Abstract
Human astroviruses have been shown in numerous studies to be an important cause of gastroenteritis in young children worldwide. The present communication addresses their characterization by use of oligonucleotide microarray hybridization. The system developed consists of an RT-PCR using primers of low degeneracy capable of detecting all eight serotypes of human astroviruses. RT-PCR products are then hybridized against a microarray consisting of short oligonucleotide probes 17 to 18 nucleotides in length. Cy3-labeled ssDNA targets are generated using a Cy3-labeled primer in the RT-PCR. The non-labeled strand is enzymatically digested, and the labeled target is rescued by column purification. This method of generating labeled target uses equimolar concentrations of the amplifying primers and does not compromise assay sensitivity for initial detection of the virus. Hybridization can be performed without the need for additional amplification. Although the amplicon spans a relatively conserved region of the astrovirus genome, the use of short probes enables type distinction despite such limited diversity. Probes differing by as little as a single nucleotide can be used to distinguish isolates. The microarray developed was capable of distinguishing representatives of the eight known serotypes of human astroviruses.
Keywords: DNA oligonucleotide microarray, human astroviruses, RT-PCR
1. Introduction
Astroviruses are 28-35 nm diameter, icosahedral viruses that have a characteristic five- or six-pointed star-like surface structure when viewed by electron microscopy (Gk. astron, star). Along with the Picornaviridae and the Caliciviridae, the Astroviridae comprise a third family of nonenveloped viruses whose genome is composed of plus-sense, single-stranded RNA. In addition to humans, astroviruses have been isolated from numerous mammalian animal species (classified as genus Mammoastrovirus) and from avian species such as ducks, chickens, and turkey poults (classified as genus Aviastrovirus). The astrovirus genome is approximately 6.8 kb in length and consists of three open reading frames. ORF1a and ORF1b overlap and encode nonstructural proteins regulating virus replication. ORF2 encodes the capsid proteins. Human astroviruses have been grouped into 8 serotypes, and phylogenetic analysis using the amino acid sequences encoded by either ORF2 or ORF1b results in a clustering of genotypes that correlates with serotype (Belliot, et al., 1997; reviewed in Matsui and Greenberg, 2001).
Human astroviruses were first detected in 1975 using direct electron microscopy (EM) on fecal samples of infants with diarrhea (Appleton and Higgins, 1975; Madeley and Cosgrove, 1975). Astrovirus replication was first detected in cell culture using immunofluorescent antibody (IFA) techniques on infected primary human embryonic kidney cultures (Lee and Kurtz, 1981). The medical importance of astrovirus in humans was established initially in large-scale studies conducted in Thailand in 1991 (Herrmann, et al., 1991) where astroviruses were found by ELISA to be the second most common cause (after rotavirus) of viral diarrhea in young children. These studies and subsequent ones based on antigen detection were made possible by use of a monoclonal antibody ELISA able to detect all known human astrovirus serotypes (Herrmann, et al., 1988). This and other improvements in detection techniques, such as better cultivation techniques (Willcocks, et al., 1990) and the application of the reverse transcription polymerase chain reaction (RT-PCR) in numerous studies have been instrumental in defining the epidemiology of this illness worldwide (Cruz, et al., 1992; Dennehy, et al., 2001; Glass, et al., 1996; Glass, 2001; Kotloff, et al., 1992; Maldonado, et al., 1998; Mitchell et al., 1999; Unicomb, et al., 1998; Walter, et al., 2001). RT-PCR is the most sensitive detection method, and in a direct comparison of RT-PCR, ELISA, and EM to monitor an outbreak, it was found that many subclinical infections could only be detected by RT-PCR. Further, use of RT-PCR revealed that several patients were infected earlier and shed virus for longer than indicated by EM or ELISA (Cubitt, et al., 1999). Astroviruses have also been quantitated by real-time RT-PCR in both sewage (Le Cann, et al., 2004) and stool samples (Zhang, et al., 2006).
