Abstract
Bacteria contain a single multisubunit RNA polymerase that is responsible for the synthesis of all RNA. Previous studies of the Escherichia coli K-12 laboratory strain identified a group of effector proteins that interact directly with RNA polymerase to modulate the efficiency of transcription initiation, elongation, or termination. Here we used a rapid affinity isolation technique to isolate RNA polymerase from the pathogenic Escherichia coli strain O157:H7 Sakai. We analyzed the RNA polymerase enzyme complex using mass spectrometry and identified associated proteins. Although E. coli O157:H7 Sakai contains more than 1,600 genes not present in the K-12 strain, many of which are predicted to be involved in transcription regulation, all of the identified proteins in this study were encoded on the “core” E. coli genome.
In all bacteria, one multisubunit RNA polymerase (RNAP) is responsible for the synthesis of RNA. In the widely studied model bacteria Escherichia coli K-12, the RNAP core enzyme is a complex of five subunits (α2ββ′ω) that is competent for DNA-directed RNA synthesis but is reliant upon the binding of a dissociable sigma factor, which forms the holoenzyme, for promoter recognition and transcription initiation. The most-abundant sigma factor in the Escherichia coli K-12 laboratory strain is σ70 (the rpoD gene product), which directs the majority of transcription during vegetative growth in rich media (4, 22). Six other sigma factors that are required for growth and survival in certain growth conditions have been identified (10).
Regulating the activity of RNAP is a key mechanism in the adaptation of E. coli to environmental stress. It is primarily achieved by more than 300 DNA binding transcription factors that modulate the activity of RNAP at specific promoters (19). In addition, several effector proteins, which interact directly with RNAP, have been identified. These effector proteins affect different processes in the transcription cycle, from modulating the binding of specific sigma factors (26) to altering the efficiency of transcription elongation or termination (15).
The genome sequences of several pathogenic E. coli strains have now been determined, and it is apparent that these strains differ greatly from the laboratory K-12 strain. For example, in a comparison of three E. coli strains, MG1655 K-12, CFT073, and O157:H7 EDL933, only 2,996 proteins out of a total of 7,638 identified proteins were found to be shared among the strains (27). Moreover, a recent comparison of 31 O157:H7 strains revealed that only 67% of open reading frames were detected in all the strains (30). Thus, it seems likely that the genetic composition of any E. coli strain consists of a “core” genome, common to all strains of the genus Escherichia, a subset of genes shared by several strains, and a number of genes specific to that particular strain. Hence, throughout the genus Escherichia, it is likely that a significant number of transcription regulatory proteins and RNAP effector proteins remain to be been identified.
In the present study, we isolated RNAP from the enterohemorrhagic E. coli strain O157:H7 Sakai, whose genome is 859 kb larger than that of K-12 and contains more than 1,600 genes that are not present in K-12 (8). Enterohemorrhagic E. coli strains are important human pathogens, capable of causing severe enteritis and hemolytic uremia syndrome (16, 21). For virulence in humans, the infectious dose of O157:H7 is very low, perhaps as low as 100 viable organisms, indicating that survival of the gastric acid barrier is key to O157:H7 virulence (13). To determine if any of the additional genes in O157:H7 Sakai encode products that are associated with RNAP, we analyzed the complex at different phases of growth and during virulence-inducing conditions. To do this, we fused the β′ subunit of RNAP with a protein A affinity tag and used a rapid affinity isolation technique to purify the RNAP enzyme from cells. The proteins were then identified by mass spectrometry. We identified several proteins previously reported to interact with RNAP and some unknown proteins. However, all the identified RNAP-associated proteins were from the core genome, and none were specific to the O157:H7 Sakai strain. By utilizing the I-DIRT (isotopic determination of interactions as random or targeted) (24) technique, we also determined which of these proteins are tightly associated with RNAP during the stationary and exponential phases of growth.
MATERIALS AND METHODS
E. coli strains.
A derivative of the E. coli strain O157:H7 Sakai with deletions of the Shiga toxin-encoding genes Stx1 and Stx2 was used in this study (a gift from M. Goldberg). To isolate RNAP, we engineered a version of this strain in which the C terminus of the rpoC gene was fused to four repeats of a sequence encoding a protein A affinity tag (4PrA) (2). RNAP from the resulting strain could then be isolated by immunoprecipitation with immunoglobulin G-tethered beads. To tag rpoC, we used the gene-gorging technique for chromosomal gene tagging (9). A derivative of pET21a containing an I-SceI meganuclease site followed by the C-terminal 200 bp of the rpoC gene fused to the 4PrA affinity tag was constructed. Immediately downstream was a kanamycin resistance gene, followed by 200 bp containing homology to the region of the chromosome immediately downstream of the rpoC gene. This plasmid was used as the donor plasmid for gene gorging. This resulted in an O157:H7 Sakai strain expressing RNAP tagged at the C terminus of the β′ subunit with a 4PrA moiety carrying a selectable kanamycin resistance cassette. We confirmed the presence of the chromosomal rpoC::4PrA fusion by PCR and the expression of the fused protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by Western blotting with an antibody specific to protein A.
Growth of strains and sample preparation.
Cells from the tagged strain were grown in either minimal salts media (MSM) supplemented with 0.4% glucose (20) or in Dulbecco's modified Eagle media (DMEM) to the mid-exponential phase (optical density at 650 nm [OD650], 0.7) of growth. Cells were collected by centrifugation, frozen as small pellets in liquid nitrogen, and stored at −80°C. Cells subjected to an acid challenge were grown in MSM plus 0.4% glucose, and the pH of the culture was lowered with HCl to pH 3.0 30 min prior to harvesting.
