GATAGATACATAGATAGATAGATAGATAGATAGATAAATAGATAGATG to GTTAAATACATAGATTAATTGTATGATCGAAACATAAATATTAAGATG. (B) Visualization of cell strains transfected with the corresponding plasmids of panel A. Cells lines, transfected with the listed plasmids, were grown to the density of 2 × 105 cells/ml and heat shocked at different temperatures as described in Materials and Methods. To verify the fluorescence emission, samples of cells were collected throughout 1 h and observed under by fluorescence microscopy. The pictures shown were taken in vivo, after 15 min of heat shock at 38°C, by a fluorescence microscope Olympus IX71 equipped with a fluorescein isothiocyanate filter.
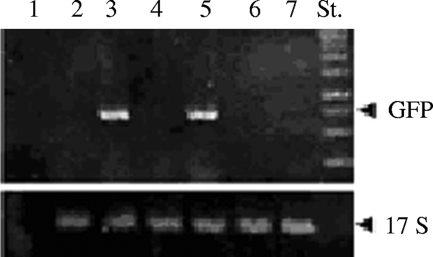
Using a similar approach, we analyzed the pΔGATA plasmids. Cell lines transfected with either pΔGATAI, which lacks the GATA sequences, or pΔGATAII, which contains mutated GATA sequences, were analyzed as for the HSE constructs. Each of the GATA constructs failed to express the reporter gene after heat shock treatment (Fig. 4). Prolonged heat shock exposure was also assayed on the GATA mutant cell lines, but no fluorescence was detected. To demonstrate that the absence of fluorescence observed with pΔGATAI, pΔGATAII, and pΔHSE-II was not attributable to artifactual effects on transcription and/or translation, we tested the presence of GFP mRNA on the transfected cells. Samples of wild-type and transfected cell strains were heat shocked at 38°C until the fluorescence was observed, and the cells collected by centrifugation and immediately processed for total RNA extraction. The RNA was retrotranscribed and used as a template to amplify the GFP coding region. As shown in Fig. 5, the cDNA obtained from the pΔGATAI, pΔGATAII and pΔHSE-II cell strains failed to generate a reverse transcription-PCR product, while the cDNA obtained from the wild-type and pΔHSE-II cell strains gave a clear amplification product. These results are in agreement with the in vivo experiments illustrated in Fig. 4 and show that the absence of fluorescence was not due to failure to translate the GFP mRNA but rather to the loss of promoter function.
FIG. 5.
Detection of the GFP mRNA on transfected T. thermophila cells. Samples of 100 ng of cDNA were used in PCR analysis with primers designed on the GFP coding region and on the sequence of the 17S rRNA to amplify fragments of 750 and 300 bp, respectively. The reaction run in lane 1 contained no cDNA, while the reaction run in lane 2 contained cDNA obtained from not transfected cells. These reactions showed the specificity of the GFP primers. The other reactions contained cDNA obtained from cells transfected with the following plasmids: pWT, pΔHSE I, pΔHSE II, pΔGATA I, pΔGATA II (in lanes from 3 to 7, respectively). St., DNA standards.
Taken together, these results support the conclusion that induction of the T. thermophila HSP70-1 gene under heat shock conditions requires both the HSE I and GATA motifs. In fact, each element alone was unable to promote the transcription of the reporter gene.
EMSA of HSE and GATA motifs.
