Abstract
The gram-negative bacterium Haemophilus influenzae is a human-restricted commensal of the nasopharynx that can also be associated with disease. The majority of H. influenzae respiratory isolates lack the genes for capsule production and are nontypeable (NTHI). Whereas encapsulated strains are known to belong to serotype-specific phylogenetic groups, the structure of the NTHI population has not been previously described. A total of 656 H. influenzae strains, including 322 NTHI strains, have been typed by multilocus sequence typing and found to have 359 sequence types (ST). We performed maximum-parsimony analysis of the 359 sequences and calculated the majority-rule consensus of 4,545 resulting equally most parsimonious trees. Eleven clades were identified, consisting of six or more ST on a branch that was present in 100% of trees. Two additional clades were defined by branches present in 91% and 82% of trees, respectively. Of these 13 clades, 8 consisted predominantly of NTHI strains, three were serotype specific, and 2 contained distinct NTHI-specific and serotype-specific clusters of strains. Sixty percent of NTHI strains have ST within one of the 13 clades, and eBURST analysis identified an additional phylogenetic group that contained 20% of NTHI strains. There was concordant clustering of certain metabolic reactions and putative virulence loci but not of disease source or geographic origin. We conclude that well-defined phylogenetic groups of NTHI strains exist and that these groups differ in genetic content. These observations will provide a framework for further study of the effect of genetic diversity on the interaction of NTHI with the host.
Haemophilus influenzae is a small (1 to 2 μm in length) gram-negative bacterium that is found only in humans. The polysaccharide-protein conjugate vaccines against serotype b H. influenzae have almost eliminated H. influenzae as a cause of pediatric meningitis in the western world. However, unencapsulated (nontypeable) H. influenzae (NTHI) remains an important pathogen, particularly in children and the elderly (5, 8, 23). NTHI infections are usually limited to respiratory mucosal sites such as the middle ear or bronchi but are occasionally systemic. It is not known whether NTHI isolates associated with localized or systemic disease are genetically distinct from each other or distinct from isolates associated with asymptomatic colonization of the nasopharynx.
Efforts to understand and control NTHI disease have been hampered by the diversity of these bacteria. Many of the surface antigens that have been studied display interstrain and intrastrain heterogeneity as a result of both sequence divergence and phase variation. It is increasingly recognized that NTHI isolates also vary in genetic content. We use the term island to refer to a genetic locus (one or more genes) that occurs in some but not all strains. As used here, the term does not imply that the locus is known to be readily transferred between strains or is thought to have been recently acquired. Islands whose function is known include the hmw, hia, and hif adhesin loci; the region flanked by infA and ksgA containing lipopolysaccharide (LPS) biosynthetic loci lic2BC or losAB; and the three loci required for the biochemical reactions used in biotyping (18, 27, 30, 34, 52). Several strains have recently been sequenced, and each contains numerous islands that are not present in the other sequenced strains (25, 47). Subtractive hybridization studies have identified other islands (55). Certain islands appear to be more common in disease isolates than in isolates from healthy subjects (17, 34, 44), suggesting that the genetic content of NTHI strains affects their likelihood of causing symptomatic infection.
Until recently, distinguishing H. influenzae strains from each other or evaluating their relatedness has been difficult. Epidemiological studies have commonly analyzed strains by gel electrophoresis of bacterial proteins, restriction fragments, or randomly amplified sequences (1, 39, 43). These methods yield patterns that vary from lab to lab. Within the last few years, two systems for typing H. influenzae have been developed, based on sequence polymorphisms of either housekeeping genes (multilocus sequence typing [MLST]) (35) or genes encoding 16S rRNA (16S typing) (46). Each of these two systems has the advantage of yielding unambiguous data, so that a database can include strains typed in several laboratories. The MLST system has so far been more widely adapted.
The primary goal of this project was to analyze the phylogenetic structure of H. influenzae, with particular focus on NTHI. Second, we sought to evaluate the extent to which phylogenetic groups of NTHI strains differ in content of genetic loci that may affect pathogenesis. A collection of clinical isolates for which we had previously used biotyping and PCR to detect several genetic islands (11) were typed by MLST in the present study, and we extended the biotyping and PCR studies to NTHI strains that were already in the MLST database.
We performed both profile-based and sequence-based analyses of the MLST database. Profile-based analyses tally the shared and differing alleles for each pair of sequence types (ST) without consideration of the actual sequences of the differing alleles. Acquisition of a new allele by genetic exchange (potentially altering several bases) has the same effect on the outcome of profile-based analyses as a single-point mutation. In contrast, for sequence-based analyses, alteration of an allele at several bases affects the outcome of the analysis to a greater extent than alteration at a single base. Because H. influenzae strains are known to undergo genetic exchange, it was not certain whether a sequence-based analysis would identify distinct phylogenetic groups, particularly within the nontypeable strains. We performed a rigorous analysis of the MLST sequences using a maximum-parsimony method and identified 13 distinct clades within the H. influenzae population. All except two of the clades corresponded closely to groups identified using eBURST, a profile-based analysis (16). Ninety-eight percent of encapsulated strains and 65 percent of the NTHI strains in the MLST database could be assigned to one of the 13 clades, while 21 percent of NTHI strains were members of a single eBURST group that did not correspond to any clade. We found that each of these phylogenetic groups had a different distribution of the genetic islands that we examined. These observations suggest that horizontal exchange has not completely blurred evidence of ancestral relationships among H. influenzae strains.
MATERIALS AND METHODS
Phylogenetic analysis. (i) MLST.
MLST typing was performed by amplifying and sequencing regions of the housekeeping genes adk, atpG, frdB, fucK, mdh, pgi, and recA as described previously (35). The complete H. influenzae database (322 NTHI strains, 244 type b strains, 84 encapsulated strains of other serotypes, and 5 strains of unknown serotype) was downloaded from the Haemophilus MLST website (http://haemophilus.mlst.net) on 18 September 2006. For sequence-based phylogenetic analyses, the seven sequences for each ST were concatenated into a single 3,057-bp sequence.
The program TNT (P. A. Goloboff, J. S. Farris, and K. Nixon, TNT: tree analysis using new technology, program and documentation, 2003 [available at http://www.zmuc.dk/public/phylogeny/tnt]) was utilized to conduct maximum-parsimony analyses of the concatenated sequences, using both constrained and random sectorial searches with search parameters optimized every 10 hits. The new technology tree drifting and tree fusing algorithms were used (21). The program was run until 4,545 equally most parsimonious trees were collected and the strict consensus tree stabilized. The majority-rule consensus of the resulting equally most parsimonious trees was calculated in TNT. The resulting majority-rule tree, plotted using ST65 as the outgroup, is shown in Fig. 1. Table S1 in the supplemental material lists the position of each ST on the tree. Frequency jackknife support values were also calculated with TNT (22), using 1,000 replications and a probability of removal for each character of 36%.
