Abstract
Conjugative transfer of the Enterococcus faecalis plasmid pCF10 is induced by the peptide pheromone cCF10 when recipient-produced cCF10 is detected by donors. cCF10 is produced by proteolytic processing of the signal sequence of a chromosomally encoded lipoprotein (CcfA). In donors, endogenously produced cCF10 is carefully controlled to prevent constitutive expression of conjugation functions, an energetically wasteful process, except in vivo, where endogenous cCF10 induces a conjugation-linked virulence factor. Endogenous cCF10 is controlled by two plasmid-encoded products; a membrane protein PrgY reduces pheromone levels in donors, and a secreted inhibitor peptide iCF10 inhibits the residual endogenous pheromone that escapes PrgY control. In this study we genetically determined the amino acid specificity determinants within PrgY, cCF10, and the cCF10 precursor that are necessary for cCF10 processing and for PrgY-mediated control. We showed that amino acid residues 125 to 241 of PrgY are required for specific recognition of cCF10 and that PrgY recognizes determinants within the heptapeptide cCF10 sequence, supporting a direct interaction between PrgY and mature cCF10. In addition, we found that a regulated intramembrane proteolysis (RIP) family pheromone precursor-processing protein Eep recognizes amino acids N-terminal to cCF10 in the signal sequence of CcfA. These results support a model where Eep directly targets pheromone precursors for RIP and PrgY interacts directly with the mature cCF10 peptide during processing. Despite evidence that both PrgY and Eep associate with cCF10 in or near the membrane, results presented here indicate that these two proteins function independently.
The gram-positive bacterium Enterococcus faecalis possesses mobile genetic elements that can rapidly spread antibiotic resistance and virulence determinants throughout bacterial populations (16, 41). A well-studied example of this is the pheromone-inducible family of conjugative plasmids that can be transferred to recipient cells at high rates upon induction by a recipient-produced oligopeptide pheromone signal (21). In this type of cell-cell signaling, communication occurs between two distinct cell types: donors (plasmid-harboring signal detectors) and recipients (plasmid-free signal producers). This is distinct from classic quorum sensing in which a homogeneous cell population serves as both the signal producer and the signal detector. In E. faecalis pheromone-inducible conjugation, donor-recipient communication allows donors to activate plasmid transfer only when recipient cells are in sufficiently close proximity for conjugation, thus minimizing the metabolic burden of the plasmid on its host cell.
The pheromone-inducible plasmids of E. faecalis have evolved a highly specific and sensitive response to peptide pheromones. These pheromones are small (7 to 8 amino acids), chromosomally encoded, and signal specifically to their cognate plasmid or family of closely related plasmids (20). Because donors produce pheromone, the plasmids have also evolved mechanisms of pheromone control that bypass a potential response to their own host cell's endogenously produced pheromone (11). The best-studied representatives of the pheromone-inducible plasmids are pCF10, pAD1, pPD1, and the genetically less-related plasmid pAM373.
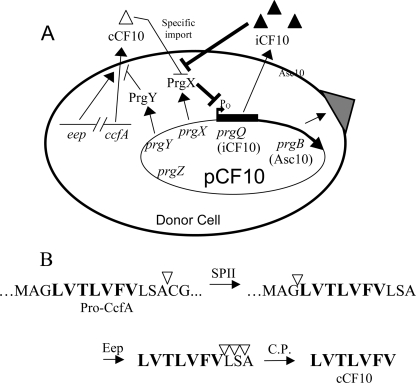
The steps of pheromone detection and response for pCF10 are outlined in Fig. 1A. The pCF10-specific pheromone cCF10 is bound at the cell surface by the pCF10-encoded PrgZ protein. After binding, cCF10 is imported into the cytoplasm via a chromosomally encoded oligopeptide permease ABC transporter (39). Inside the cytoplasm pheromone binds and destabilizes tetramers of a plasmid-encoded intracellular regulator PrgX (35, 47). This relieves PrgX-dependent repression of the prgQ promoter (PQ) in pCF10 and ultimately drives the expression of downstream conjugation functions (6). The upstream sequence of prgQ contains two weak binding sites for PrgX dimers, one of which overlaps PQ (5, 35). Protein-protein interactions between pairs of PrgX dimers occupying the binding sites results in the formation of a DNA loop, which favors cooperative binding of PrgX to both sites (35, 47). PrgX tetramer destabilization by cCF10 disrupts the DNA loop and thus abolishes PrgX-mediated repression at PQ (5). This ultimately results in expression of a large number of genes involved in conjugation (4, 6, 27, 35). One of these genes encodes a surface-expressed protein, Aggregation Substance (Asc10), which is involved in the formation of mating aggregates in broth culture (32). Asc10 also enhances E. faecalis virulence (40) and is induced in the absence of recipient cells in the mammalian bloodstream (28) by donor-produced pheromone that escapes plasmid control (13). While the genetic determinants for pheromone production and control have been identified, the molecular details of these processes are not completely understood.
FIG. 1.
(A) Model of pheromone response and control in a donor cell. cCF10 is imported into the cytoplasm, where it interacts with PrgX, causing derepression of PQ and subsequent induction of downstream conjugation genes. PrgY reduces endogenous cCF10, and iCF10 neutralizes remaining cCF10. (B) Predicted proteolytic processing of cCF10. cCF10 is processed from CcfA in several steps. SPII, signal peptidase II; Eep, membrane protease; C.P., carboxy peptidase. Triangles indicate cleavage sites.
pCF10 encodes two elements that control self-pheromone in donor cells; a membrane protein PrgY and an inhibitor peptide iCF10. PrgY reduces pheromone activity in both cell culture supernatants and cell wall fractions of donor cells (9), possibly by binding or degrading endogenous cCF10 as it is released from the donor cell. However, PrgY does not encode any known proteolytic or peptide interaction motifs and the possibility that PrgY interacts directly with cCF10 has not yet been explored. iCF10 is similar to cCF10 in size and hydrophobicity and competes with cCF10 for intracellular PrgX binding (36). Although iCF10 and cCF10 bind the same pocket of PrgX, they have different effects; iCF10 binding enhances PrgX tetramer formation and repression at PQ (36), whereas cCF10 binding has the opposite effect, as described above. The balance of iCF10 and cCF10 is carefully regulated so that iCF10 is produced at a level just sufficient to neutralize cCF10 in a donor cell (13). It is thought that iCF10 neutralizes cCF10 that escapes control by PrgY.
All pheromones analyzed to date appear to be encoded within the N-terminal signal sequences of predicted surface lipoproteins (17), and the inhibitor peptides are similarly encoded within signal sequence-like elements on the plasmid. All of the pheromone precursors have a cysteine residue downstream of the pheromone sequence that is within a known lipoprotein signal peptidase processing site (17). This cysteine residue is the site where signal peptidase II cleaves, liberating the signal peptide from the lipoprotein (18). In the case of the putative lipoprotein CcfA, whose signal peptide contains the mature cCF10 amino acid sequence, the cysteine residue is three amino acids downstream from the C terminus of the mature peptide and therefore may require further processing by a host exoprotease. The current working model for posttranslational processing of cCF10 is illustrated in Fig. 1B.
A putative membrane protease Eep has been shown to be involved in production of the cCF10, cAD1, and cPD1 pheromones (2). Eep is predicted to liberate the pheromone peptides from their cognate lipoprotein signal sequences by endoproteolytic cleavage (2). Eep is a predicted metalloprotease belonging to the site-2 protease (S2P) family of intramembrane-cleaving proteases (I-Clips) that cleave their substrate by regulated intramembrane proteolysis (RIP) (7). Because Eep appears to be a membrane protein, Eep-mediated cleavage of pheromone precursors is predicted to occur there and is possibly accompanied by active export of pheromone across the membrane to the extracytoplasmic environment. The prepeptides of pheromones cOB1 and cAM373 do not appear to be processed by Eep (2), implying the existence of an alternative processing system.