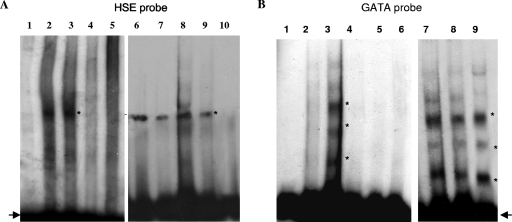
Since the expression of the T. thermophila HSP70-1 gene depends on the presence of the canonical HSE I and GATA motifs, we used an electrophoretic mobility shift assay (EMSA) to assess whether Tetrahymena possesses transcription factors that specifically bind these genetic elements. Nuclear proteins, extracted from unshocked and heat-shocked T. thermophila cells, were assayed with 32P-labeled double-stranded targets corresponding to the sequences of the HSE I or GATA motifs. When the nuclear extracts were incubated with the HSE target, a single DNA-protein complex was observed both in unshocked and heat-shocked cell extracts (Fig. 6A, lanes 2 and 3), which suggest that the putative HSE transcription factor is constitutively positioned on the HSE. The specificity of binding was demonstrated by the addition of 50- and 100-fold molar excesses of unlabeled target, which dramatically reduced complex formation (Fig. 6A, lanes 4 and 5). The specificity of protein binding was further shown by using a mutated HSE target as a competitor. The sequence of the HSE I element in the target was changed from GAAGAGTTCCAGAA to GATGAGTACCACAA, while the length of the target was kept constant. As shown in Fig. 6A, when 50-fold (lane 8) and 100-fold (lane 9) molar excesses of unlabeled mutant target were incubated with nuclear protein extracts from heat-treated cells, the quantity of bound labeled target was unaffected (cf. lanes 8 and 9 with lanes 6 and 7). Thus, the unlabeled mutant target cannot compete with the labeled wild-type target for binding to nuclear proteins. In contrast, when the mutant target was labeled and used in place of the wild-type target (lane 10), complex formation was not observed, which indicates that the mutant HSE I element was unable to interact specifically with nuclear proteins.
FIG. 6.
EMSA of nuclear protein extracts from heat shocked and unshocked cells performed with targets containing HSE (A) or GATA elements (B). In both panels, binding reactions containing only the labeled wild-type target or the labeled wild-type target with the nuclear extracts from unshocked cells or from heat shocked cells are shown in lanes 1 to 3, respectively; 50- or 100-fold molar excesses of unlabeled wild-type target as cold competitor were added to binding reactions containing nuclear extracts from heat shocked cells (lanes 4 and 5, respectively). Lanes from 6 to 10 show binding reactions from experiments involving mutant targets as competitors. Specifically, in panel A binding reactions with nuclear extracts from shocked cells in the presence of labeled wild-type target with 50- or 100-fold molar excesses of the HSE mutated unlabeled target are shown in lanes 8 and 9, respectively; the binding reaction of HSE mutated labeled target with nuclear extracts from shocked cells is shown in lane 10; in lanes 6 and 7, binding reactions in conditions identical to those in lanes 2 and 3 were run as a reference. In panel B, binding reactions with nuclear extracts from shocked cells in the presence of GATA labeled wild-type target with 50- or 100-fold molar excesses of the GATA mutated unlabeled target are shown in lanes 8 and 9, respectively. The binding reaction of GATA mutated labeled probe with nuclear extracts from shocked cells is shown in lane 6; a binding reaction in conditions identical to those for lane 3 was run as a reference in lane 7. Asterisks indicate the DNA-protein complexes, and arrows indicate the free target.
The GATA probes were assayed in EMSA experiments using nuclear extracts and binding conditions identical to those used with the HSE probes. Three DNA-protein complexes formed in the sample corresponding to the heat-shocked nuclear extract in the presence of wild-type labeled probe, whereas no complexes were detected when the same target was incubated with nuclear proteins extracted from unshocked cells. (Fig. 5C, lanes 2 and 3, respectively). To confirm these results, competition analyses were performed with 50- and 100-fold molar excesses of an unlabeled wild-type GATA target and with mutant GATA sequences (for the sequences, see Materials and Methods). The unlabeled wild-type target was able to compete with the labeled wild-type probe for binding of the nuclear proteins of the complex (Fig. 6B, lanes 4 and 5). Furthermore, complex formation was not observed when the labeled mutant target was used in place of the wild type. In contrast, when nuclear extracts were incubated with the labeled wild-type target in the presence of 50- and 100-fold molar excesses of unlabeled mutant target (Fig. 6B, lanes 8 and 9), the DNA-protein complex formed, thus indicating that the mutant target does not compete with the labeled wild-type target sequence.