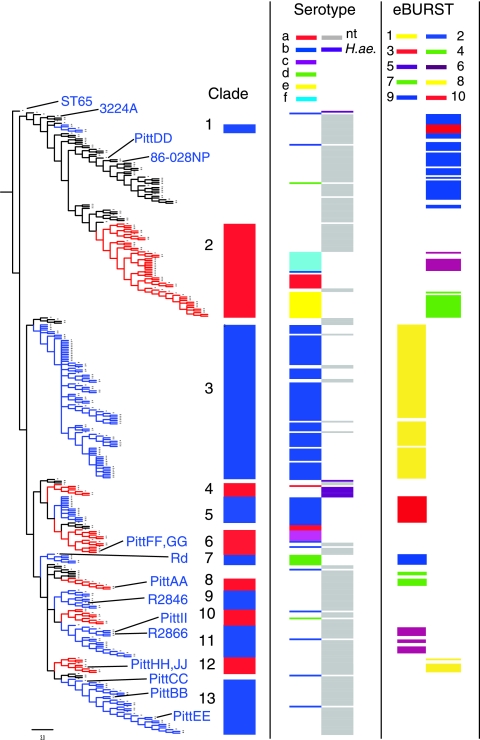
FIG. 1.
Maximum-parsimony majority-rule consensus tree for 359 ST, plotted with ST65 as the outgroup. Colored regions of the tree numbered 1 through 13 indicate branches (clades) that were found in 100% of trees, with the exception of clades 9 and 11, which were found in 92% and 81% of trees, respectively. The tree is aligned with a plot showing the serotype and the eBURST group for each ST. Each ST corresponded either to a single serotype or to one or more NTHI strains. H. influenzae biogroup aegyptius (H. ae.) strains are plotted separately from other NTHI strains. The ST for 15 sequenced strains are indicated on the tree. Table S1 in the supplemental material lists the position of each ST on the tree as well as its eBURST group and the serotype of associated strains.
The MLST profiles (i.e., the set of seven allele numbers for each ST) were analyzed using eBURST v3 (16), available at http://eburst.mlst.net/default.asp. The database was also analyzed using both sequence-based and profile-based lineage assignment algorithms in the START2 suite of sequence typing software (29) available at http://pubmlst.org/software/analysis/start2/.
(ii) 16S rRNA typing.
The 16S rRNA genes were amplified using external primers, and the 1,595-bp products were sequenced as described previously (46). Maximum-parsimony analysis was performed on the complete set of 16S sequences, as described above for MLST sequences, yielding 2,230 equally most parsimonious trees.
Bacterial strains.
The MLST database includes 322 NTHI isolates, of which 296 were included in our phylogenetic analyses. (When more than one strain with the same ST was obtained from the same subject, we excluded all except one isolate.) Of the 296 independently isolated strains, 206 were available for laboratory studies. These included 52 pediatric isolates of diverse sources used in an earlier study (11), 25 of the 27 otitis isolates from Finland that were used to develop the H. influenzae MLST scheme and have been used in several other studies (2, 7, 28, 35), 88 isolates submitted to the Active Bacterial Core Surveillance program at the Centers for Disease Control and Prevention that had been used to develop the 16S typing system (46), and several isolates for which partial genome sequences have recently been reported. All strains were confirmed to be H. influenzae based on the requirement for β-NAD+ and hemin and were judged not to be H. haemolyticus based on recently described criteria (37). Serotyping was performed by PCR as described previously (13). bexA primers yielded a PCR product from control strains of each serotype and failed to amplify a product from any of the NTHI strains.
Genetic and phenotypic heterogeneity. (i) Biotyping.
Biotypes were assigned according to Kilian's system (30), using API 20E strips to detect biochemical reactions as previously described (11).
(ii) PCR of genetic islands.
The hic, hif, hia, hmw, lic2BC, and losAB loci were detected by PCR as described previously (11). For these loci, PCR was initially carried out using flanking primers, and positive results were confirmed by partial sequencing of the PCR product or by amplification with combinations of internal and external primers, as described previously (11). To confirm the presence of the hia and hmw loci, PCR was performed using flanking primers (Table 1) as well as the internal primers described previously (11). For hmw, a multiplex PCR with primers flanking both the hmw1 and hmw2 loci was performed. The two products were not clearly resolved by gel electrophoresis, so the lack of one hmw locus would not have been detected. Both hmw and hia are known to differ in sequence from strain to strain (4, 33). We did not attempt to assess the sequence heterogeneity of these loci; rather the internal primers that we used were selected to hybridize with conserved regions of the hia and hmwA sequences.
TABLE 1.
Primers used for PCR detection of genetic islands
| Locus | Primer sequencea
|
|
|---|---|---|
| Forward | Reverse | |
| hia | 5′-CCGAAAGCACAATGGATATGGACG-3′ | 5′-CAGATAAATCCTGACCTCGCTCTC-3′ |
| hmw1 | 5′-CAAAGCCATCAGGTTGTTGTGC-3′ | 5′-CCTATTTGGTCTTGCTACGAGTGG-3′ |
| hmw2 | 5′-CCGCACTTTCTTCTCGTTCTTCT-3′ | 5′-GCTATTCGGTTAGGTAATGCAGATCC-3′ |
| ahpC (tsaA) | 5′-CAGGGTTTACCGATCCTTGTGA-3′ | 5′-AGATAGCCATCTAGCCAGTCAGTG-3′ |
| HaeII | 5′-CATCCTTGGTTCTTATGGACAGCG-3′ | 5′-CTCTAATGGATCATCTCCGCTACG-3′ |
| R2846.1179-80 | 5′-CGGATCCACTCAATCTACTGCAAG-3′ | 5′-GAGTATCCCAACAAGATCTCAGCG-3′ |
| R2866.503-507 | 5′-CTGCGACGCATTTATTACAGGG-3′ | 5′-GTGGAATGCAAGGCTTAATGGG-3′ |
| R2866.1857-58 | 5′-CTGGAGCTTCATCTACCAATACGC-3′ | 5′-CAAGCAGAAACCCGTGAAATTATCG-3′ |
The primers shown for hia, hmw1, and hmw2 bind to sequences flanking the respective loci and were used to confirm the results of PCR using the internal primers described previously for these loci (11). The loci R2846.1179-80, R2866.503-507, and R2866.1857-58 encode putative restriction-modification systems of types II, I and III, respectively.
Primers flanking the ahpC locus (tsaA; NTHI0212) and HaeII locus (NTHI1786 and NTHI1787) were designed based on the published sequences of these regions in strain 86-028NP (25). A search of the genome sequences of strains R2846 and R2866, available as contigs through the NCBI microbial genome database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=9621) identified three novel putative restriction-modification systems; the presence of these loci was evaluated by PCR using primers listed in Table 1. All primers were synthesized by Integrated DNA Technologies (Coralville, IA).
Statistics.
Frequency data were evaluated by Fisher's exact probability test, calculated online at the VassarStats website for statistical computation (http://faculty.vassar.edu/lowry/VassarStats.html). The resulting P values were subjected to the Bonferroni adjustment for multiple comparisons on a set of data (42).
Nucleotide sequence accession numbers.
The sequences of the novel restriction-modification loci have been submitted to GenBank and assigned accession numbers EU285588 (R2846.1179-80, putative type II restriction system), EU296480 (R2866.503-507, probable type I restriction system), and EU296479 (R2866.1857-58, probable type III restriction system).