In this report, we describe the results of experiments that address cCF10 production and control in donor cells. We confirm and extend previous studies showing that iCF10 can neutralize endogenous cCF10 but is insufficient to completely control self-induction in donors. We genetically characterize a cCF10 specificity domain between amino acids 125 and 241 of PrgY, providing evidence for cCF10 interaction through this domain. We demonstrate that the amino acid sequence specificity determinants for PrgY recognition lie within the mature cCF10 peptide, whereas those involved in Eep recognition are located in the amino-terminal region of the signal peptide that comprises the cCF10 precursor. We also report that PrgY acts independently of Eep.
MATERIALS AND METHODS
Culture conditions.
E. faecalis was grown at 37°C in Todd-Hewitt broth (THB; Difco) or in M9-YE glucose medium (22), an M9-based medium supplemented with 0.3% yeast extract, 1% Casamino Acids, 0.1% glucose, 1 mM MgSO4, and 0.1 mM CaCl2. Antibiotics were used at the following concentrations: tetracycline at 10 μg/ml, erythromycin at 10 μg/ml, chloramphenicol at 20 μg/ml, rifampin at 200 μg/ml, spectinomycin at 1,000 μg/ml, kanamycin at 1,000 μg/ml, and fusidic acid at 25 μg/ml. For growth of strains overexpressing pheromone or inhibitor from the pDL278-derived vectors, antibiotics were used at 1/10 the above concentrations. Nisin (Sigma) was used at 25 ng/ml in broth. Synthetic iCF10 and cCF10 (Mimotopes, Australia) were dissolved at 50 μg of dimethyl formamide/ml and used at the indicated concentrations. Escherichia coli DH5α was used for cloning purposes and was grown in Luria-Bertani broth at 37°C with antibiotics at the following concentrations: erythromycin at 200 μg/ml, chloramphenicol at 20 μg/ml, carbenicillin at 50 μg/ml, kanamycin at 50 μg/ml, and spectinomycin at 50 μg/ml.
Bacterial strains.
Strains JRC107 and JRC110 were made by allelic exchange using conjugative delivery according to the method of Kristich et al. (37). Mobilizable and counterselectable donor plasmids were constructed for introducing gene deletions (in the case of eep, sprE, and gelE) or point mutations introducing stop codons (in the case of ccfA) into E. faecalis strain OG1RF. These donor plasmids were made from the pORI280-derived mobilizable vector carrying the counterselectable marker pheS* (33) and were transformed via electroporation into the donor strain CK111(pCF10-101) (37). Subsequently, the donor plasmid was transferred into strain OG1RF via conjugation and plated on selective medium to select for transconjugants harboring chromosomally integrated copies of the donor plasmid. These single-crossover strains were grown for approximately 20 generations at 37°C and plated on para-chlorophenyalanine to select for excision of the integrated plasmid. Colonies that grew in the presence of para-chlorophenyalanine were checked to verify that they did not acquire donor chromosomal DNA conferring resistance to 5-fluorouracil (encoded by upp) or to streptomycin and that they remained sensitive to the antibiotics fusidic acid and rifampin (Table 1). After marker verification, potential recombinant strains were screened for each strain-specific allele by phenotypic or PCR-based assays as described previously (37).
TABLE 1.
Strains used in this study
| Strain | Relevant characteristic or descriptiona | Source or reference |
|---|---|---|
| E. coli DH5α | E. coli cloning host | Invitrogen |
| E. faecalis | ||
| OG1RF | Rifr Far; derived from clinical strain OG1 | 21 |
| OG1Sp | Spectr | 37 |
| CK111 | Spectr 5-FUr; OG1Sp upp4::P23repA4 | 37 |
| TX5128 | GelE− SprE−; mγδ gelE insertion in OG1RF | 48 |
| JRC107 | cCF10-, GelE-, and SprE-deficient OG1RF derivative | 37 |
| JRC110 | Eep-deficient JRC107 derivative | 37 |
Rifr, rifampin resistance; Far, fusidic acid resistance; Spectr, spectinomycin resistance; 5-FUr, 5-fluorouracil resistance.
DNA manipulation.
Plasmids were isolated with a Qiagen miniprep kit as recommended by the manufacturer. Digested DNA products were purified by using a Qiaquick gel extraction kit (Qiagen), and PCR products were purified using a Qiaquick PCR purification kit (Qiagen). Plasmid constructs were verified using restriction enzyme digestion. Restriction enzymes were purchased from Promega and New England Biolabs. PCR was performed with a Perkin-Elmer Gene Amp PCR system or an Eppendorf Mastercycler using either BioXact DNA polymerase (Bioline), Platinum Pfx polymerase (Invitrogen), or Platinum Taq polymerase (Invitrogen). All sequencing was done by the Advanced Genetic Analysis Center at the University of Minnesota. Primers were purchased from Integrated DNA Technologies, Inc. E. faecalis was transformed with DNA as described previously (5). For all plasmids constructed using a PCR-generated fragment, the DNA was sequenced to confirm that no mutation was incorporated by PCR.
Plasmids.
The plasmids used in the present study are listed in Table 2. Plasmids pPCR4 and pJRC2, -3, and -4 were constructed using the gram-positive shuttle vector pDL278 (23). pJRC2, -3, and -4 were constructed from pPCR4 (45). pJRC2 and pJRC3 were made by overlap extension PCR essentially as described by Horton et al. (29). Upstream and downstream amplicons with overlapping termini were separately amplified by PCR and then combined in equimolar proportions and used as a template for overlap extension PCR, which spliced them together. The upstream amplicon for both pJRC2 and pJRC3 was amplified from plasmid pPCR4 (45) and spanned the prgQ promoter and upstream multicloning site in pPCR4. The downstream amplicon was amplified from chromosomal E. faecalis DNA and spanned the signal sequence encoding the pheromone cAM373 in the camE gene (25). For pJRC3, the 3′ primer for the downstream amplicon included base changes in the coding sequence for cAM373 pheromone so that this region instead encoded cCF10. These upstream and downstream amplicons were used as a template for overlap extension PCR, and the resulting PCR-amplified product was cloned into pGEM-T-Easy (Promega, Madison, WI) to make pGEM pJRC2 or pGEM pJRC3. pJRC2 and pJRC3 were constructed by excising the ∼250-bp fragments from pGEM pJRC2 or pGEM pJRC3 with EcoRI and cloning them into EcoRI digested pDL278. Both fragments contained an EcoRI site internal to the cloned piece at the 5′ end and an EcoRI site 10 bases external to the cloned piece at the 3′ end within pGEM-T-Easy. The resulting pJRC2 and pJRC3 constructs were screened for the same insert orientation as that of pPCR4. For pJRC3, none were found of the 12 that were screened, and a clone was picked that has the opposite insert orientation from pPCR4 and pJRC2 in the multicloning site of pDL278. Because expression of the cAM373 pheromone in pJRC2 is driven by the adjacent prgQ promoter and not a vector-supplied promoter, we believe the orientation of the insert has no effect on expression levels of the cAM373 pheromone. Plasmid pJRC4, bearing modification in the cCF10 coding region of pPCR4 that resulted in iCF10 expression, was constructed by using the QuikChange mutagenesis strategy (Stratagene) with pPCR4 as the template. The nucleotide sequence of the prgQ promoter and peptide coding sequence of the resulting pJRC4 plasmid was determined to verify the desired changes to the cCF10 coding region.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic or descriptionc | Source or reference |
|---|---|---|
| pCF10 | cCF10-inducible conjugative plasmid | 19 |
| pCF10-101 | pCF10 ΔoriT2 | 49 |
| pPD1 | cPD1-inducible conjugative plasmid | 52 |
| pBK2 | pCI3340 vector containing prgX-Q region of pCF10 with a pheromone-inducible lacZ reporter gene | 34 |
| p043lacZ | pAT18 vector containing prgX-Q region of pCF10 with a pheromone-inducible lacZ reporter gene | 35 |
| pDL278 | Gram-positive cloning shuttle vector; Spectr | 23 |