DISCUSSION
The HSP70 genes and the expression of their products have been well characterized in some eukaryotic microbes, such as yeasts. In contrast, in ciliates these genes have been largely studied for their phylogenetic or ecological characteristics (22) and less with respect to their encoded proteins and regulation of expression (23). T. thermophila, for example, has long served as a valuable model for studying the regulation of gene expression by mechanisms of histone acetylation and deacetylation (40) and linker histone H1 phosphorylation (14, 36), but very little information is available on the regulatory elements of specific genes. To some extent the A/T prevalence in the Tetrahymena genome makes it difficult to identify regulatory elements by sequence homology to the promoters of other organisms. Although TATA and CG boxes seem to be absent in Tetrahymena promoters (7), a few regulatory elements have been identified by deletion and mutational analysis. For example, a PSE element was found to be conserved in the promoters of ciliate telomerase genes, and three elements were identified in the promoter of the RAD-51 gene (17, 35). Among the latter, only the UV1-UV2 repeats are homologous to the damage-responsive elements of yeasts (20). Clearly, it can be difficult to detect regulatory elements in ciliates by reference to other eukaryotes.
In the present study, we identify by comparative analysis two well-conserved cis-acting sequences in the 5′ untranslated regulatory region of the T. thermophila HSP70-1 gene: the HSEs and GATA elements. By means of in vivo expression of various reporter constructs, we also define the functional significance of these elements in heat shock-mediated gene activation. In vivo mutational analysis indicates a tight functional correlation between the HSE I and the GATA motifs in driving the heat-induced expression of a reporter gene (GFP), while the presence of HSE II (gapped-like) appears to be dispensable for the induction of gene expression. The functional HSE I is organized in the typical arrangement of three inverted repeats with short gaps, which suggests that it interacts with a trimeric HSF. Our results also demonstrate that the GAA/TTC motifs that are widespread downstream of the GATA elements do not function by themselves as HSEs, although their potential cooperative interaction with HSE I cannot be ruled out.
The combinatorial cooperation of the GATA element with other regulatory elements has been well characterized in other organisms (21). In the human β-globin locus control region, the NF-E2 and GATA transcription factors, when bound to their respective regulatory elements, control the position-independent expression of β-globin genes (16, 32). In C. elegans the GATA transcription factor (ELT-2) constitutively activates the metallothionein gene expression by binding specific GATA elements, whereas a second metal-responsive factor prevents transcription in the absence of metals (28). Our results constitute the second report that GATA elements regulate HSP70 genes, the first being the description of their role in the regulation of the HSP70 gene of chronic myelogenous leukemia cells (33). In this pathological case, upregulation of the HSP70 gene is coordinated by GATA-1 and the fusion oncoprotein (p210 BCR-ABL) in a pathway independent of heat stress (33).
EMSA experiments showed that HSE elements form a DNA-protein complex under both stress and nonstressed conditions, while GATA elements combine with proteins to form multiple DNA-protein complexes only under conditions of heat shock. The formation of multiple complexes may be dependent on the cooperative interactions between GATA elements and their binding proteins. These results provide evidence that HSFs are constitutively present on the HSEs, as has been described in the yeast S. cerevisiae (15) and in Tetrahymena pyriformis (8), whereas the GATA-recognizing proteins bind the target sequences only after stress.
On the basis of our experimental results, we propose a model in which the uninduced T thermophila HSP70-1 promoter is occupied by the heat shock transcription factors and the GATA binding proteins are recruited only after thermal stress. This combinatorial binding may alter the packaging of the chromatin such that other members of the transcriptional complex enter the promoter and induce gene transcription. In fact, the GATA-1 erythroid transcription factor is involved in the generation of active chromatin, thereby facilitating the access of other transcription factors (6, 30). GATA factors in general are capable of temporarily altering the nucleosome structure, which reforms once the GATA factors are removed (4).