RESULTS
Division of the H. influenzae population into clades.
In order to generate phylogenetic trees in which branches have a high probability of reflecting ancestral relationships among strains, we performed a maximum-parsimony analysis. To allow detection of NTHI clusters that are closely related to encapsulated strains, this analysis was carried out using the complete H. influenzae MLST database (359 ST) rather than only the 195 ST that are associated with NTHI strains. Maximum-parsimony analysis of the 359 concatenated sequences yielded 4,545 equally parsimonious trees of length 4,761. The tree shown in Fig. 1 is a majority-rule consensus tree, showing only branches that were found in over 50% of the equally most parsimonious trees. The shaded regions numbered 1 through 8 and 10, 12, or 13 indicate branches of six or more ST that were found in 100% of trees. Branches 9 and 11 were found in 92% and 81% of trees, respectively. Together these 13 branches (referred to below as clades) contain 77.7% of ST, representing 79% of the H. influenzae strains in the MLST database. Three of the clades are serotype specific, consisting entirely or almost entirely of type b strains (clades 3 and 5) or type d strains (clade 7). Clades 2 and 6 each include serotype-specific clusters as well as clusters containing only NTHI, while in the remaining eight clades almost all strains are NTHI. The position of each ST on the tree is listed in Table S1 in the supplemental material.
Jackknife resampling using frequency differences (22) indicated low support (<50%) for most of the clades identified in the majority-rule consensus tree. Notable exceptions include clades 1, 5, and 8, with jackknife values of 66%, 72%, and 87%, respectively. While resampling support was low for most of the tree, many of the clades identified using the MLST housekeeping genes also correlate with other characteristics, including serotype, biochemical traits, and other genetic traits (see below), indicating that the clades have biological relevance. In addition, the bootstrap and jackknife resampling methods are considered conservative, erring toward low confidence values (15, 48).
We also utilized eBURST, an alternative method of analyzing MLST data, to define groups in which each ST is identical at five or more of the seven sequenced loci to at least one other ST in the group. Ten such groups contained five or more ST. Each eBURST group mapped to a single region of the majority-rule tree, in most cases corresponding closely to a clade or to a serotype-specific cluster within a clade (Fig. 1; see Table S1 in the supplemental material). In contrast, the 40 ST in eBURST group 2 are not in any of the clades identified by the parsimony analysis.
These data provide the first evidence that the NTHI population contains several distinct genetic groups: of 296 independently isolated NTHI strains in the MLST database, 80% have ST in one of 10 clades (192 strains) or in eBURST group 2 (67 strains).
Clustering of encapsulated strains within serotype-specific clusters.
The clustering of encapsulated strains on the tree shown in Fig. 1 is consistent with that observed in previous studies. A large multilocus enzyme electrophoretic study by Musser et al. (40) and a subsequent smaller MLST study by Meats et al. (35) each identified two major clusters of type b strains, with much smaller numbers of type b strains elsewhere on the phylogenetic tree. Each study also found type a strains to be clustered in three sites on the tree and a single cluster each for types c, d, e, and f. Since publication of the original H. influenzae MLST database, 194 type b strains have been added, with 96 new ST. We found that the vast majority of these new ST fall into the same two type b-specific clusters described previously but that 12 type b strains are outside of the clusters. The presence of type b capsular genes was confirmed by PCR for the two of those strains that were available to us. Twenty-nine strains of types a, c, d, e, and f have been added to the MLST database, and with the exception of two type d strains (confirmed by PCR), all fall into the previously described clusters. It was proposed previously that strains of types e and f and certain type a strains have a common ancestor (35). This was confirmed in the present study, in which these strains were all in clade 2. Nine NTHI strains have ST outside of the main NTHI clusters. The lack of capsular biosynthetic genes in each of these strains was confirmed by PCR. These strains may have lost capsule biosynthesis genes relatively recently, as seven of these NTHI strains are within the large type b group (clade 3), and the other two are most closely related to type e strains.
Overall, the addition of 525 strains to the MLST database since its original publication has not significantly altered our understanding of the genetic relationships among encapsulated H. influenzae strains. In contrast, the NTHI-specific regions of the MLST tree are now sufficiently developed that distinct groups can be defined.
Genetic and phenotypic comparison of NTHI clades.
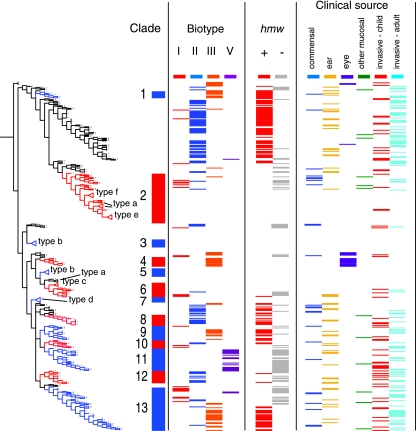
We sought evidence for common features among NTHI strains within a phylogenetic group (i.e., within one of the 13 clades or in eBURST group 2). NTHI strains are known to be heterogeneous in the presence and structure of adhesins and other outer membrane proteins, in LPS structure, and in metabolism. Of 296 independently isolated NTHI strains in the MLST database, 206 were available to us for laboratory study. This collection, which includes 52 strains studied previously (11), was evaluated by biotyping and by performing PCR to detect the hmw adhesin loci and the LPS biosynthetic loci flanked by infA and ksgA. PCR results were confirmed using a second set of primers and in some cases by partial sequencing of the product. For each of these traits, we found significant differences in distribution across the phylogenetic groups (Fig. 2; Table 2). For example, 7 strains of clade 1 were all biotype III; of 12 clade 2 strains studied, seven were biotype I; clade 8 strains (n = 7) were all biotype II; and clade 11 strains (n = 15) were all biotype V. Of 49 strains in eBURST group 2, all except 5 were biotype II. Each of these phylogenetic groups was equally uniform with regard to the hmw loci (Fig. 2; Table 2) and nearly so for the content of the infA-ksgA locus (Table 2). These data suggest that there may be biological differences among clades, resulting in biochemical differences as well as in the presence of genes affecting the structure of bacterial surface components, including both outer membrane proteins and LPS.
FIG. 2.
Characterization of NTHI strains and correlation of biotype and hmw locus with clustering on the majority-rule consensus tree. The tree differs from that in Fig. 1 in that each ST found in more than one NTHI strain was expanded to allow plotting of data for each strain, while serotype-specific clusters were collapsed (indicated by triangles). The tree is aligned with a plot showing clade (as in Fig. 1), biotype, the presence of the hmw loci (determined by PCR), and the type of specimen from which the strain was cultured. Commensal, strains cultured from oropharyngeal or nasopharyngeal swabs from healthy children; otitis, strains from middle ear aspirates from children with otitis media; eye, strains from children with conjunctivitis; mucosal, strains from other mucosal samples, primarily sputum or endotracheal aspirates; invasive, strains from blood or cerebrospinal fluid. The biotype and PCR data, including the results of PCR of the LPS biosynthetic locus flanked by infA and ksgA, are summarized in Table 1.
TABLE 2.