Knowledge of the molecular epidemiology of a virus is now considered to be a key step in understanding its burden in human health (Guix, et al., 2005). Although RT-PCR provides a very sensitive means of detecting astrovirus genomic material in feces, microarrays have been shown to be particularly useful as an adjunct to viral detection methods involving nucleic acid target amplification (Wang, et al., 2002). This communication describes RT-PCR of all eight serotypes of human astroviruses using a single set of primers of low degeneracy. Amplified products are identified to type by hybridization to a microarray consisting of short oligonucleotides. The use of short oligonucleotides as probes enables type identification using amplified sequences of limited diversity.
2. Materials and methods
2.1. Cell culture and virus strains
Viral isolates representative of the 8 known astrovirus serotypes were tested. Original seed viruses for astroviruses types 1-7 were obtained from Dr. John B. Kurtz, John Radcliffe Hospital, Oxford, England. These were passed four times in Caco-2 cells for use as stock viruses. Caco-2 cells were obtained from the American Type Culture Collection, Manassas, VA. The cells were grown in D-MEM medium with 10% fetal bovine serum added. Additionally, a stool sample containing astrovirus type 8 was obtained from Prof. Ian Brierley, University of Cambridge, Cambridge, U.K. This sample was used for the present study directly, as the virus was unable to be passaged in culture. For virus passage, cells were rinsed twice with serum-free D-MEM and inoculated with 100μl of original seed or passaged virus stocks at 37° C for one hour. The inoculum was removed and 1.0 ml of D-MEM containing 100 units penicillin, 100 μg streptomycin, 10 μg gentamicin, 1.0 μg amphotericin B, and 20 μg porcine trypsin 1:250 (Gibco BRL, Grand Island, N.Y) per ml was added. The porcine trypsin used contained a minimum of 225 USP U/mg BAEE units of activity.
Turkey astrovirus (TAstV1987) was obtained from Dr. Y. M. Saif, The Ohio State University, Wooster, OH. A stool sample containing a genogroup I norovirus has been described elsewhere (Herrmann, et al., 1985) and a stool sample containing a genogroup II norovirus was obtained from Dr. J. Vinje, Centers for Disease Control and Prevention, Atlanta, GA. Rotavirus SA-11 was obtained from the American Type Culture Collection, Rockville, MD.
2.2. Isolation of viral RNA
Viral RNA was purified from supernates of 10% fecal suspensions or cell cultures using Qiagen's QIAamp Viral RNA Mini Kit using the manufacturer's instructions.
2.3. Design of primers for RT-PCR
ClustalW analysis of astrovirus ORF1b genomic sequences was used to produce an alignment for subsequent primer selection. Primers were selected using Premier Biosoft International's Primer Premier Version 5.0. A selection bias for low degeneracy and an optimum annealing temperature in the range of 49°C to 52°C was applied to the search.
2.4. Microarray probe design
The design concept was based on single nucleotide polymorphism (SNP) probe designs. SNP-probes typically contain a centrally located single point of variation that allows discrimination based on length of contiguous stretches of nucleotide identity, typically 25 nucleotides for perfect-match and 12 nucleotides for mismatch. Since the Astrovirus serotype sequences do not differ from each other at either the same point or a single point, any probe designed for one serotype sequence will have variable asymmetric points of variation relative to the different family member sequences. Consequently, short (17 nucleotide) oligonucleotide probes were designed to three different regions within the astrovirus amplicon that contained different degrees of genetic variability among eight serological strains of the virus (see Fig. 1). This allows for the potential discrimination between perfectly matched hybridization domains of 17-contiguous nucleotides and mismatched hybridization domains of shorter contiguous stretches, ranging from 2 to 11 nucleotides. In two cases, two astroviruses had the same sequence at the selected probe design sites (astrovirus 1 & 5 at site 4, and astrovirus 2 & 4 at site 3), so the respective oligonucleotide probes were excluded from this study.
Fig. 1.

Sequences of the probes in the astrovirus oligonucleotide microarray. The probe layout is provided in Fig. 3.
2.5. Microarray production
Short array probes (17-18mers, see Figure 1) were each synthesized as a standard desalt purified oligonucleotide with a 5' I-Linker™ modification (Integrated DNA Technologies, Coralville, IA). The oligonucleotide probe set was then printed at 40uM in ESB, Epoxide Spotting Buffer, (Integrated DNA Technologies, Coralville, IA) on Corning Epoxide Slides using a BioRobotics MicroGrid 610 spotter equipped with Telechem 946MP3 pins. Each oligonucleotide probe was spotted in duplicate per array with each slide containing 12 replicate arrays in a format compatible with the 16-chamber mask of the Grace Bio-Labs ProPlate™ Multi-Array Slide System. Printed slides were then treated for 1 hour in a humidity chamber with 84% humidity followed by 1 hour of drying in a desiccator. The slides were stored at room temperature until ready to hybridize.