| pPCR4a | cCF10 expression construct derived from prgQ region cloned into pDL278 | 45 |
| V2L | V→L change at position 2 of cCF10 | 24 |
| T3S | T→S change at position 3 of cCF10 | 24 |
| L4V | L→V change at position 4 of cCF10 | 24 |
| L4I | L→I change at position 4 of cCF10 | 24 |
| F6Y | F→Y change at position 6 of cCF10 | 24 |
| V7I | V→I change at position 7 of cCF10 | 24 |
| pJRC2 | pPCR4 construct altered to express cAM373 signal peptide and pheromone from prgQ promoter | This study |
| pJRC3 | pJRC2 construct altered to express cCF10 | This study |
| pJRC4 | pPCR4 construct altered to express native iCF10 peptide | This study |
| pGEM-T-Easy | AmprlacZ cloning vector | Promega |
| pGEM pJRC2 | pGEM-T-Easy containing ∼250-bp PCR product used for cloning pJRC2 | This study |
| pGEM pJRC3 | pGEM-T-Easy containing ∼250-bp PCR product used for cloning pJRC3 | This study |
| pDL276 | Gram-positive cloning shuttle vector; Kanr | 23 |
| pMSP6043 | prgN, -O, -P, -W, -Z, and -Y cloned into pDL276 | 26 |
| pMSP6049 | prgN, -O, -P, -W, and -Y cloned into pDL276 | 26 |
| pMSP6043-1 | prgN, -O, -P, -W, and -Z cloned into pDL276 | 12 |
| pMSP6043-2 | prgN, -O, -P, and -W cloned into pDL276 | This study |
| pMSP3545 | Nisin-inducible cloning vector | 8 |
| pMSP3545Y | prgY cloned into pMSP3545 | This study |
| pMSP3545Sb | Spectr derivative of pMSP3545 | 12 |
| pMSP3545S-1 | Cloned E. faecalis pCF10 prgY | 12 |
| pMSP3545S-2 | Cloned E. faecalis pPD1 traB | 12 |
| pB/Y1 | Fusion of prgY after pPD1 traB base 726 | This study |
| pB/Y2 | Fusion of pPD1 traB after prgY base 723 | This study |
| pB/Y3 | Fusion of prgY after pPD1 traB base 234 | This study |
| pB/Y4 | Fusion of prgY after pPD1 traB base 375 | This study |
| pB/Y5 | Fusion of pPD1 traB after prgY base 960 | This study |
| pB/Y6 | Fusion of pPD1 traB after prgY base 1050 | This study |
| pB/Y9 | pPD1 traB fusion between bases 234 and 375 of prgY | This study |
| pB/Y10 | pPD1 traB fusion between bases 375 and 1098 of prgY | This study |
Constructs expressing mutant cCF10 peptides V2L, T3S, L4V, L4I, F6Y, and V7I were made by oligonucleotide-directed random mutagenesis of the cCF10 coding sequence of the pPCR4 construct, and their construction is described by Fixen et al. (24).
Genes and chimeric gene fusions in pMSP3545S-1 and -2 and B/Y1-10 were cloned into NcoI and XbaI of pMSP3545S.
Spectr, spectinomycin resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Plasmids pMSP6043, pMSP6049, pMSP6043-1, and pMSP6043-2 were constructed by using the gram-positive shuttle vector pDL276 (23). pMSP6043 encodes the pCF10 region including prgN, -O, -P, -W, -Z, and -Y (26). pMSP6049 encodes the same region but has a spectinomycin resistance cassette cloned into the middle of prgZ, resulting in abolished PrgZ expression (26). Plasmids pMSP6043-1 encoding prgN, -O, -P, -W, and -Z (12) and pMSP6043-2 encoding prgN, -O, -P, and -W were derived from pMSP6043. pMSP6043-2 was made by ligating the EcoRI and PstI fragment from prgN through the middle of prgZ from pINY6023 (46) into the EcoRI and PstI sites of pDL276 (23) and is in the same orientation as pMSP6043-1. pMSP6043-1 and pMSP6043-2 were checked by restriction digestion and Western blotting and found to express PrgZ but no detectable PrgY (in the case of pMSP6043-1) and neither PrgZ nor PrgY (in the case of pMSP6043-2) (data not shown).
Plasmids pMSP3545Y, pMSP3545S, and pMSP3545S-1 and -2, and pB/Y1-10 were made from pMSP3545, a nisin-inducible expression vector (8). pMSP3545S, pMSP3545S-1, and pMSP3545S-2 were constructed as described previously (12), pMSP3545Y was constructed by cloning PCR-amplified prgY into the XbaI/NcoI sites of pMSP3545, and pB/Y1 to pB/Y10 were constructed using overlap extension PCR essentially as described by Horton et al. (29) (Table 2). For pB/Y1 to pB/Y10, the coding sequences for the desired portions of pPD1 traB and pCF10 prgY were each PCR amplified using primer pairs that resulted in products with overlapping termini. These PCR products were combined in equimolar proportions and used as a template for overlap extension PCR to splice them together. For overlap extension PCR, five cycles were done without primers to allow the overlapping termini to anneal for extension of the recessed termini via Pfx polymerase. After five cycles, primers corresponding to the DNA sequence of the 5′ and 3′ ends of the desired chimeric gene product with flanking NcoI/XbaI sites were added, and the PCR was continued for an additional 35 cycles. The resulting amplified chimeric PCR product was subsequently purified, digested with NcoI/XbaI, and ligated into pMSP3545S cut with the same restriction enzymes.
Determination of pheromone activity in cell culture supernatants.
Cell culture supernatants were prepared for pheromone activity assays by first growing strains overnight at 37°C without shaking in THB with appropriate antibiotics and 25 ng of nisin/ml. Overnight cultures were immediately harvested (for strains overexpressing pheromone from the pDL278-derived plasmids) or diluted 1:5 or 1:10 in fresh THB plus 25 ng of nisin/ml and grown an additional 4 to 6 h before harvesting (for strains expressing pheromone from its native promoter). The optical density at 600 nm was determined for each sample, and equivalent cell volumes were harvested by centrifugation at 8,000 rpm (Beckman J2-21 centrifuge; JA20 rotor) for 10 min. For measuring pheromone from native promoters, the supernatant from each sample was autoclaved for 15 min at 121°C and 15 lb/in2. Prepared supernatants were either immediately assayed for pheromone activity or kept at 4°C for no more than 24 h before the pheromone activity was measured.
The pheromone activity from cell culture supernatants was determined one of two ways. High levels of pheromone activity produced from plasmids pJRC3 or pPCR4 were determined by β-galactosidase assay using OG1RF(p043lacZ) as an indicator strain. Cell culture supernatants of overnight cultures grown in THB plus 25 ng of nisin/ml was added to exponentially growing OG1RF(p043lacZ) indicator cultures in M9-YE medium and incubated for 1.5 h before the β-galactosidase activity was determined. Cell culture supernatants were added to indicator cultures at a 1:100 dilution (for three cCF10 variants with low activity [V2L, L4V and F6Y]), at a 1:400 dilution (for strains harboring pJRC3), or at a 1:200 dilution (for all other strains). All other cell culture supernatants were subjected to a clumping assay (9) to detect pheromone activity. Briefly, twofold serial dilutions of prepared supernatant samples were made in M9-YE medium in a round-bottom-well microtiter plate. A total of 10 μl of a 15-h OG1RF(pCF10) indicator culture was added to each well. Samples were incubated shaking at 37°C for 2 h, and positive clumping reactions were scored. The titer is reported as the reciprocal of the highest dilution that demonstrated a positive clumping reaction. The results reported are representative of at least two independent experiments done on separate days.
Determination of inhibitor activity in cell culture supernatants.
To determine inhibitor activity, cell culture supernatants were prepared as for determination of pheromone activity. Inhibitor assays were done as described previously (30) with some modifications. A known concentration of synthetic cCF10 (50 ng/ml) was serially diluted (1:2) through 100-μl aliquots of the culture filtrate being tested for inhibitor activity, and 10 μl of responder cells was added to each well before incubation at 37°C with shaking. The inhibitor activity was measured as the extent of the reduction of the pheromone titer relative to a parallel control when broth was substituted for culture filtrates. The inhibitor titer was therefore defined as the log2 of the number of wells that clumped in the presence of the culture filtrate subtracted from the log2 of the number of wells that clumped in the absence of the culture filtrate.