The HSP70 protein family is known to consist of several members with a similar molecular sizes (27), encoded by both inducible and constitutive genes. Therefore, we searched the T. thermophila genome for the presence of other HSP70s (http://www.tigr.org/tdb/e2k1/ttg) and found five genes with high sequence similarity to T. thermophila HSP70-1. None of the five other genes have GATA elements in their 5′ UTRs, but they do have many HSE elements. The use of a molecular approach such as the one described here will help to elucidate whether the GATA elements confer unique functional properties to T. thermophila HSP70-1 that are absent in genes lacking these elements. A search of the genome bank for the presence of putative transcription factors (HSFs and GATA factors) revealed two predicted amino acid sequences annotated as putative HSFs (TTHERM_00571940 and TTHERM_00794590) but no GATA-like factors with the typical zinc finger organization (24). Because the spatial organization and sequences of the T. thermophila HSP70-1 GATA elements are unlikely to be casual, the characterization of the GATA-binding proteins, revealed by the EMSA experiments, may contribute to the discovery of a novel pattern of gene transcription regulation. Moreover, our results demonstrate that HSEs do not represent the minimal elements required for heat shock activation in the T. thermophila HSP70-1 gene and show for the first time the essential role of GATA motifs in the heat shock induction of one HSP70 gene.
In conclusion, although a more extensive analysis is required to fully elucidate the interplay between HSE and GATA motifs, we provide the first demonstration by in vivo analysis of the function of HSEs in ciliates and GATA elements in protozoa. This simple system of cell transformation via electroporation opens the way to the characterization of other cis-acting elements and of their pathways of transcriptional gene regulation in a model ciliate whose genome has been completely sequenced. Future comparison of gene regulation in ciliates and in higher eukaryotes will undoubtedly contribute to our understanding of the evolution of this vital process.
Acknowledgments
This study was supported by a grant from the Ministry of University and Research (PRIN).
We thank the anonymous reviewers for helpful comments and P. Luporini and H. W. Detrich for insightful discussions.
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Amaral, M. D., L. Galego, and C. Rodrigues-Pousada. 1988. Stress response of Tetrahymena pyriformis to arsenite and heat shock: differences and similarities. Eur. J. Biochem. 171463-470. [DOI] [PubMed] [Google Scholar]
- 2.Amaral, M. D., L. Galero, and C. Rodrigues-Pousada. 1993. Heat-shock-induced protein synthesis is responsible for the switch-off of hsp 70 transcription in Tetrahymena. Biochim. Biophys. Acta 1174133-142. [DOI] [PubMed] [Google Scholar]
- 3.Boorstein, W. R., T. Ziegelhoffer, and E. A. Craig. 1994. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 381-17. [DOI] [PubMed] [Google Scholar]
- 4.Boye, J., J. Omichinsky, D. Clark, M. Pikaart, and G. Felsenfeld. 1998. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J. Mol. Biol. 279529-544. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bresnick, E. H., M. L. Martowicz, S. Pal, and K. D. Johnson. 2005. Developmental control via GATA factor interplay at chromatin domains. J. Cell Physiol. 2051-9. [DOI] [PubMed] [Google Scholar]
- 7.Brunck, C. F., and L. A. Sadler. 1990. Characterization of the promoter region of Tetrahymena genes. Nucleic Acids Res. 18323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmo Avides, M., C. E. Sunkel, P. Moradas-Ferreira, and C. Rodrigues-Pousada. 1990. Properties and partial characterization of the heat-shock factor from Tetrahymena pyriformis. Eur. J. Biochem. 194331-336. [DOI] [PubMed] [Google Scholar]