2.6. RT-PCR
The astrovirus RT-PCR was performed using Qiagen's OneStep RT-PCR Kit. The sense primer (5'-ACTGCCTRTCWCGGACTG-3') and a modified Cy3-labeled antisense primer (5'-Cy3-TGTGACACCYTGTTTCCT-3') were used at equimolar concentrations (final concentrations of 600nM each in a 30 μl reaction volume). Following reverse transcription at 50°C for 30 minutes, HotStarTaq DNA polymerase was activated by heating to 95°C for 15 min. Ten cycles of denaturation, annealing , and extension at 94°C, 51°C, and 72°C, respectively were followed by an additional 10 cycles at 93°C, 52°C, and 72°C, and a final 20 cycles at 93°C, 53°C, and 72°C. Amplification ended with a 10 minute extension at 72°C.
2.7. Preparation of single stranded Cy3-labeled astrovirus target cDNA
Following RT-PCR with the labeled antisense primer, single stranded Cy3-labeled targets were isolated using a SNP-Chip ssDNA Target Preparation protocol (Integrated DNA Technologies, Coralville, IA). Briefly, RT-PCR products were digested with a strand specific enzyme followed by column purification of the protected Cy3-labeled target strands using Promega ChipShot purification columns.
The strand specific digestion reaction consisted of 33 μl molecular biology grade water, 6 μl 10X digestion buffer, 1 μl enzyme (ten units), and 20 μl RT-PCR reaction product. Following a 2 hour incubation at room temperature, 6 μl sodium acetate (3M, pH 5.2) and 337.5 μl of binding solution were added to each 60 μl digestion reaction volume. Each mixture was gently mixed and applied to a Promega ChipShot purification column, incubated at room temperature for five minutes, and centrifuged at 10,000 × g for 1 minute. The flow-through was discarded, and the column was washed with 500 μl 80% ethanol. Following centrifugation at 10,000 × g for 1 minute, the flow-through was again discarded. The wash was repeated twice for a total of three washes. An additional centrifugation at 10,000 × g for 1 minute was performed to remove residual ethanol.
For elution of target, the column was placed in a clean 2 ml collection tube, and Cy3-labeled ssDNA was eluted by adding 60 μl of elution buffer to each column. After two minutes incubation at room temperature, the column was centrifuged at 10,000 × g for one minute. The eluted sample was dried down in a Speed-Vac. The dried Cy3-labeled ssDNA target was resuspended in 55 μl of 1X hybridization buffer, which consisted of 11 μl molecular biology grade water plus 44 μl 1.25X SNP Hybridization Buffer (Integrated DNA Technologies, Coralville, IA).
2.8. Microarray Hybridization
Prior to use, the slides were washed for 5 minutes with agitation using filtered, de-ionized water, rinsed for 1 minute in fresh water, and spun dry. The 16-chamber hybridization mask from the Grace Bio-Labs ProPlate™ Multi-Array Slide System was assembled onto the microarray slide. The resuspended Cy3-labeled ssDNA targets were heated for five minutes at 80°C and pulse spun. 25 μl of each target/hybridization mix was applied to a single well of the 16-chamber mask on the array slide, covered with plastic film, and hybridized for 2 hours 15 minutes at 50°C (in a humidity chamber in a water bath). The hybridization reaction was then removed by pipetting from each well. The hybridization mask was disassembled, and the slide immediately washed for 15 minutes in 200 of 1X SNP Wash Buffer 1 (Integrated DNA Technologies, Coralville, IA) that had been preheated to 50°C. The wash buffer was maintained at 50°C during the 15 minute wash. A second (200 ml 2X SSC buffer at room temperature) and third wash (200 ml 0.2X SSC buffer at room temp) were performed, and the slide was centrifuged at 1500 × g to remove excess fluid.