β-Galactosidase assay.
Strains were grown overnight in M9-YE medium and then diluted 1:5 into fresh M9-YE medium and grown an additional 1 h before addition of iCF10 or supernatant from a cCF10-expressing strain grown overnight in THB. After induction the cultures were incubated for an additional 90 min. β-Galactosidase assays were performed as described previously (15). The results were averaged from technical duplicates and are shown as representative experiments or as averaged results from two or more independent experiments that were each done in duplicate (specified in the figure legends).
RESULTS
iCF10 is insufficient to neutralize endogenous cCF10 that is reimported by PrgZ.
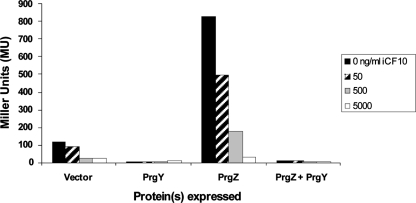
PrgY and iCF10 are two distinct gene products that control endogenously produced cCF10 activity. In a donor strain harboring a prgY disrupted pCF10 derivative, iCF10 cannot control self-induction of donor aggregation (9). A more sensitive method to detect self-induction by cCF10 was utilized to determine whether iCF10 could independently control some endogenous pheromone in the absence of PrgY. This was done by using the reporter plasmid pBK2. pBK2 carries the pCF10 regulatory genes encoding PrgX and iCF10 and also has a prgQ::lacZ fusion that sensitively reports cCF10 induction (34). As observed previously for a strain harboring prgY-disrupted pCF10 (14), a strain harboring pBK2 constitutively expressed prgQ-dependent reporter gene activity due to self-induction by endogenous cCF10 (Fig. 2, Vector, black bar). Exogenously added iCF10 decreased this basal level of induction significantly (Fig. 2, Vector), indicating that iCF10 can control self-cCF10 independent of PrgY. We also used this system to address whether the pCF10-encoded surface-expressed pheromone-binding protein PrgZ affects donor cell sensitivity to self-cCF10. PrgZ significantly increases pCF10 sensitivity to cCF10 (39); thus, we hypothesized that PrgZ expression would also increase sensitivity to self-cCF10 even when iCF10 is exogenously added. Indeed, PrgZ expression increased pBK2 reporter activity ∼5-fold in the absence or presence of 50 or 500 ng of iCF10/ml (Fig. 2, PrgZ). Because PrgZ protein levels may be higher in this system (due to its expression from a multicopy cloning vector) than in wild-type donor cells, the PrgZ-dependent increase in cCF10 sensitivity may not be as high as in cells carrying pCF10. However, these results support that PrgZ expression increases donor sensitivity to self-cCF10 and that iCF10 is insufficient to control self-cCF10 that is reimported by PrgZ. iCF10. PrgY expression reduced reporter activity levels nearly to zero even when PrgZ was coexpressed (Fig. 2, PrgY and PrgY+PrgZ). The addition of the solvent dimethyl formamide used for dissolving iCF10 and cCF10 did not significantly affect induction levels (data not shown). These results demonstrate that iCF10 functions independent of PrgY but is not sufficient to control high levels of PrgZ-dependent cCF10 activation. Therefore, both PrgY and iCF10 are necessary for the control of endogenous pheromone in the presence of PrgZ.
FIG. 2.
iCF10 is insufficient to control endogenous pheromone in the absence of PrgY. Pheromone induction by endogenous pheromone was measured in OG1RF(pBK2) strains coharboring a plasmid expressing PrgZ+PrgY (pMSP6043), PrgY (pMSP6049), PrgZ (pMSP6043-1), or a vector control (pMSP6043-2). Cultures grown overnight in M9 medium were diluted 1:5, and β-galactosidase levels were determined after 2.5 h of growth. iCF10 was added at 0, 50, 500, or 5,000 ng/ml to overnight cultures and again to diluted cultures. The data shown are representative of three independent experiments, each performed in duplicate.
The membrane protease Eep is involved in both cCF10 and iCF10 production.
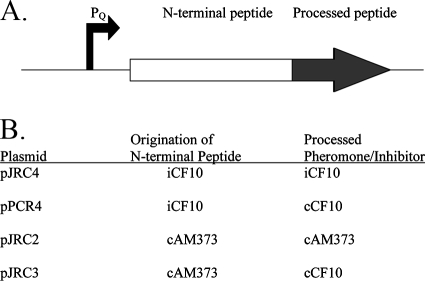
cCF10 is processed from the 22-amino-acid signal sequence of the chromosomally encoded CcfA lipoprotein (3) (Fig. 1B). Eep was previously shown to be involved in production of the pheromones cCF10, cPD1, cAD1, and the inhibitor iAD1 but was not involved in processing of the pheromone cAM373 (1, 2). To test whether Eep is also involved in processing iCF10, a strain was made with a disruption in eep using markerless allelic exchange (see Materials and Methods). Plasmid pJRC4 was constructed that carries the PQ-driven prgQ open reading frame (ORF) encoding iCF10 (Fig. 3), and this plasmid was moved into strain JRC110 to test for iCF10 production in the absence of Eep. iCF10 production from pJRC4 was undetectable in strain JRC110,compared to a titer of 32 in the Eep+ strain JRC107 (Table 3). A similar plasmid, pPCR4, encodes cCF10 within the 3′ end of the prgQ Orf (45), and pPCR4-produced cCF10 activity was similarly much higher in the Eep+ strain than in the Eep− strain (Table 3). These results show for the first time that Eep is involved in iCF10 production from the prgQ Orf. Attempts to clone PQ driven cCF10 and its native N-terminal peptide (the CcfA signal sequence) were unsuccessful, possibly due to a toxic effect of such a high level of cCF10 expression by this construct; however, in the Eep− strain JRC110 cCF10 activity from chromosomally encoded CcfA was undetectable (data not shown). Of note, JRC110(pPCR4) exhibited a low level of cCF10 production (Table 3), suggesting that another protease in this Eep-deficient strain may mediate processing of cCF10 at a greatly reduced efficiency.
FIG. 3.
PQ-driven peptide expression constructs used in these studies. (A) Diagram of the peptide-coding portion of the plasmids. Transcription of each peptide is driven by the prgQ promoter (PQ), shown at the left. Within the 22- to 23-amino-acid peptide of each plasmid is an N-terminal peptide sequence derived from the iCF10 or cAM373 preprocessed sequence (N-terminal peptide) and a pheromone or inhibitor at the C-terminal end (in gray). (B) N-terminal peptide and pheromone/inhibitor that is encoded within each plasmid. For details regarding the construction of these plasmids, see Materials and Methods.
TABLE 3.
Eep involvement in the production of iCF10, cCF10, and cAM373
| Straina | Eep | Terminusb
|
Titerc | |
|---|---|---|---|---|
| N | C | |||
| JRC107(pJRC4) | + | iCF10 | iCF10 | 32 |
| JRC110(pJRC4) | - | iCF10 | iCF10 | <1 |
| JRC107(pPCR4) | + | iCF10 | cCF10 | 4,096 |
| JRC110(pPCR4) | - | iCF10 | cCF10 | 64 |
| JRC107(pJRC2) | + | cAM373 | cAM373 | 8,192 |
| JRC110(pJRC2) | - | cAM373 | cAM373 | 8,192 |
| JRC107(pJRC3) | + | cAM373 | cCF10 | 8,192 |
| JRC110(pJRC3) | - | cAM373 | cCF10 | 8,192 |
Strain JRC107 is cCF10 and GelE/SprE deficient but Eep positive (+); strain JRC110 is derived from JRC107 but is Eep deficient (−). See Materials and Methods for details.
The N terminus of the peptide is native to either the iCF10 or the cAM373 propheromone sequence and is designated as such (see Fig. 2 and 3). The C terminus of the peptide is the active portion and is either cCF10, iCF10, or cAM373, as designated (see Fig. 2 and 3).