Phylogenetic groups differ in distribution of genetic islands
| Locus or biotypea | No. of strains positive/no. examined (% positive), P valueb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All strains | Clade 1 | Clade 2 | Clade 3 | Clade 6 | Clade 8 | Clade 9 | Clade 10 | Clade 11 | Clade 12 | Clade 13 | eBURST group 2 | No group | |
| hmw | 121/206 (58.7) | 8/8 (100) | 0/8, 0.0054 | 2/3 (66.7) | 1/5 (83.3) | 7/7 (100) | 10/13 (76.9) | 2/4 (50) | 0/22, 4.8 × 10−9 | 2/9 (22.2) | 22/35 (62.9) | 51/57 (89.5), 9.6 × 10−8 | 16/34 (47.0) |
| LPS biosynthetic locus (infA/ksgA) | |||||||||||||
| lic2BCc | 104/201 (51.7) | 0/7 | 6/8 (75) | 3/3 (100) | 5/5 (100) | 7/7 (100) | 5/13 (38.5) | 1/4 (25) | 0/21 | 1/9 (11.1) | 31/35 (88.6), 1.2 × 10−6 | 23/55 (41.8) | 22/34 (64.7) |
| losAB | 35/201 (17.4) | 7/7 (100), 2.2 × 10−5 | 0/8 | 0/3 | 0/5 | 0/7 | 7/13 (53.8), 0.016 | 0/4 | 0/21 | 0/9 | 2/35 (5.7) | 12/55 (21.8) | 7/34 (20.6) |
| No insert | 62/201 (30.8) | 0/7 | 2/8 (25) | 0/3 | 0/5 | 0/7 | 1/13 (7.7) | 3/4 (75) | 21/21 (100), 8 × 10−12 | 8/9 (88.9), 0.032 | 2/35 (5.7) | 20/55 (36.4) | 5/34 (14.7) |
| Biotype | |||||||||||||
| I | 30/208 (14.4) | 0/7 | 7/12 (62.5), 0.0025 | 1/2 (50) | 3/4 (75), 0.079 | 0/7 | 1/11 (9.1) | 2/4 (50) | 0/15 | 0/10 | 8/37 (21.6) | 2/49 (4.1) | 6/41 (14.6) |
| II | 92/208 (44.2) | 0/7 | 2/12 (16.7) | 0/2 (50) | 1/4 (25) | 7/7 (100), 0.023 | 0/11 | 2/4 (50) | 0/15 | 10/10 (100), 0.0016 | 4/37 (10.8) | 44/49 (89.6), 3.8 × 10−13 | 22/41 (53.6) |
| III | 61/208 (29.3) | 7/7 (100), 0.0011 | 2/12 (16.7) | 1/2 (50) | 0/4 | 0/7 | 8/11 (72.7), 0.022 | 0/4 | 0/15 | 0/10 | 21/37 (56.8), 0.0009 | 3/49 (6.1) | 10/41 (14.4) |
| IV | 6/208 (2.9) | 0/7 | 1/12 (8.3) | 0/2 | 0/4 | 0/7 | 2/11 (18.2) | 0/4 | 0/15 | 0/10 | 2/37 (5.4) | 0/49 | 1/41 (2.4) |
| V | 19/208 (9.1) | 0/7 | 0/12 | 0/2 | 0/4 | 0/7 | 15/15 (100), 1.1 × 10−18 | 0/10 | 2/37 (5.4) | 0/49 | 2/41 (4.9) | ||
The hmw and LPS biosynthetic loci were evaluated by PCR as described in the text,and biotypes were determined biochemically. Included are data for 52 strains described previously (11). The LPS biosynthetic locus is the locus flanked by infA and ksgA, containing lic2BC, losAB, or no genes. Biotype data include data reported to the MLST database by others (n = 45) as well as biotypes determined by ourselves (n = 163). Biotypes: I, positive for indole production, urease, and ornithine decarboxylase reactions; II, negative for ornithine decarboxylase only; III, positive for urease only; IV, negative for indole only; V, negative for urease only (30). No biotype VI, VII, or VIII strains were identified.
Clade 4 strains were not examined in the laboratory in this study. All clade 4 NTHI strains are H. influenzae biogroup aegyptius and are biotype III (31). Clades 5 and 7 are also omitted from this table, as they do not contain any NTHI strains. No group, NTHI strains with ST that are not in any of the 13 clades and not in eBURST group 2. For each of the eight phylogenetic groups for which five or more strains were studied, the frequency of the genotype (or biotype) among strains in each group was compared to the frequency in the remaining population using the Fisher exact test (two tailed). The resulting P value was multiplied by eight to apply the Bonferroni adjustment for multiple hypotheses on a set of data (42). P values of <0.10 are reported.
Strains listed as containing lic2BC include five strains that contain lic2C only. One of these is in eBURST group 2 (ST258); the others are not in any of the phylogenetic groups (two strains in ST2 and one each in ST162 and ST316).
These observations were extended by evaluating additional loci in a subset of strains. We used PCR to seek evidence of loci encoding surface structures (hia, lav, hif, and hic) or other putative virulence determinants (ahpC/tsaA, encoding a peroxiredoxin). In addition, we evaluated the presence of the HaeII locus (25) and of three novel restriction-modification systems identified in the genome sequence of strain R2846 or R2866 by PCR using primers flanking each locus. Each of these loci was evaluated in 80 strains and was found to be distributed nonrandomly across the phylogenetic groups (Table 3). For the restriction systems and for ahpC, strains were scored positive if the primers amplified an appropriately sized product. We would not have detected the locus at a different site, and we did not evaluate sequence heterogeneity in the PCR products. However, the results are striking. For example, approximately one-third of the strains examined (23 of 67 strains) yielded a product consistent with HaeII. All except one of these strains were in eBURST group 2 (13 of 13 strains examined; P = 2 × 10−7), clade 11 (5 of 7 strains), or clade 1 (2 of 2 strains). This suggests that restriction barriers between phylogenetic groups may help to maintain their differences in genetic content.
TABLE 3.