2.9. Scanning of the microarray
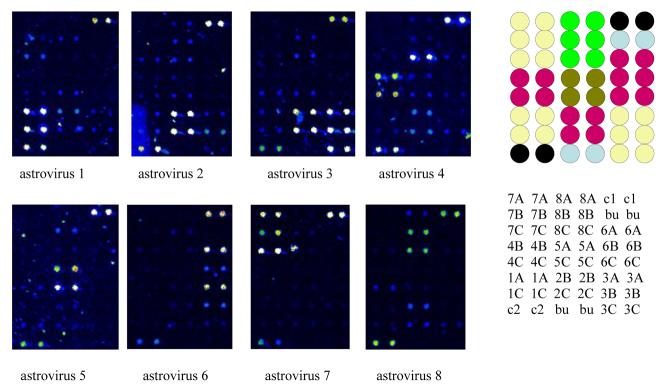
Hybridized slides were scanned using an Affymetrix 418 Scanner at an excitation wavelength of 532 nm and an emission wavelength of 570 nm. Laser power and gain for Figure 3 were 80% and 50%, respectively.
Fig. 3.
Oligonucleotide microarray for distinguishing the eight different types of human astrovirus. RT-PCR was performed using a single pair of primers at equimolar concentrations. The antisense primer was labeled with Cy3. The RT-PCR products were enzymatically digested to remove the unlabeled (and unprotected) sense strands, and the remaining labeled antisense targets were column purified and applied to the microarray consisting of predominantly 17mer positive sense probes. Duplicate dots in the upper right and lower left of each array are two conserved sequences in common to all the astroviruses. Type specific probes are clustered as two to three pairs of duplicate dots on the array. The pseudocolor scale used to indicate signal intensity is as follows: black < blue < green < yellow < orange < red < white. Locations of probes on the microarray are provided to the right of the scans. A = site 3. B = site 4. C = site 5. c1 = common sequence, site 1. c2 = common sequence, site 2. bu = buffer control.
2.10. Sequencing of astrovirus RT-PCR products
Astrovirus RT-PCR products were sequenced at the Tufts University Core Facility using an ABI 3100 automated DNA sequencer.
3. Results
3.1. Primers for RT-PCR
The primers used for RT-PCR were characterized by low degeneracy; the antisense primer contained one variable nucleotide and the sense primer contained two variable nucleotides. The specificity of the primers was demonstrated by their inability to amplify RNA sequences from a turkey astrovirus or from other RNA viruses commonly associated with gastroenteritis (rotavirus, genogroup I and II noroviruses) (data not shown). RT-PCR products in relation to astrovirus sequences of eight serotypes are shown in Fig. 2. An asterisk under the sequences being compared indicates conserved nucleotides.
Fig. 2.
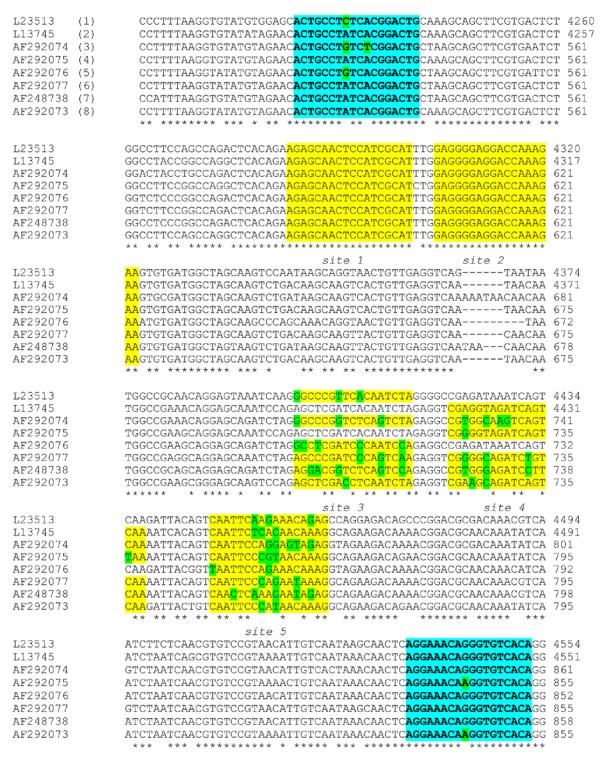
Astrovirus sequences from eight serotypes in the region amplified by the RT-PCR primers used for detection and generation of labeled targets for microarray hybridization. The GenBank accession number for each sequence is listed to the left. In parentheses are the serotype designations. Primers used for RT-PCR are indicated in aqua. Probe sequences at conserved sites (1 and 2) and sites used for type identification (3, 4, and 5) are indicated in yellow. Nucleotides that differ from the consensus are highlighted in green. Astroviruses 2 and 4 are identical at site 3, and astroviruses 1 and 5 are identical at site 4. Probes for these were not included in the microarray.