Supernatants (grown 6 h from a 10% overnight inoculum induced) were diluted twofold, and cCF10 or cAM373 activity is reported as the inverse of the largest dilution that was able to aggregate an OG1RF(pCF10) or OG1RF(pAM373) indicator strain. The iCF10 activity is reported as the extent of reduction of the ability to aggregate OG1RF(pCF10) relative to a parallel 50-ng/ml cCF10 control (see Materials and Methods). The 50-ng/ml control had a cCF10 titer of 512, and culture filtrates with iCF10 activity were diluted 1:50 in THB prior to determination of the iCF10 activity. The relative activities are representative of at least two independent experiments. A titer of <1 indicates that no active pheromone or inhibitor was observed at the dilutions examined.
Eep functions posttranslationally through recognition of a precursor peptide sequence N-terminal to the mature pheromone or inhibitor sequence.
With the exception of Eep, S2P proteases generally are involved in the cleavage and release of membrane-bound transcription factors (50). Thus, it is formally possible that Eep could regulate such a factor and thereby control the activity of promoters for genes encoding pheromones and inhibitors. To determine whether Eep could act indirectly on pheromone production by cleavage of a transcription factor regulating expression of the pheromone, we made use of the prgQ promoter (PQ) driving pheromone expression in plasmid pPCR4. If this promoter was regulated by Eep-mediated signaling, then any gene expressed from PQ would show similar changes in expression in the eep knockout strain. A previous study demonstrated that Eep is not involved in the production of cAM373 (2); therefore, plasmid pJRC2 was constructed, which encodes the cAM373 pheromone and signal sequence downstream of PQ (Fig. 3). Plasmid pJRC2 was moved into strains JRC107 and JRC110 to determine whether Eep is involved in cAM373 production when it is expressed from the PQ promoter. cAM373 production from plasmid pJRC2 was the same for both the Eep+ and Eep− strains (Table 3), indicating that Eep acts posttranscriptionally and supporting the notion that Eep functions as a protease that directly processes pheromone/inhibitor precursors.
To elucidate the peptide sequence recognized by Eep, a chimeric construct was made to determine whether the N or C terminus of the peptide substrate is important for Eep processing. Plasmid pJRC3 was derived from pJRC2, but the cAM373 pheromone sequence was changed to encode cCF10 (Fig. 3). The production of cCF10 from pJRC3 was Eep independent (Table 3), whereas cCF10 production from an iCF10-derived N-terminal peptide was greatly enhanced by Eep (pPCR4, Table 3). This demonstrates that the N-terminal sequence of the signal peptide is important for recognition by Eep.
PrgY recognizes cCF10 peptide sequence regardless of sequence N-terminal to cCF10.
Like Eep, PrgY specifically recognizes endogenously produced pheromone (12). The potential specificity determinants recognized by PrgY are currently unknown. To elucidate whether PrgY targets the cCF10 heptapeptide or adjacent amino acids that are part of the precursor peptide, the PQ-driven peptide expression constructs pPCR4 and pJRC3 (Fig. 3) were coexpressed with PrgY or a vector control, and the PrgY ability to decrease pheromone activity in each strain was measured. Because PQ-driven pheromone activity is very high in cell culture supernatants (Table 3) the effect of PrgY expression on this pheromone activity was difficult to measure using clumping of an OG1RF(pCF10) strain as a biological assay. Therefore, a more sensitive and quantitative assay was developed. We used a cCF10-inducible β-galactosidase reporter construct p043lacZ (4) for measuring pheromone activity in the cell culture supernatants (see Materials and Methods).
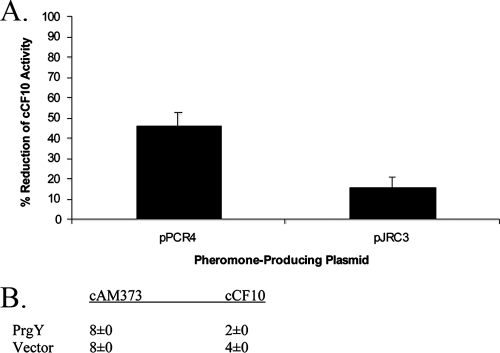
In strains harboring pPCR4 and pJRC3, cCF10 is processed from a non-native peptide sequence. To determine whether PrgY targets cCF10 produced from non-native signal sequences, we measured the cCF10 activity in strains coharboring pPCR4 or pJRC3 with pMSP3545Y (expressing PrgY) or pMSP3545 (vector). PrgY reduced the activity of cCF10 produced by both pPCR4 and pJRC3 (Fig. 4A). It is unclear why PrgY more efficiently reduced pPCR4-produced cCF10 than pJRC3-produced cCF10. Possibly, PrgY targets the former better than the latter. Alternatively, PrgY function could be saturated in these strains due to very high cCF10 production. In any case, these results support that PrgY targets the cCF10 heptapeptide and not the N-terminal amino acids of the cCF10 precursor. PrgY did not reduce cAM373 activity when it was produced endogenously from its native signal sequence (Fig. 4B), whereas PrgY can decrease endogenous cCF10 activity by ≥50% (Fig. 4B) (12).
FIG. 4.
(A) PrgY reduces the activity of cCF10 processed from both the iCF10 and the cAM373 signal peptides. Supernatant from JRC107 or JRC110 (Eep−) strains harboring pPCR4 or pJRC3 and pMSP3545Y (expressing PrgY) or pMS3545 (vector) were used to induce exponentially growing OG1RF(p043lacZ) strains, and the ability of supernatant cCF10 to induce LacZ expression of this strain was measured by β-galactosidase assay and expressed in Miller units. The PrgY-dependent percent reduction of cCF10 activity (+PrgY/Vector) was averaged from three independent experiments (error bars represent the standard deviation of three independent experiments). (B) PrgY does not decrease endogenous cAM373 activity. Cell culture supernatants prepared as for Table 3 were diluted twofold, and the pheromone activity is represented as the inverse of the largest dilution able to aggregate an OG1RF(pCF10) or OG1RF(pAM373::pAD2) indicator strain. The titers represent the results of two independent experiments; the same results were seen for both assays.
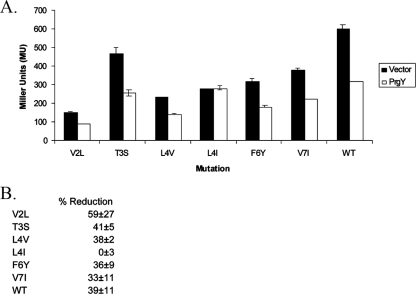
The fourth residue of the cCF10 peptide is important for PrgY recognition.
To further define the PrgY-targeted specificity determinants within the seven amino acids that comprise cCF10, we made use of a library of functional cCF10 variants (24). These variants were identified in a previous study using random oligonucleotide-directed mutagenesis of plasmid pPCR4 (24). The resulting peptide variants were screened for reduced ability to induce expression of pCF10 conjugation machinery [measured by clumping of OG1RF(pCF10)]. Although 14 amino acid substitutions were originally identified in the screen that influence biological activity of cCF10, only peptide variants that retained relatively high biological activity as inducing pheromones were tested for PrgY recognition to ensure that PrgY-dependent reductions in activity would be measurable. We tested cCF10 peptide variants with the mutations V2L, T3S, L4V, L4I, F6Y, and V7I, representing changes in most of the amino acids of cCF10 (amino acid substitutions at positions 1 and 5 were not tested here due to their low pheromone activity). Strains were made expressing each of these peptide variants with PrgY or a vector control, and the PrgY-dependent reduction of peptide activity was measured by using a β-galactosidase reporter assay to measure the pheromone activity in culture supernatants. The results demonstrate that PrgY reduced the supernatant activity of each peptide variant except one with an L4I substitution (Fig. 5). This L4I peptide variant had the same activity in supernatants of PrgY+ and PrgY− strains, suggesting that this sequence change abolished targeting by PrgY. These results also support that PrgY interacts with the cCF10 peptide.
FIG. 5.