Further comparison of phylogenetic groups
| Genetic islanda | No. of strains positive/no. examined (% positive), P valueb
|
||||||
|---|---|---|---|---|---|---|---|
| All NTHI strains | Clade 2 | Clade 9 | Clade 11 | Clade 13 | eBURST group 2 | No group | |
| Islands potentially affecting pathogenesis | |||||||
| ahpC | 63/78 (80.8) | 4/4 (100) | 3/5 (60) | 7/7 (100) | 6/14 (42.8), 0.003 | 19/19 (100), 0.096 | 16/19 (84.2) |
| hia | 30/80 (37.5) | 5/5 (100), 0.036 | 1/5 (20) | 9/9 (100), 0.00036 | 4/15 (26.7) | 0/19, 0.00036 | 9/19 (47.4) |
| hic | 49/77 (63.6) | 1/4 (25) | 2/5 (40) | 4/7 (57.1) | 13/14 (92.9), 0.078 | 17/19 (89.5), 0.072 | 8/19 (42.1) |
| hif | 12/79 (15.2) | 1/5 (20) | 1/5 (20) | 2/7 (28.6) | 5/15 (33.3) | 0/19, 0.06 | 2/19 (10.5) |
| lav | 23/78 (29.5) | 2/5 (40) | 0/5 | 5/7 (71.4) | 0/15, 0.072 | 13/18 (72.2), 0.00012 | 1/19 (5.3), 0.048 |
| Islands potentially encoding restriction systems | |||||||
| HaeII | 23/68 (33.8) | 0/4 | 0/5 | 5/7 (71.4) | 0/14, 0.018 | 13/13 (100), 2.2 × 10−7 | 2/18 (11.1) |
| R2846.1179-80 | 10/75 (13.3) | 0/4 | 4/5 (80), 0.0048 | 0/7 | 4/12 (33.3) | 0/18 | 2/19 (10.5) |
| R2866.503-07 | 61/74 (82.4) | 0/4, 0.0036 | 4/5 (80) | 7/7 (100) | 11/14 (78.6) | 16/16 (100) | 17/18 (94.4) |
| R2866.1857-58 | 13/73 (17.8) | 0/3 | 0/5 | 5/6 (83.3), 0.003 | 1/14 (7.1) | 0/17 | 4/19 (21.0) |
The presence of the islands in NTHI strains was evaluated by PCR as described in the text. Included are previously reported data on the hia, hic, hif, and lav loci for 52 strains (11). Additional strains included in this table were 18 Finnish isolates, the recently sequenced strains 3224A and 86-028NP, and 8 additional clinical isolates.
Phylogenetic groups are included if more than two NTHI strains in the group were examined for the indicated loci. “All NTHI strains” includes strains of clades 1, 3, 6, 8, 10, and 12, for which one or two strains per clade were analyzed. No group, strains with ST that are not in any of the 13 clades and not in eBURST group 2. For each of the six phylogenetic groups for which five or more strains were studied, the frequency of the genotype (or biotype) among strains in each group was compared to the frequency in the remaining population using the Fisher exact test (two tailed). The resulting P value was multiplied by six to apply the Bonferroni adjustment for multiple hypotheses on a set of data (42). P values of <0.10 are reported.
Separation of strains into phylogenetic groups allows detection of patterns that were not previously apparent. For example, we previously noted that in a cluster of biotype V strains (now identified as clade 11), the autotransporter gene lav was more frequently present than in the remaining strains in the study (11). At that time we could not identify any common feature among lav-containing strains that were not biotype V. It is now evident that nearly all of the lav-containing strains not in clade 11 are in eBURST group 2 (P = 0.00012). We previously noted an association between hmw and the hif-contiguous locus hicAB. Consistent with that observation, the hic locus is common in eBURST group 2, in which nearly all strains contain hmw. However, hic is also common in clade 13, which contains both hmw-positive and hmw-negative strains. The hif fimbrial locus was found in only 15% of strains examined. It was absent from the largest group of strains, eBURST group 2, and was found at increased frequency in clade 13.
In contrast, there was no apparent correlation between phylogeny and the clinical or geographic source of strains, with the exception of H. influenzae biogroup aegyptius. H. influenzae biogroup aegyptius strains from the Brazilian purpuric fever clone are all ST65 (not in any of the 13 clades), while epidemiologically unrelated conjunctivitis strains are ST70 to -77, clustered in or near clade 4. Of the remaining NTHI strains in the MLST database, the clinical source is known for all except 17. Isolates cultured from middle ear aspirates (n = 52) or from systemic infections in children (n = 57) or adults (n = 100) were all broadly distributed across the phylogenetic tree (Fig. 2). Consistent with a previous report that commensal strains are less likely than otitis isolates to contain hmw (10), we noted that clade 2 isolates, which are uniformly hmw negative, were more frequently commensal than other isolates. Correspondingly, strains of eBURST group 2, 90% of which are hmw positive, are less likely to be commensal. These differences are not statistically significant. Moreover, as the database contains relatively small numbers of nasopharyngeal isolates from healthy children (n = 35) or from sputum or other nonconjunctival mucosal sites (n = 17), it would be premature to draw any firm conclusions about phylogenetic differences between commensal isolates and those from disease.
Comparison of maximum-parsimony analysis with rapid MLST analyses.
As a practical matter, the maximum-parsimony analysis used to generate the majority-rule tree required hundreds of hours of computer time, so it is unlikely to be repeated routinely as new ST are added to the database. We sought to determine whether any of the readily available tools for analyzing MLST data would cluster strains in a way that approximates the results of the maximum-parsimony analysis. A comparison between eBURST and maximum parsimony is shown in Fig. 1. As noted above, while several eBURST groups correspond closely to one of the 13 clades, the two sets of groups are not equivalent. eBURST group 2 contains a set of 36 ST that are not in any of the 13 clades. Conversely, clades 4, 6, 9, and 13 do not contain eBURST groups larger than five ST. The NTHI-specific cluster within clade 2 is also not in an eBURST group. Thus, while analysis of the MLST database by eBURST will identify several clusters of closely related strains, it will not identify all such clusters.
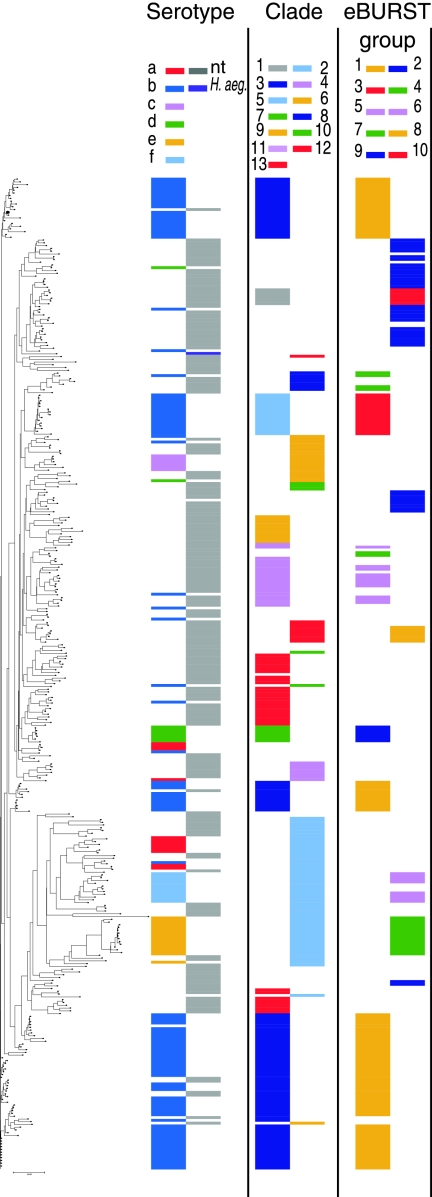
The lineage assignment algorithms in the START2 suite of sequence typing software (29) allow rapid generation of four trees from MLST data: neighbor-joining trees and unweighted-pair group method using average linkages trees based on either concatenated sequences or profiles (i.e., the set of seven allele numbers for each ST). We used the START2 algorithms to analyze the complete MLST database. Of the four resulting trees, the sequenced-based neighbor-joining tree was the one in which clustering of ST was most similar to the tree generated by maximum-parsimony analysis (Fig. 3). The main differences between the two were that the neighbor-joining analysis divided clade 3 (the large type b cluster) into three regions of the tree and clade 13 (NTHI) into two clusters. This method should allow tentative assignment of a newly typed strain to a cluster of strains that are likely to have a similar set of genetic islands.