3.2. Probes
The probes used for microarray analysis were 17 nucleotides in length (type-specific probes) or 18 nucleotides in length (conserved sequence probes). Their relative positions in the microarray are shown in Fig. 3, and their location relative to the amplified RT-PCR products are shown in Figure 2.
3.3. Hybridization
In Figure 3, Cy3-labeled antisense targets obtained from amplification products of eight different serotypes of astrovirus were hybridized to the astrovirus microarray. Not all probes bound target efficiently. Probes for astroviruses 6 (site 4) and 8 (site 3) were associated with weak binding and thus were not useful for typing. However, distinct patterns of hybridization were obtained for each of the eight viruses. For astrovirus 3, substantial hybridization was observed with the two astrovirus 2 probes. Level of binding to the astrovirus 2 probes suggested potential cross contamination with an astrovirus 2 sequence. A repeat RT-PCR and hybridization resulted in the expected binding pattern to astrovirus 3-specific probes.
The astrovirus 4 target bound at high levels to the astrovirus 8, site 5 probe. Based on the GenBank sequences used for probe design, the site 5 probes for astroviruses 4 and 8 should have differed by a single nucleotide (5'-CAATTCCCGTAACAAAG-3' for astrovirus 4 versus 5'-CAATTCCCATAAACAAAG-3' for astrovirus 8). Sequencing of the RT-PCR products for astroviruses 1 through 7 revealed that all sequences at sites 3, 4, and 5 were as expected except for site 5 of the astrovirus 4 isolate. This isolate was identical to astrovirus 8 as a result of a change of a single nucleotide from G to A. Additional astrovirus 4 sequences listed with GenBank reveal more variability at this site, which will require the addition of more probe variants to the array. The astrovirus 8 labeled target bound as expected to the astrovirus 8, site 5 probe, demonstrating that a single nucleotide difference is sufficient to substantially impact target binding.
4. Discussion
RT-PCR provides a very sensitive means of detecting astrovirus genomic material. For maximum sensitivity, primer degeneracy should be kept to a minimum. This necessitates the targeting of conserved viral sequences in the initial amplification step. Such an approach lowers the level of primer degeneracy required to account for sequence variation. Conserved sequences, although essential for sensitive detection of a majority of viral isolates, can pose some problems for distinguishing different isolates using microarray analysis. By using a series of short probes, however, conserved regions of the genomes can still be sufficiently variable for characterization of enteric viruses. For example, Chizhikov, et al. (2002) and Lovmar, et al. (2003) used short probes (about 20 nucleotides in length) to successfully distinguish different rotavirus isolates.
Detection of enteric viral genomes in feces presents a particular challenge because of the great amount of genomic material present from the bacterial flora of the GI tract, from cells shed from the lining of the GI tract, and from ingested material. Non-specific amplification techniques suffer from a lack of sensitivity due to the amplification of non-target sequences, and are more appropriate for detection of genomic material in fluids such as CSF, serum, water, and possibly respiratory secretions in which the amounts of competing non-target sequences are limited. A single microarray for a comprehensive panel of pathogens coupled with a non-specific amplification technique, although potentially valuable for screening samples such as serum or CSF, is likely to suffer substantially in sensitivity in the presence of great excesses of non-target sequences, as would be present in feces. For most enteric viruses, fecal samples are the best source of virus, since most enteric viral infections remain localized.