- 9.Dignani, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dondero, F., M. Cavaletto, A. R. Ghezzi, A. La Terza, M. Benni, and A. Viarengo. 2004. Biochemical characterization and quantitative gene expression analysis of the multi-stress inducible metallothionein from Tetrahymena thermophila. Protist 155157-168. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, J. A., R. S. Coyne, M. Wu, D. Wu, M. Thiagarajan, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PloS Biol. 4e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaertig, J., and G. Kapler. 2000. Transient and stable DNA transformation of Tetrahymena thermophila by electroporation. Methods Cell Biol. 62485-500. [DOI] [PubMed] [Google Scholar]
- 13.Gaertig, J., and M. A. Gorovsky. 1992. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc. Natl. Acad. Sci. USA 899196-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, B. A., S. Joshi, E. C. Thomas, R. K. Chiotta, R. L. Diaz, S. A. Busb, P. C. Andrews, R. R. Ogorzalek Loo, J. Shabanowitz, N. L. Kelleher, C. A. Mizzen, D. Allis, and D. F. Hunt. 2006. Comprehensive phosphoprotein analysis of linker histone H1 from Tetrahymena thermophila. Mol. Cell Proteomics 9293-1609. [DOI] [PubMed] [Google Scholar]
- 15.Giardina, C., and J. T. Lis. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 152737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, A. J., J. M. McInerney, M. A. Glander, O. Pomerantz, and C. H. Lowrey. 2001. In vivo formation of a human beta-globin locus control region core element requires binding sites for multiple factors including GATA-1, NF-E2, erythroid Kruppel-like factor, and Sp1. J. Biol. Chem. 27626883-26892. [DOI] [PubMed] [Google Scholar]
- 17.Hargrove, B. W., A. Bhatacharyya, A. M. Domitrovich, J. M. Kapler, K. Kirk, D. E. Shippen, and G. R. Kunkel. 1999. Identification of an essential proximal sequence element in the promoter of the telomerase RNA gene of Tetrahymena thermophila. Nucleic Acids Res. 274269-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hightower, L. E., S. E. Sadis, and I. M. Takenaka. 1994. Interactions of vertebrate hsc70 and hsp70 with unfolded proteins and peptides, p. 179-207. In R. I. Morimoo, A. Tissieres, and C. Georgopoulos (ed.), The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 19.Howard-Till, R. A., and M. C. Yao. 2006. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol. Cell. Biol. 268731-8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang, Y. K., Y. H. Jin, Y. S. Shim, M. J. Kim, E. J. Yoo, I. S. Choi, J. S. Lee, R. H. Seong, S. H. Hong, and S. D. Park. 1996. Identification of the DNA damage-responsive elements of the rhp51+ gene, a recA and RAD51homolog from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 251167-175. [DOI] [PubMed] [Google Scholar]
- 21.Kiela, P. R., J. LeSueur, J. F. Collins, and F. K. Ghishan. 2003. Transcriptional regulation of the Rat NH3 gene functional interactions between GATA-5 and Sp family transcription factors. J. Biol. Chem. 2785659-5668. [DOI] [PubMed] [Google Scholar]
- 22.La Terza, A., C. Miceli, and P. Luporini. 2004. The gene for the heat-shock protein 70 of Euplotes focardii, an Antarctic psychrophilic ciliate. Antarctic Sci. 1623-28. [Google Scholar]
- 23.La Terza, A., G. Papa, C. Miceli, and P. Luporini. 2001. Divergence between two Antarctic species of the ciliate Euplotes, E. focardii and E. nobilii, in the expression of heat-shock protein 70 genes. Mol. Ecol. 101061-1067. [DOI] [PubMed] [Google Scholar]
- 24.Lowry, J. A., and W. R. Atchley. 2000. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 50103-115. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Martindale, D. W., C. D. Allis, and P. J. Bruns. 1982. Conjugation in Tetrahymena thermophila: a temporal analysis of cytological stages. Exp. Cell Res. 140227-236. [DOI] [PubMed] [Google Scholar]