FIG. 3.
Comparison between different analyses of the complete MLST database. The dendrogram was generated from MLST sequences by the neighbor-joining method, using software in the START2 suite of MLST analytic tools. In alignment with each ST on the tree is plotted the serotype of associated strains, and the assignment of the ST to clade and eBURST group as previously described is indicated. Table S1 in the supplemental material lists the position of each ST on this tree and on three other trees generated using other programs in START2.
Comparison with 16S rRNA typing.
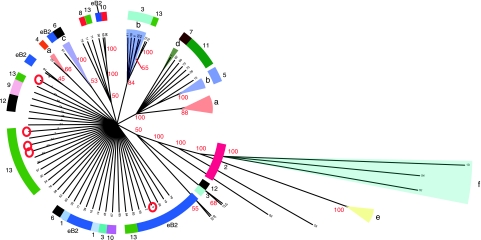
The phylogenetic clusters identified by maximum-parsimony analysis were also compared to the results of 16S typing. Forty-five NTHI strains were typed by 16S rRNA sequencing, resulting in the identification of 21 new ST. Maximum-parsimony analysis was used to compare these sequences with the 65 16S sequences obtained previously for 91 NTHI and 239 encapsulated strains (46) and to generate a majority-rule consensus tree (Fig. 4). As seen previously, strains were clustered on the tree by serotype, and NTHI strains were found in several regions on the tree. There was less diversity of 16S type than of MLST type. For example, 40 strains in eBURST group 2, of 22 different MLST types, were found to have only 9 different 16S types. Of these, 12 strains were 16S type 3 and 11 were 16S type 29. Certain clusters were very similar in both the 16S and MLST analyses. For example, 17 strains from MLST clade 2 (5 type e, 9 type f, and 2 NTHI) were all in a single 16S cluster that was found in 100% of trees. Clade 11 strains (n = 14) were in a monophyletic 16S cluster found in 100% of trees; the cluster also contained clade 5 (the small type b cluster), clade 7 (type d), and five type a strains. Taken as a whole, the 16S tree was not inconsistent with MLST analysis. However, most branches of the tree had very little support. As previously concluded (46), 16S typing can be useful for tracking both NTHI strains and serotypeable H. influenzae strains. Our current data suggest that it is less useful than MLST for inferring phylogenetic relationships of NTHI strains.
FIG. 4.
Comparison between 16S typing and MLST. The dendrogram is a majority-rule tree derived from maximum-parsimony analysis of 84 16S sequences. Each branch is labeled with a red numeral representing the percentage of trees contributing to the majority-rule consensus tree for that branch. Branches representing encapsulated strains are shaded in color and labeled by serotype. For each branch, the MLST phylogenetic group (clade or eBURST group 2) for the same strains is indicated by a colored arc. Strains with 16S ST 3, 4, 5, 31, and 80 (indicated by red circles) each belonged to more than one MLST phylogenetic group.
DISCUSSION
In an earlier study we identified a cluster of closely related NTHI strains that appeared to be similar in genetic content (11). It is now apparent that this cluster was only one of approximately 10 distinct phylogenetic groups within the NTHI population. Comparative genomic studies have suggested that H. influenzae, like several other pathogens, may have a species-wide “pan-genome” or “supra-genome” that is substantially larger than the genome of any single isolate (20). It has been further suggested that recombination occurs frequently, particularly in biofilms, so that each strain has a different combination of genetic islands (47). Our data do not contradict this suggestion, but they indicate that the distribution of genetic islands across the NTHI population is far from random. The relative uniformity of NTHI strains within each phylogenetic group with regard to these islands was unexpected. Humans are colonized with a series of NTHI strains throughout life, often carrying more than one strain at a time (12, 14, 36, 38). This would appear to provide ample opportunity for transfer of genes between strains, as H. influenzae is known to be naturally transformable. Indeed, horizontal genetic exchange between strains has been demonstrated in patients (26). Recent comparative genomic studies using genome sequencing and subtractive hybridization have revealed many more genetic islands than were previously known (25, 47, 55). Determination of the distribution of some of these islands across the H. influenzae phylogenetic tree is in progress (A. L. Erwin et al., unpublished data) and will add to our understanding of the differences between genetic groups. While in this study we have scored genetic islands simply as present or absent, we recognize that there is substantial sequence heterogeneity for many islands and for several other genes encoding outer membrane proteins (3, 6, 19, 24). Our goal in studying these islands was not to characterize differences between clades in detail but rather was to evaluate the hypothesis that clades differ in genetic content.
The finding that there are several distinct genetic groups within the NTHI population raises the question of how these groups are maintained. There is no obvious effect of geography, as most clades have members that were isolated in more than one European country as well as in several regions of North America. The finding that hmw-containing strains are clustered in certain clades suggests that clades may differ in their interaction with epithelial cells. It is known that many strains lacking hmw contain the gene for a different adhesin, Hia (10, 11, 51, 52). HMW adhesins bind sialylated glycoproteins, while the ligand(s) of Hia has not been identified. It is known that hmw-containing strains differ from hmw-negative strains in the specificity of binding to several epithelial cell lines (54). It is thus conceivable that strains in different clades occupy different niches within the nasopharynx. Finally, there may be restriction barriers between clades. Several restriction endonucleases in H. influenzae strains are known (45, 49), and the recently sequenced genomes revealed the existence of additional restriction-modification systems. For example, the 86-028NP genome contains three restriction systems not found in the Rd KW20 genome sequence, including the HaeII system (25). PCR analysis in the present study suggested that the HaeII locus and three novel restriction-modification loci are each limited to strains in a small number of phylogenetic clusters. Such heterogeneity in restriction systems may limit genetic exchange between randomly chosen strains.
We analyzed the MLST database using two methods that differ in their assumptions. TNT is a parsimony method that assumes descent with modification (50), uses full sequences (which makes it more sensitive to recombination), and quickly explores tree space to find the most parsimonious trees (41). eBURST additionally assumes evolution by clonal complex formation (16), uses allelic profiles (which makes it more sensitive to stochastic error [9]), and forms groups based on single-locus variants in alleles, with founders dependent on the number of single-locus variants. eBURST does not link ST that differ by more than two alleles, whereas TNT will construct their entire evolutionary history. Despite the differences between these methods in assumptions about the evolutionary process, in the data used, and in their sensitivity to recombination and stochastic errors, the results were surprisingly similar (Fig. 1). As expected, the eBURST analysis focused on the recently diverged tips of the tree. Most of the differences between the analyses were that TNT linked some strains that eBURST did not link. This most likely occurred where there were small differences between more than two alleles.