A DNA microarray consisting of long oligonucleotides (70mers) has been described for the detection of a broad range of viral pathogens (Wang, et al., 2002). A related technology (reverse line blotting) has been reported for the characterization of human noroviruses (Vinje and Koopmans, 2000). An advantage of microarrays over line blotting is the smaller size of the microarrays, enabling the use of smaller quantities of samples and reagents and the inclusion of a greater number of probes. Jaaskelainen and Maunula (2006) reported the use of a microarray for the detection of noroviruses and astroviruses. Their assay for astroviruses consisted of reverse transcription followed by PCR amplification and subsequent generation of ssRNA transcripts. The RNA transcripts were applied to the microarray and primer extension with labeled nucleotides was used to label bound transcripts. This approach entails more manipulations, deals with the more labile RNA as the hybridization target material, and their results indicated that a majority of isolates (15 of 25) were untypable. Although a large number of isolates have not yet been tested with this system, all representatives of the eight serotypes were typable.
This diagnostic approach enables rapid detection and characterization of human astrovirus isolates. The assay can be performed using direct labeling of RT-PCR products with a single fluorophore per target molecule without the need for a second target amplification step or enzyme-based signal amplification. Enzymatic digestion of the non-labeled strand enables production of labeled ssDNA targets without compromising the optimum primer concentrations for initial detection of the virus (as would occur with asymmetric amplification procedures). Use of conserved primers for the initial RT-PCR should improve chances of detecting uncharacterized isolates. By using short nucleotides (17-mers) as probes in the oligonucleotide microarray, single nucleotide changes can be detected, thus improving chances of identifying isolates differing at the sites of the probe sequences. As more isolates are characterized, the microarray can be expanded to account for greater diversity as such diversity is encountered.
In conclusion, an RT-PCR, a target labeling system, and a microarray of short oligonucleotides for detection and characterization of human astroviruses has been developed. Use of short oligonucleotides offers a sensitive means of distinguishing closely related amplicons. Proof of principle was demonstrated with the current array distinguishing eight known serotypes of human astroviruses.
Acknowledgement
This work was supported by contract NIAID N01 AI30050.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Appleton H, Higgins PG. Viruses and gastroenteritis in infants [Letter] Lancet. 1975;1:1297. doi: 10.1016/s0140-6736(75)92581-7. [DOI] [PubMed] [Google Scholar]
- Belliot G, Laveran H, Monroe SS. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 1997;142:1323–1334. doi: 10.1007/s007050050163. [DOI] [PubMed] [Google Scholar]
- Brinker JP, Blacklow NR, Herrmann JE. Human astrovirus isolation and propagation in multiple cell lines. Arch. Virol. 2000;145:1847–1856. doi: 10.1007/s007050070060. [DOI] [PubMed] [Google Scholar]
- Chizhikov V, Wagner M, Ivanshina A, Hoshino Y, Kapikian AZ, Chumakov K. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 2002;40:2398–2407. doi: 10.1128/JCM.40.7.2398-2407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JR, Bartlett AV, Herrmann JE, Caceres P, Blacklow NR, Cano F. Astrovirus-associated diarrhea among Guatemalan ambulatory rural children. J. Clin. Microbiol. 1992;30:1140–1144. doi: 10.1128/jcm.30.5.1140-1144.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy PH, Nelson SM, Spangenberger S, Noel JS, Monroe SS, Glass RI. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 2001;184:10–15. doi: 10.1086/321007. [DOI] [PubMed] [Google Scholar]
- Glass RI, Noel J, Mitchell D, Herrmann JE, Blacklow NR, Pickering LK, Dennehy P, Ruiz-Palacios G, de Guerrero ML, Monroe SS. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch. Virol. 1996;(Suppl 12):287–300. doi: 10.1007/978-3-7091-6553-9_31. [DOI] [PubMed] [Google Scholar]