PrgY targets L4 of cCF10. Wild-type (WT) cCF10 peptide from pPCR4 or cCF10 variants were coexpressed with PrgY (pMSP3545Y) or a vector control (pMSP3545). The peptide produced by each strain during overnight growth in THB was collected in the supernatant and used to induce an exponentially growing responder strain OG1RF(p043lacZ) for 1.5 h, and the β-galactosidase activity was subsequently determined. Cell culture supernatants were added to the responder strain at a 1:100 dilution (for three cCF10 variants with low activity [V2L, L4V, and F6Y]) or at a 1:200 dilution (for all others). (A) Absolute values and (B) the PrgY-dependent percent reduction of cCF10 activity are shown. For both panels A and B, the error range represents the standard deviation of two independent experiments. MU, Miller units.
Amino acids 125 to 241 of PrgY comprise a domain specific for cCF10 interaction.
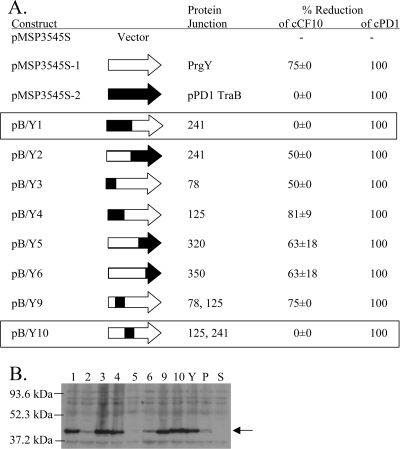
Our previous genetic data indicate that the functional domain for PrgY lies within a predicted extracellular domain including the first 150 amino acids of the 317-residue polypeptide. We believe that the primary function of the C-terminal segment (residues 250 to 317) is to anchor the protein with the proper topology in the cell membrane. Previous studies suggest that PrgY recognition of cCF10 is specific (12); thus, we hypothesized that there is also a domain in PrgY that mediates cCF10 specificity. Such findings would further support the notion that there is a specific interaction between PrgY and cCF10. To test this hypothesis, genetic fusions were made between prgY and a homolog traB encoded by plasmid pPD1; TraB controls its cognate pheromone cPD1 but does not efficiently control cCF10 (12). These chimeras were used to characterize PrgY-encoded cCF10 specificity determinants by testing their ability to reduce supernatant cCF10 activity. PrgY and TraB share high similarity (92% at the amino acid level), supporting that chimeric fusions would retain functional activity. In addition, PrgY has overlapping specificity for cPD1 and can reduce supernatant cPD1 activity to comparable levels as TraB (12). Thus, the functional activity of each chimera could be determined by testing their ability to reduce supernatant cPD1 activity.
Chimeras were constructed from pPD1 traB and pCF10 prgY genes by reciprocal exchange of the coding sequences using overlap extension PCR. Eight chimeric traB/prgY genes were generated and cloned into the multicopy vector pMSP3545S (12) to yield the plasmids pB/Y1 to pB/Y6, pB/Y9, and pB/Y10 (Table 2). Because the GelE protease can degrade pheromones (51), chimeric proteins were expressed in the E. faecalis GelE-deficient strain TX5128 (48) and tested for their ability to reduce cCF10 or cPD1 activity in the supernatant compared to wild-type PrgY or TraB or a vector control. As previously seen (12), strains expressing PrgY or TraB had reduced cPD1 levels in cell culture supernatants, and PrgY expression similarly reduced cCF10 levels (Fig. 6). TraB expression did not affect cCF10 levels (Fig. 6). Previously, TraB partially reduced cCF10 levels (12), but this partial phenotype was most likely masked in the present experiments because cCF10 levels were slightly different than previously observed. To determine whether cCF10 specificity was retained in the N- or C-terminal half of PrgY, we tested the chimeric plasmids pB/Y1 or pB/Y2 that each encoded PrgY fused to the N- or C-terminal half of TraB, respectively. The chimeric protein B/Y2 reduced supernatant cCF10 levels by 50% compared to the vector control, whereas B/Y1 did not reduce cCF10 levels (Fig. 6A), suggesting that B/Y2 retained specificity for cCF10 and that the cCF10-specific domain is localized to the N terminus of PrgY.
FIG. 6.
Specific reduction of cCF10 activity is conferred by PrgY amino acids 125 to 241. (A) cCF10 or cPD1 activity was measured in supernatants of TX5128 strains harboring the indicated construct. Cell culture supernatants (grown 6 h from a 10% overnight inoculum induced with 25 ng of nisin/ml) were diluted twofold, and the pheromone activity was determined in a clumping induction assay using OG1RF(pCF10) or (pPD1) indicator strains. The PrgY-dependent percent reduction of cCF10 activity averaged from two independent experiments (the error represents the standard deviation of two independent experiments; the cCF10 titer in the absence of PrgY, TraB, or a chimeric protein was 16 in the first experiment and 32 in the second). cPD1 was similarly assayed twice with the same results each time; the cPD1 titer was 4 in the absence of either PrgY or TraB, and all of the chimeras completely reduced all cPD1 activity. (B) The expression of pPD1 TraB (P), PrgY (Y) and TraB/PrgY chimeras 1 to 6, 9, and 10 (labeled 1 to 6, 9, and 10) in whole-cell extracts was analyzed by Western blotting with a polyclonal anti-PrgY antibody (8). TX5128 was the strain background in these experiments. The arrow at 43 kDa corresponds to the expected size of PrgY. All strains were treated with 25 ng of nisin/ml for 2.5 h prior to harvesting. An equivalent amount of protein was loaded for each sample.
To more precisely define the PrgY domain responsible for cCF10 specificity, chimeric constructs were tested that encoded smaller fragments of pPD1 TraB fused to PrgY. All of the chimeric proteins except B/Y10 reduced supernatant cCF10 activity to a level similar to that of PrgY (Fig. 6A), demonstrating that each of these chimeras retained PrgY-encoded cCF10 specificity determinants. In contrast, B/Y10 did not affect cCF10 levels, and B/Y10 has the TraB amino acid sequence internal to the N terminus of PrgY spanning amino acids 125 to 241. These results indicate that a cCF10 specificity domain may be localized to amino acids 125 to 241.
The functional activity and stability of each of the chimeric proteins was determined by ensuring that the chimeras retained the ability to reduce cPD1 activity and by Western blot analysis (Fig. 6). These results support that the chimeras were sufficiently stable to retain functional pheromone control activity, although it is formally possible that cCF10 control could be more sensitive to minor reductions in stability than pPD1 control. To test protein expression levels, a Western blot with a polyclonal anti-PrgY antibody was used to detect chimeric proteins in cell extracts (Fig. 6B). The chimeric proteins B/Y1 and B/Y10 were unable to reduce cCF10 activity but were detectable with a PrgY antibody at about the same level as PrgY, supporting that their expression and stability is similar to PrgY. These combined results indicate that PrgY amino acids 125 to 241 comprise a domain that is important for cCF10 specificity. It is noteworthy that when the amino acid sequence identity and similarity of residues 180 to 240 of both proteins are compared to that of the entire 250 residue extracellular domain, the divergence between the two proteins is highest in residues 180 to 240 (not shown), which would not be unexpected based on the functional studies of the chimeras.
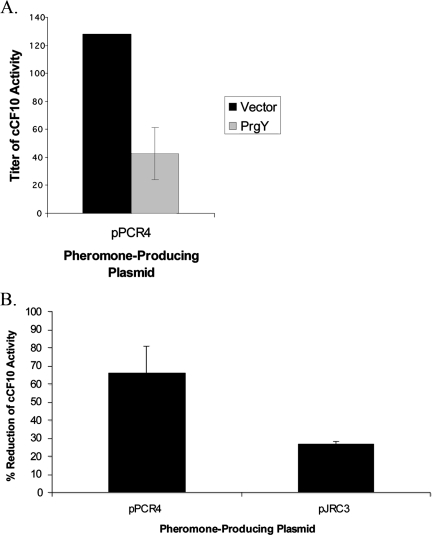
PrgY function is independent of Eep.