Comparison of the results of eBURST and TNT maximum-parsimony analyses provides some evidence for the relative importance of recombination and mutation in different parts of the H. influenzae population. Turner et al. recently described the outcome of eBURST analysis in three simulated populations differing in rates of mutation and recombination (53). For a clonal complex in which recombination is rare or is only moderately frequent, a plot of eBURST relationships yields a radial pattern (53). In the H. influenzae MLST database, the ST in eBURST group 1 form this type of pattern (see Fig. 7 in reference 53), suggesting descent from the founder, ST6. The fact that essentially the same strain cluster (clade 3) (Fig. 1) was identified by the sequence-based TNT analysis is consistent with relatively rare recombination among this group of type b strains.
A second type of eBURST pattern has no clear founder and is described as “straggly.” This occurs when eBURST groups strains that differ at only a single MLST locus even though they are not descended from a recent common ancestor. This pattern occurs in populations with frequent recombination (53). eBURST group 2, which contains approximately 23% of the NTHI strains in the database, has a straggly pattern (not shown). The interpretation that these strains do not form a radial clonal complex is consistent with their not forming a single monophyletic branch in the TNT analysis. Interestingly, we found that strains in eBURST group 2 are as similar in genetic content as strains within the TNT-defined phylogenetic groups (clades). The strains in this group may be closely related but undergo frequent intragroup recombination. More research on the effects of sampling on eBURST patterns will help determine if straggly groups in poorly sampled databases are indeed the result of recombination. Alternatively, the eBURST group can be explained as a large monophyletic group beginning at the top strain in eBURST group 2 and ending at the bottom strain in parsimony clade 2. The ancestral strains are included in eBURST group 2, but as lineages diverged, they may have accumulated mutations in more than two genes so that they could no longer be grouped by eBURST.
The third eBURST pattern described by Turner et al. (53) occurs in highly divergent populations, where each eBURST group contains only a very small number of ST. In H. influenzae, clade 13 does not correspond to an eBURST group; in other words, it is rare for strains in this group to share more than four MLST alleles with any other strain. However, the ST in clade 13 were shown by TNT analysis to be closely related, as they were clustered together in 100% of equally parsimonious trees. One interpretation is that in clade 13, mutation is frequent relative to recombination, resulting in large numbers of differences at the allele level, caused by a relatively few differences at the sequence level. These conclusions are limited by the potential differences between the simulations (53) and the actual content of the MLST database. The MLST database contains strains sampled with no consistent plan. It is highly likely that many intermediates connecting the MLST strains have not been sampled, which may affect eBURST patterns.
Our data have implications for the study of H. influenzae pathogenesis. The role of a genetic locus in disease is assessed in part by evaluating its distribution among strains from different clinical sources (10, 32, 44, 55). Recognition that many loci characteristically occur in certain phylogenetic clusters adds a different dimension to the distribution data. For example, the observation that hmw is more frequent in otitis media isolates than in isolates from healthy children was previously interpreted as indicating that the HMW adhesins have a specific role in pathogenesis of otitis media (10). The recognition that hmw-positive strains are clustered in certain phylogenetic groups may lead to identification of other loci that are common in those groups and are also candidates for virulence factors in otitis media. Otitis media strains in phylogenetic groups that are predominantly hmw negative may have a different set of virulence factors. Discovery of genes affecting complex phenotypes such as serum resistance will be aided by comparing strains within phylogenetic groups rather than by seeking genes common to all strains with a given phenotype. In some cases, such as evaluation of a vaccine candidate, it is desirable to study strains that are known to be genetically diverse. The phylogenetic tree thus provides a framework for design and interpretation of studies of H. influenzae biology.
An obvious question is whether clades differ in the frequency of association with disease or in its clinical presentation. For the NTHI strains currently in the database, we did not observe any correlation between clinical source and phylogenetic clustering. However, a limitation of this study is its sampling bias. Although asymptomatic colonization with NTHI is far more common than symptomatic infection, nearly 90% of the NTHI strains in the MLST database were isolated from patients with symptomatic infections. Although most NTHI infections are limited to the respiratory tract, strains from these mucosal infections are underrepresented in the database. Sixty percent of the NTHI strains in the database were cultured from normally sterile sites and typed as part of bacteremia surveillance studies carried out by public health laboratories in the United States and Scotland. While there have been epidemiologic studies characterizing NTHI strains from subjects who were not experiencing acute infections (10, 14, 17), few of those strains have been typed by MLST. Addition of these and other clinically diverse strains to the MLST database may result in development of new branches of the tree. We do not propose that the phylogenetic groups we describe should constitute an NTHI typing system. The majority-rule tree shown in Fig. 1 is our current best estimate of ancestral relationships among H. influenzae strains, but it is unlikely that this tree is complete.
In summary, a rigorous phylogenetic analysis of the MLST database showed for the first time that most NTHI strains are members of phylogenetic groups that are as distinct from each other as the six capsular serotypes. The clades that we identified are unexpectedly uniform in possession of genetic islands affecting metabolism, outer membrane protein composition, and LPS structure.
Supplementary Material
Acknowledgments
This work was funded by grants AI 44002 and AI 46512 (to A.L.S.), grants HL 083893 and DC 05840 (to J.R.G.), and a grant to the Seattle Biomedical Research Institute from the M.J. Murdock Charitable Trust.
This work would not have been possible without the Haemophilus influenzae MLST website, hosted at Imperial College, London, and funded by the Wellcome Trust.