- Guix S, Bosch A, Pinto RM. Human astrovirus diagnosis and typing: current and future prospects. Lett. Appl. Microbiol. 2005;41:103–105. doi: 10.1111/j.1472-765X.2005.01759.x. [DOI] [PubMed] [Google Scholar]
- Herrmann JE, Hudson RW, Perron-Henry DM, Kurtz JB, Blacklow NR. Antigenic characterization of cell-cultivated astrovirus serotypes and development of astrovirus-specific monoclonal antibodies. J. Infect. Dis. 1988;158:182–185. doi: 10.1093/infdis/158.1.182. [DOI] [PubMed] [Google Scholar]
- Herrmann JE, Nowak NA, Blacklow NR. Detection of Norwalk virus by enzyme immunoassay. J. Med. Virol. 1985;17:127–133. doi: 10.1002/jmv.1890170205. [DOI] [PubMed] [Google Scholar]
- Herrmann JE, Taylor DN, Echeverria P, Blacklow NR. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 1991;324:1757–1760. doi: 10.1056/NEJM199106203242501. [DOI] [PubMed] [Google Scholar]
- Jaaskelainen AJ, Maunula L. Applicability of microarray technique for the detection of noro- and astroviruses. J. Virol. Meth. 2006;136:210–216. doi: 10.1016/j.jviromet.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Herrmann JE, Blacklow NR. The frequency of astrovirus as a cause of diarrhea in Baltimore children. Pediatr. Infect. Dis. J. 1992;11:587–589. [PubMed] [Google Scholar]
- Le Cann PL, Ranarijaona S, Monpoeho S, Le Guyader FL, Ferre V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 2004;155:11–15. doi: 10.1016/j.resmic.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Lee TW, Kurtz JB. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J. Gen. Virol. 1981;57:421–424. doi: 10.1099/0022-1317-57-2-421. [DOI] [PubMed] [Google Scholar]
- Lovmar L, Fock C, Espinoza F, Bucardo F, Syvanen A-C, Bondeson K. Microarrays for genotyping human group A rotavirus by multiplex capture and type-specific primer extension. J. Clin. Microbiol. 2003;41:5153–5158. doi: 10.1128/JCM.41.11.5153-5158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeley CR, Cosgrove BP. 28 nm particles in faeces in infantile gastroenteritis. Lancet. 1975;2:451–452. doi: 10.1016/s0140-6736(75)90858-2. [DOI] [PubMed] [Google Scholar]
- Maldonado Y, Cantwell M, Old M, Hill D, Sanchez ML, Logan L, Millan-Velasco F, Valdespino JL, Sepulveda J, Matsui S. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J. Infect. Dis. 1998;178:334–339. doi: 10.1086/515625. [DOI] [PubMed] [Google Scholar]
- Matsui SM, Greenberg HB. Astroviruses. In: Fields BN, Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2001. pp. 875–893. [Google Scholar]
- Mitchell DK, Matson DO, Jiang X, Berke T, Monroe SS, Carter MJ, Willcocks MM, Pickering LK. Molecular epidemiology of childhood astrovirus infection in child care centers. J. Infect. Dis. 1999;180:514–517. doi: 10.1086/314863. [DOI] [PubMed] [Google Scholar]
- Unicomb LE, Banu NN, Azim T, Islam A, Bardhan PK, Faruque AS, Hall A, Moe CL, Noel JS, Monroe SS, Albert MJ, Glass RI. Astrovirus infection in association with acute, persistent and nosocomial diarrhea in Bangladesh. Pediatr. Infect. Dis. J. 1998;17:611–614. doi: 10.1097/00006454-199807000-00007. [DOI] [PubMed] [Google Scholar]
- Vinje J, Koopmans MP. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 2000;38:2595–601. doi: 10.1128/jcm.38.7.2595-2601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JE, Mitchell DK, Guerrero ML, Berke T, Matson DO, Monroe SS, Pickering LK, Ruiz-Palacios G. Molecular epidemiology of human astrovirus diarrhea among children from a periurban community of Mexico City. J. Infect. Dis. 2001;183:681–686. doi: 10.1086/318825. [DOI] [PubMed] [Google Scholar]
- Willcocks MM, Carter MJ, Laidler FR, Madeley CR. Growth and characterisation of human faecal astrovirus in a continuous cell line. Arch. Virol. 1990;113:73–81. doi: 10.1007/BF01318354. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Mitchell DK, Afflerbach C, Jakab F, Walter J, Zhang Y-J, Staat MA, Azimi P, Matson DO. Quantitation of human astrovirus by real-time reverse-transcription-polymerase chain reaction to examine correlation with clinical illness. J. Virol. Methods. 2006;134:190–196. doi: 10.1016/j.jviromet.2006.01.009. [DOI] [PubMed] [Google Scholar]