PrgY and Eep are both transmembrane proteins that target the CcfA signal peptide that encodes cCF10. Given this, it is possible that PrgY and Eep interact in the membrane and that PrgY functions specifically through Eep (e.g., by specifically inhibiting Eep-dependent cCF10 processing). To address this possibility, we tested whether PrgY can reduce cCF10 activity in the Eep-deficient strain JRC110. In JRC110 supernatants, cCF10 activity is low but detectable when it is overexpressed from plasmid pPCR4 (presumably because pro-cCF10 can be processed inefficiently by another protease) (Table 3). The cCF10 activity in this strain was very low, and PrgY coexpression further reduced cCF10 activity in this strain by 67% (Fig. 7), demonstrating PrgY functions in the absence of Eep. We also the tested PrgY ability to reduce the activity of pJRC3-produced cCF10, which is Eep independent (see Table 3). PrgY reduced activity of pJRC3-produced cCF10 by 27%, supporting that the function of PrgY is Eep independent (Fig. 7B). We have not ruled out the possibility that Eep and PrgY interact or are colocalized in the membrane (this could account for the more effective control of cCF10 produced from the Eep-dependent precursor), but these results indicate that such an interaction is not necessary for PrgY function.
FIG. 7.
PrgY reduces the activity of cCF10 in the Eep-deficient strain JRC110. (A) cCF10 activity produced by pPCR4 expressed with pMSP3545Y (PrgY) or pMSP3545 (vector), as indicated, in strain JRC110 was measured by clumping assay as described in Table 3 and Materials and Methods. (B) PrgY-dependent percent reduction of cCF10 activity for JRC110 strains harboring pPCR4 (cCF10 processed from the iCF10 leader) or pJRC3 (cCF10 processed from the cAM373 leader). For both panels A and B, the results are averaged from three independent experiments done in duplicate (error bars represent the standard deviation of three independent experiments).
DISCUSSION
In this work, we genetically characterized several of the important steps that take place during cCF10 processing and control in a pCF10-containing donor cell. Two proteins involved in this process were the focus of the present study: PrgY, a pCF10-encoded membrane protein involved in donor-mediated control of self-produced cCF10, and Eep, a chromosome-encoded membrane metalloprotease involved in the production of cCF10 and several related pheromones. The nature and specificity of the association of each of these proteins with cCF10 had not previously been addressed and, prior to the initiation of these studies, the steps of cCF10 processing and release were based primarily on sequence analysis (3).
Eep was previously shown to be involved in the production of cAD1, cPD1, and cCF10 (2), as well as of the pAD1 inhibitor peptide iAD1 (1). The results presented here demonstrate that Eep is also involved in production of iCF10 (Table 3). Inhibitor peptides are expressed from plasmid-encoded signal sequence-like peptides whose expression is controlled at the transcriptional level. Eep is closely related to the S2P SpoIVFB of Bacillus subtilis, which transcriptionally regulates genes involved in sporulation through the proteolysis and release of a membrane-bound σ factor σK (44). Prior to the present study, it was unclear whether Eep regulated pheromone and inhibitor production by posttranscriptional peptide processing or at the transcriptional level by a mechanism similar to SpoIVFB. To address whether Eep transcriptionally regulates iCF10, we used the iCF10 prgQ promoter (PQ) to drive expression of the Eep-independent peptide cAM373 (Fig. 3). Production of cAM373 from the non-native promoter PQ was also Eep independent (Table 3), indicating that Eep does not transcriptionally regulate pheromone production. Instead, these results suggest that Eep acts posttranscriptionally to process the pheromone and inhibitor precursors as they pass through the membrane.
The recognition site of the Eep proteases is not known, and the mechanism of intramembrane cleavage by S2P proteases such as Eep is poorly understood. These proteases often target single-transmembrane substrates that have been preprocessed by another protease (7). In the case of the pheromone precursor substrates, preprocessing may occur by signal peptidase II to release the signal peptide, which is most likely the substrate recognized by Eep. Our findings support that Eep indeed recognizes the mature signal peptide; we demonstrated that Eep releases pheromone or inhibitor from the signal-sequence-like polypeptides used for these studies (Table 3). These polypeptides are not cleaved by signal peptidase II, indicating that Eep acts downstream and independently of signal peptidase II during proteolytic processing of the pheromones from their lipoprotein precursors.
The genetic determinants required for Eep recognition were also characterized. We found that Eep recognizes the N-terminal (nonpheromone or inhibitor) portion of the signal-sequence-like precursor peptides (amino acids 1 to 16 of the iCF10 precursor sequence) (Table 3). SpoIVFB requires the N-terminal 117 amino acids of its substrate for RIP-mediated cleavage (42). The region recognized by Eep is much smaller and is more similar to that of the E. coli S2P protein RseP (YaeL), which requires the N-terminal 28 amino acids of its substrate pro-σE for RIP-mediated processing (10, 31). It has been proposed that the substrate requirements for RIP cleavage are based on a secondary or tertiary structure of the peptide substrate in the membrane rather than the primary sequence (38). Indeed, primary sequence analysis of known Eep targets did not reveal a common motif (data not shown). Possibly, the N terminus of the Eep-recognized polypeptides determines the structure in the bacterial cell membrane and therefore directs Eep targeting.
The finding that PrgY does not require Eep (Fig. 7) was important for developing a model for PrgY function. Prior to these studies, one possible model involved PrgY inhibition of Eep-mediated peptide cleavage. Our results indicate that this is not the case. Rather, the results reported here suggest that PrgY interacts directly with the cCF10 peptide, perhaps after Eep proteolysis and release from the membrane. A direct interaction between PrgY and the cCF10 peptide was supported by the following genetic results from this work: (i) the finding that PrgY recognizes the cCF10 peptide sequence itself rather than the surrounding N-terminal signal sequence residues (Fig. 4); (ii) the identification of a residue (L4) in cCF10 that is required for PrgY function (Fig. 5, L4I); and (iii) identification of a cCF10-specific domain in PrgY (Fig. 6). Like PrgX, PrgY possibly binds cCF10 in a pocket (47) that can tolerate some amino acid substitutions in cCF10 (24). Substitutions that were tolerated by PrgY include all of those tested except L4I (Fig. 5). At position L4, two changes (L4I versus L4V) resulted in very different phenotypes, supporting that some changes in cCF10 may be more tolerated by PrgY than others. More severe changes to the other cCF10 residues may be less tolerated by PrgY, but these types of changes could not be detected since they most likely caused the loss of cCF10 activity.
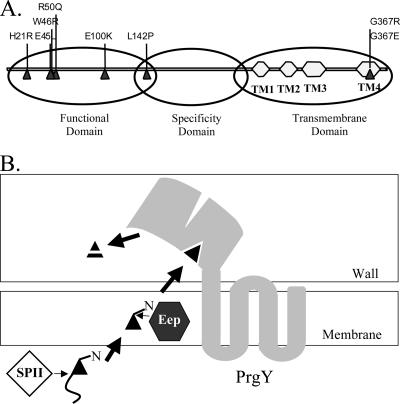
Previously, a conserved functional domain was mapped to the N-terminal half of PrgY based on mutations that abolish function (12). These mutations primarily localize to a conserved region of the first 150 amino acids of PrgY. In the present study, an additional domain was identified from amino acids 125 to 241 that confers cCF10 specificity. The C terminus of PrgY from amino acids 250 to 384 is a predicted transmembrane domain (http://www.enzim.hu/hmmtop/). These three PrgY domains are illustrated in Fig. 8A. The predicted topology of PrgY and a preliminary experimental analysis of topology involving β-galactosidase and alkaline phosphatase fusions to PrgY both suggest that this N-terminal domain is on the outside of the cell membrane, as shown in Fig. 8B. Such an orientation of PrgY would support a model wherein PrgY interacts with cCF10 after it is released from the cell membrane. This type of interaction would require that PrgY is precisely positioned in the cell membrane so that cCF10 is trapped immediately upon release and before reimport by PrgZ (Fig. 8B). Our results support that this type of positioning does not require a direct interaction with Eep because PrgY function is Eep independent, although a direct interaction between these proteins could increase the efficiency of PrgY control. Possibly, both PrgY and Eep are directed to the same site in the cell membrane, such as the ExPortal microdomain identified in Streptococcus pyogenes wherein Sec-dependent export of several characterized proteins is directed (43).