- 27.Michiaki, Y., H. Kazonuri, and Kazuhiro. 2004. Characterization of multiple members of the HSP 70 family in platyfish culture cells: molecular evolution of stress protein HSP 70 in vertebrates. Gene 336207-218. [DOI] [PubMed] [Google Scholar]
- 28.Moilanen, K. H., T. Fukushige, and J. H. Freedman. 1999. Regulation of metallothionein gene transcription. J. Biol. Chem. 27429655-29665. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto, R. I. 1993. Cell in stress: transcriptional activation of heat shock genes. Science 2591409-1410. [DOI] [PubMed] [Google Scholar]
- 30.Muro-Pastor, M. I., R. Gonzalez, J. Strauss, J. Narendja, and Scazzocchio, C. 1999. The GATA factor AreA is essential for chromatin remodeling in a eukaryotic bidirectional promoter. EMBO J. 181584-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orias, E., E. P. Hamilton, and J. D. Orias. 2000. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62189-211. [DOI] [PubMed] [Google Scholar]
- 32.Pomeratz, O., A. J. Goodwin, T. Joice, and C. H. Lowrey. 1998. Conserved elements containing NF-E2 tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human (beta)-globin locus control region. Nucleic Acids Res. 265684-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray, S., Y. Lu, S. H. Kaufmann, W. C. Gustafson, J. E. Karpa, I. Boldogh, A. P. Fields, and A. R. Brasier. 2004. Genomic mechanisms of p210BCR-ABL signaling: induction of heat shock protein 70 through the GATA response element confers resistance to paclitaxel-induced apoptosis. J. Biol. Chem. 27935604-35615. [DOI] [PubMed] [Google Scholar]
- 34.Santoro, N., N. Johansson, and D. J. Thiele. 1998. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol. Cell. Biol. 186340-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. J., E. S. Cole, and D. P. Romero. 2004. Transcriptional control of RAD 51 expression in the ciliate Tetrahymena thermophila. Nucleic Acids Res. 324313-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song, X., and M. A. Gorovsky. 2007. Unphosphorylated H1 is enriched in a specific region of the promoter when CDC2 is downregulated during starvation. Mol. Cell. Biol. 271925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuart, R. K., and E. S. Cole. 2000. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62291-311. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana, T., S. Astumi, R. Shiod, M. Uenom, M. Uritani, and T. Ushimaru. 2002. A novel non-conventional heat shock element regulates expression of MDJ1 encoding a DnaJ homolog in Saccharomyces cerevisiae. J. Biol. Chem. 22722140-22146. [DOI] [PubMed] [Google Scholar]
- 39.Turkewits, A. P., E. Orias, and G. Kapler. 2002. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 1835-40. [DOI] [PubMed] [Google Scholar]
- 40.Vavra, K. J., C. D. Allis, and M. A. Gorovsky. 1982. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J. Biol. Chem. 2572591-2599. [PubMed] [Google Scholar]
- 41.Yamamoto, A., Y. Mizukami, and H. Sakuray. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factors in Saccharomyces cerevisiae. J. Biol. Chem. 28011911-11919. [DOI] [PubMed] [Google Scholar]
- 42.Yao, M. C., C. Yao, and B. Monks. 1990. The controlling sequence for site-specific chromosome breaking in Tetrahymena. Cell 63763-772. [DOI] [PubMed] [Google Scholar]
- 43.Yao, M. C., and C. H. Yao. 1989. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injection into developing nuclei of Tetrahymena thermophila. Mol. Cell. Biol. 91092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, G. L., and E. H. Blackburn. 1989. Transformation of Tetrahymena thermophila with a mutated circular ribosomal DNA plasmid vector. Genetics 868487-8491. [DOI] [PMC free article] [PubMed] [Google Scholar]