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Augustynowicz, E., A. Gzyl, L. Szenborn, D. Banys, G. Gniadek, and J. Slusarczyk. 2003. Comparison of usefulness of randomly amplified polymorphic DNA and amplified-fragment length polymorphism techniques in epidemiological studies on nasopharyngeal carriage of non-typable Haemophilus influenzae. J. Med. Microbiol. 521005-1014. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss, C. D., M. J. Callaghan, and E. R. Moxon. 2006. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 344046-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolduc, G. R., V. Bouchet, R. Z. Jiang, J. Geisselsoder, Q. C. Truong-Bolduc, P. A. Rice, S. I. Pelton, and R. Goldstein. 2000. Variability of outer membrane protein P1 and its evaluation as a vaccine candidate against experimental otitis media due to nontypeable Haemophilus influenzae: an unambiguous, multifaceted approach. Infect. Immun. 684505-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buscher, A. Z., K. Burmeister, S. J. Barenkamp, and J. W. St. Geme, III. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol. 1864209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardines, R., M. Giufre, P. Mastrantonio, M. L. Ciofi degli Atti, and M. Cerquetti. 2007. Nontypeable Haemophilus influenzae meningitis in children: phenotypic and genotypic characterization of isolates. Pediatr. Infect. Dis. J. 26577-582. [DOI] [PubMed] [Google Scholar]
- 6.Clemans, D. L., C. F. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 357-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cripps, A. W., and D. C. Otczyk. 2006. Prospects for a vaccine against otitis media. Expert Rev. Vaccines 5517-534. [DOI] [PubMed] [Google Scholar]
- 9.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2002. Retraction: a link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 295971. [DOI] [PubMed] [Google Scholar]
- 10.Ecevit, I. Z., K. W. McCrea, M. M. Pettigrew, A. Sen, C. F. Marrs, and J. R. Gilsdorf. 2004. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 423065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erwin, A. L., K. L. Nelson, T. Mhlanga-Mutangadura, P. J. Bonthuis, J. L. Geelhood, G. Morlin, W. C. Unrath, J. Campos, D. W. Crook, M. M. Farley, F. W. Henderson, R. F. Jacobs, K. Mühlemann, S. W. Satola, L. van Alphen, M. Golomb, and A. L. Smith. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 735853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faden, H., L. Duffy, A. Williams, D. A. Krystofik, and J. Wolf. 1996. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first two years of life. Acta Otolaryngol. Suppl. 523128-129. [PubMed] [Google Scholar]
- 13.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 322382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 2341-46. [DOI] [PubMed] [Google Scholar]
- 15.Farris, J. S. 1997. The future of phylogeny reconstruction. Zoologica Scripta 26303-311. [Google Scholar]
- 16.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernaays, M. M., A. J. Lesse, S. Sethi, X. Cai, and T. F. Murphy. 2006. Differential genome contents of nontypeable Haemophilus influenzae strains from adults with chronic obstructive pulmonary disease. Infect. Immun. 743366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geluk, F., P. P. Eijk, S. M. van Ham, H. M. Jansen, and L. van Alphen. 1998. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect. Immun. 66406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giufre, M., M. Muscillo, P. Spigaglia, R. Cardines, P. Mastrantonio, and M. Cerquetti. 2006. Conservation and diversity of HMW1 and HMW2 adhesin binding domains among invasive nontypeable Haemophilus influenzae isolates. Infect. Immun. 741161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gladitz, J., K. Shen, P. Antalis, F. Z. Hu, J. C. Post, and G. D. Ehrlich. 2005. Codon usage comparison of novel genes in clinical isolates of Haemophilus influenzae. Nucleic Acids Res. 333644-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goloboff, P. A. 1999. Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15415-428. [DOI] [PubMed] [Google Scholar]
- 22.Goloboff, P. A., J. S. Farris, M. Källersjö, and B. Oxelman. 2003. Improvements to resampling measures of group support. Cladistics 19324-332. [Google Scholar]
- 23.Gotfried, M. H. 2001. Epidemiology of clinically diagnosed community-acquired pneumonia in the primary care setting: results from the 1999-2000 respiratory surveillance program. Am. J. Med. 111(Suppl. 9A)25S-29S. [DOI] [PubMed] [Google Scholar]
- 24.Haase, E. M., A. A. Campagnari, J. Sarwar, M. Shero, M. Wirth, C. U. Cumming, and T. F. Murphy. 1991. Strain-specific and immunodominant surface epitopes of the P2 porin protein of nontypeable Haemophilus influenzae. Infect. Immun. 591278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 1874627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiltke, T. J., A. T. Schiffmacher, A. J. Dagonese, S. Sethi, and T. F. Murphy. 2003. Horizontal transfer of the gene encoding outer membrane protein P2 of nontypeable Haemophilus influenzae, in a patient with chronic obstructive pulmonary disease. J. Infect. Dis. 188114-117. [DOI] [PubMed] [Google Scholar]
- 27.Hood, D. W., M. E. Deadman, A. D. Cox, K. Makepeace, A. Martin, J. C. Richards, and E. R. Moxon. 2004. Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 1502089-2097. [DOI] [PubMed] [Google Scholar]
- 28.Hood, D. W., K. Makepeace, M. E. Deadman, R. F. Rest, P. Thibault, A. Martin, J. C. Richards, and E. R. Moxon. 1999. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33679-692. [DOI] [PubMed] [Google Scholar]
- 29.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 171230-1231. [DOI] [PubMed] [Google Scholar]
- 30.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 939-62. [DOI] [PubMed] [Google Scholar]
- 31.Kilian, M., C. H. Mordhorst, C. R. Dawson, and H. Lautrop. 1976. The taxonomy of haemophili isolated from conjunctivae. Acta Pathol. Microbiol. Scand. B 84132-138. [DOI] [PubMed] [Google Scholar]
- 32.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme, III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46731-743. [DOI] [PubMed] [Google Scholar]
- 34.Martin, K., G. Morlin, A. Smith, A. Nordyke, A. Eisenstark, and M. Golomb. 1998. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J. Bacteriol. 180107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 411623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, T. F., A. L. Brauer, A. T. Schiffmacher, and S. Sethi. 2004. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 170266-272. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 19581-89. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, T. F., S. Sethi, K. L. Klingman, A. B. Brueggemann, and G. V. Doern. 1999. Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J. Infect. Dis. 180404-409. [DOI] [PubMed] [Google Scholar]
- 39.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 857758-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nixon, K. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15407-414. [DOI] [PubMed] [Google Scholar]
- 42.Olsen, C. H. 2003. Review of the use of statistics in Infection and Immunity. Infect. Immun. 716689-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peerbooms, P. G., M. N. Engelen, D. A. Stokman, B. H. van Benthem, M. L. van Weert, S. M. Bruisten, A. van Belkum, and R. A. Coutinho. 2002. Nasopharyngeal carriage of potential bacterial pathogens related to day care attendance, with special reference to the molecular epidemiology of Haemophilus influenzae. J. Clin. Microbiol. 402832-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 703551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piekarowicz, A., and R. Brzezinski. 1980. Cleavage and methylation of DNA by the restriction endonuclease HinfIII isolated from Haemophilus influenzae Rf. J. Mol. Biol. 144415-429. [DOI] [PubMed] [Google Scholar]
- 46.Sacchi, C. T., D. Alber, P. Dull, E. A. Mothershed, A. M. Whitney, G. A. Barnett, T. Popovic, and L. W. Mayer. 2005. High level of sequence diversity in the 16S rRNA genes of Haemophilus influenzae isolates is useful for molecular subtyping. J. Clin. Microbiol. 433734-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen, K., P. Antalis, J. Gladitz, S. Sayeed, A. Ahmed, S. Yu, J. Hayes, S. Johnson, B. Dice, R. Dopico, R. Keefe, B. Janto, W. Chong, J. Goodwin, R. M. Wadowsky, G. Erdos, J. C. Post, G. D. Ehrlich, and F. Z. Hu. 2005. Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect. Immun. 733479-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sitnikova, T., A. Rzhetsky, and M. Nei. 1995. Interior-branch and bootstrap tests of phylogenetic trees. Mol. Biol. Evol. 12319-333. [DOI] [PubMed] [Google Scholar]
- 49.Smith, H. O., and G. M. Marley. 1980. Purification and properties of HindII and HindIII endonucleases from Haemophilus influenzae Rd. Methods Enzymol. 65104-108. [DOI] [PubMed] [Google Scholar]
- 50.Sober, E. 1988. The conceptual relationship of cladistic phylogenetics and vicariance biogeography. Syst. Zool. 37245-253. [Google Scholar]
- 51.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 623881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner, K. M., W. P. Hanage, C. Fraser, T. R. Conner, and B. G. Spratt. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 684658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie, J., P. C. Juliao, J. R. Gilsdorf, D. Ghosh, M. Patel, and C. F. Marrs. 2006. Identification of new genetic regions more prevalent in nontypeable Haemophilus influenzae otitis media strains than in throat strains. J. Clin. Microbiol. 444316-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.