FIG. 8.
(A) Linear diagram of PrgY denoting the location of functional and specificity domains. The functional domain was identified through random and site-directed mutagenesis strategies (12) (random mutations are shown), and the specificity domain (amino acids 125 to 241) was identified in the present study (Fig. 5). Predicted transmembrane domains (TM1 to TM4) are noted. (B) Model of cCF10 processing and export in a donor cell. Mature cCF10 is encoded within the signal sequence of host-encoded lipoprotein CcfA (3). The CcfA signal peptide is most likely removed by signal peptidase II (SPII). The membrane protease Eep then recognizes the N-terminal domain of the signal peptide and further processes it to enable the release of mature cCF10. Active export or further proteolytic processing by host-encoded proteins may also occur at this stage. The topology of PrgY (center) is shown as predicted by the HMMTOP program (http://www.enzim.hu/hmmtop/). PrgY specifically targets cCF10 through amino acids 125 to 241 in PrgY and inhibits the activity of cCF10 through an N-terminal functional domain (12); this inhibition may occur through degrading or modifying newly processed or preprocessed cCF10 in the cell wall (as shown) or by blocking release into the cell medium.
Our results provide the first genetic evidence for substrate recognition by Eep, as well as important insight into the mechanism of PrgY in endogenous pheromone control. Ongoing experiments are aimed at biochemically characterizing cCF10 inactivation by PrgY and confirming a direct interaction between PrgY and cCF10. PrgY is representative of a novel family of proteins with a potentially uncharacterized enzymatic function, thus understanding the biology of this intriguing protein may be relevant to the entire protein family. Questions about the S2P family member Eep also remain, including its substrate target site, the mechanism for peptide release from the membrane, and the motifs that are involved in S2P-mediated substrate recognition.
Acknowledgments
We thank Thinh Le, Dawn Manias, and Chris Kristich for helpful discussions.
This study was supported by NIH grant GM49530 awarded to G.M.D. J.R.C. was a trainee under the University of Minnesota MinnCResT program supported by NIH training grant T32DE07288.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 1841880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 1815915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 1841155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, T., and G. M. Dunny. 2001. Dominant-negative mutants of prgX: evidence for a role for PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 391307-1320. [DOI] [PubMed] [Google Scholar]
- 5.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315995-1007. [DOI] [PubMed] [Google Scholar]
- 6.Bae, T., B. K. Kozlowicz, and G. M. Dunny. 2004. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol. Microbiol. 51271-281. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100391-398. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44183-190. [DOI] [PubMed] [Google Scholar]
- 9.Buttaro, B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 1824926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson, H. C., S. Lu, L. Kroos, and W. G. Haldenwang. 1996. Exchange of precursor-specific elements between Pro-σE and Pro-σK of Bacillus subtilis. J. Bacteriol. 178546-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 251377-1388. [DOI] [PubMed] [Google Scholar]
- 12.Chandler, J. R., A. R. Flynn, E. M. Bryan, and G. M. Dunny. 2005. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J. Bacteriol. 1874830-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler, J. R., H. Hirt, and G. M. Dunny. 2005. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc. Natl. Acad. Sci. USA 10215617-15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie, P. J., and G. M. Dunny. 1986. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid 15230-241. [DOI] [PubMed] [Google Scholar]
- 15.Chung, J. W., and G. M. Dunny. 1992. cis-Acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 899020-9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clewell, D. B. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 739-12. [DOI] [PubMed] [Google Scholar]
- 17.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35246-247. [DOI] [PubMed] [Google Scholar]
- 18.Dev, I. K., and P. H. Ray. 1990. Signal peptidases and signal peptide hydrolases. J. Bioenerg. Biomembr. 22271-290. [DOI] [PubMed] [Google Scholar]
- 19.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6270-278. [DOI] [PubMed] [Google Scholar]
- 20.Dunny, G. M., M. H. Antiporta, and H. Hirt. 2001. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides 221529-1539. [DOI] [PubMed] [Google Scholar]
- 21.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 753479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 571194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fixen, K. R., J. R. Chandler, T. Le, B. K. Kozlowicz, D. A. Manias, and G. M. Dunny. 2007. Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in the interactions with the pheromone sensing machinery. J. Bacteriol. 1891399-1406, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44803-817. [DOI] [PubMed] [Google Scholar]
- 26.Hedberg, P. J., B. A. Leonard, R. E. Ruhfel, and G. M. Dunny. 1996. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid 3546-57. [DOI] [PubMed] [Google Scholar]
- 27.Hirt, H., D. A. Manias, E. M. Bryan, J. R. Klein, J. K. Marklund, J. H. Staddon, M. L. Paustian, V. Kapur, and G. M. Dunny. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J. Bacteriol. 1871044-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt, H., P. M. Schlievert, and G. M. Dunny. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 7761-68. [DOI] [PubMed] [Google Scholar]
- 30.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 805369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju, J., T. Luo, and W. G. Haldenwang. 1997. Bacillus subtilis Pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 1794888-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, S. M., S. B. Olmsted, A. S. Viksnins, J. C. Gallo, and G. M. Dunny. 1991. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J. Bacteriol. 1737650-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kast, P., and H. Hennecke. 1991. Amino acid substrate specificity of Escherichia coli phenylalanyl-tRNA synthetase altered by distinct mutations. J. Mol. Biol. 22299-124. [DOI] [PubMed] [Google Scholar]
- 34.Kozlowicz, B. K. 2005. Ph.D. thesis. University of Minnesota, Minneapolis.
- 35.Kozlowicz, B. K., T. Bae, and G. M. Dunny. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 54520-532. [DOI] [PubMed] [Google Scholar]
- 36.Kozlowicz, B. K., K. Shi, Z. Y. Gu, D. H. Ohlendorf, C. A. Earhart, and G. M. Dunny. 2006. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 62958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristich, C. J., J. R. Chandler, and G. M. Dunny. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemberg, M. K., and B. Martoglio. 2004. On the mechanism of SPP-catalyzed intramembrane proteolysis; conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 564213-218. [DOI] [PubMed] [Google Scholar]
- 39.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCormick, J. K., H. Hirt, G. M. Dunny, and P. M. Schlievert. 2000. Pathogenic mechanisms of enterococcal endocarditis. Curr. Infect. Dis. Rep. 2315-321. [DOI] [PubMed] [Google Scholar]
- 41.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 42.Prince, H., R. Zhou, and L. Kroos. 2005. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK. J. Bacteriol. 187961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in gram-positive bacteria. Science 3041513-1515. [DOI] [PubMed] [Google Scholar]
- 44.Rudner, D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 9614765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhfel, R. E., B. A. Leonard, and G. M. Dunny. 1996. Pheromone-inducible conjugation in Enterococcus faecalis: mating interactions mediated by peptide signals and direct contact, p. 53-68. In J. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, NY.
- 46.Ruhfel, R. E., D. A. Manias, and G. M. Dunny. 1993. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J. Bacteriol. 1755253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, K., C. K. Brown, Z. Y. Gu, B. K. Kozlowicz, G. M. Dunny, D. H. Ohlendorf, and C. A. Earhart. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 10218596-18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 1781416-1420. [DOI] [PubMed] [Google Scholar]
- 49.Staddon, J. H., E. M. Bryan, D. A. Manias, Y. Chen, and G. M. Dunny. 2006. Genetic characterization of the conjugative DNA processing system of enterococcal plasmid pCF10. Plasmid 56102-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urban, S., and M. Freeman. 2002. Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev. 12512-518. [DOI] [PubMed] [Google Scholar]
- 51.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 1853613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yagi, Y., R. E. Kessler, J. H. Shaw, D. E. Lopatin, F. An, and D. B. Clewell. 1983. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J. Gen. Microbiol. 1291207-1215. [DOI] [PubMed] [Google Scholar]