To prepare samples for I-DIRT analysis, the tagged strain was grown in MSM supplemented with 0.4% glucose. The parent strain was grown in media in which the (NH4)2SO4 was replaced with (15NH4)2SO4 (Cambridge Isotopes). Growth rates for the two strains were tested and found to be identical. Both strains were grown at 37°C to mid-log phase (OD650, 0.7) and stationary phase (OD650 for 24-h culture, 3.0), and cells were collected by centrifugation, frozen as small pellets in liquid nitrogen, and stored at −80°C.
Affinity isolation of affinity-tagged RNAP.
To attempt to maintain protein interactions during the isolation of RNAP:4PrA, we used nonstringent conditions and a rapid purification protocol (6, 28), ensuring minimal loss of interacting proteins while reducing the likelihood of contamination. For growth in MSM, MSM plus acid, and DMEM, we resuspended 1 g of cell pellets in 10 ml of extraction buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 2 mM MgCl2, 0.1% Tween 20, 200 μg/ml phenylmethylsulfonyl fluoride, 4 μg/ml pepstatin, protease inhibitor cocktail [1 tablet; Roche]). DNase I (20 μg/ml), RNase A (300 μg/ml), and lysozyme (200 μg/ml) were then added, the mixture was sonicated three times for 1 min each, and the lysate was cleared by centrifugation.
To prepare I-DIRT samples, an equal weight of cell pellets from the parent strain (isotopically heavy) and the tagged strain (isotopically light) were mixed and cryogenically lysed in a grinding mill (Retsch MM301) maintained at liquid nitrogen temperature. Two grams of the lysed mixture from the parent and tagged strains was then resuspended in 20 ml of extraction buffer. DNase I and RNase A were added, and the lysate was cleared by centrifugation.
For each of the lysis methods, the supernatant was removed and incubated with 20 mg of Dynabeads (M270-epoxy; Dynal) coated with rabbit immunoglobulin G for 3 min. The beads were collected with a magnet and washed with buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 0.1% Tween 20). The proteins were then eluted from the beads by incubation for 5 min with elution buffer (0.5 M NH4OH-0.5 mM EDTA). Coeluted proteins were vacuum dried and reduced and alkylated by resuspension in SDS-PAGE loading buffer containing 10 mM tris(2-carboxyethyl)phosphine-HCl (Sigma) and 50 mM iodoacetamide (Sigma). The proteins were resolved by SDS-PAGE in 4% to 12% gradient gels (Invitrogen) and visualized with Coomassie blue staining. The entire lane was sliced, and the proteins in each slice were digested with trypsin for 8 h at 37°C. The peptides from each gel slice were purified and analyzed by mass spectrometry.
Mass spectrometry and I-DIRT analyses.
The samples derived from exponential-phase cells grown in DMEM were analyzed using an LTQ FT Ultra hybrid mass spectrometer (Thermo Scientific), and the data were analyzed using BioWorks and the SEQUEST algorithm (Thermo Scientific). The samples derived from MSM plus acid were analyzed using a MALDI QqTOF mass spectrometer (12) and then subjected to MALDI ion trap tandem mass spectrometry (11). The data were analyzed using the programs ProFound (29) and XProteo.
The I-DIRT samples were analyzed using a MALDI QqTOF mass spectrometer, and ion peak masses were assigned using the program MoverZ. A list of peptide masses was obtained for each gel slice and searched for proteins by using ProFound and XProteo. For each protein identified, the assigned peptides were validated by searching for each assigned peptide in a mass spectrum derived from an affinity isolate from tagged, isotopically light cells. Next, the number of nitrogen atoms in each peptide was calculated, and the mass spectrum for the light/heavy mix was searched for the presence of the heavy peptides. Heavy peptides were validated by their absence in a mass spectrum derived solely from affinity isolates from tagged, isotopically light cells. All detected ion peaks corresponding to light and heavy peptides were then subjected to MALDI ion trap tandem mass spectrometry, the resulting fragmentation masses were searched for proteins, using XProteo, and each peptide was validated, using the program ProteinProspector (prospector.ucsf.edu). The nature of the interaction was investigated by analyzing the isotopic ratio of the identified peptides (I-DIRT analysis). For each peptide identified, peak areas were assigned for the light and heavy species, using the program MoverZ, and light/heavy ratios were determined as previously described (24).
RESULTS
RNAP-associated proteins.
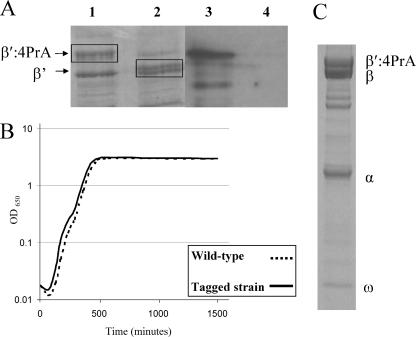
To identify proteins from the O157:H7 Sakai strain that interact with RNAP, we affinity isolated RNAP, using a chromosomally tagged β′:4PrA subunit. The 4PrA tag (2) was located at the C terminus of the β′ subunit; a previous report showed that tagging at this location had no detrimental effects on the function of RNAP (17). To confirm the presence of tagged β′, we analyzed a whole-cell lysate by SDS-PAGE and visualized β′:4PrA by Western blotting, which was probed with antibodies specific for protein A (Fig. 1A). Figure 1B shows that the presence of the 4PrA tag had little or no effect on cell growth compared to the growth of the parent strain.
FIG. 1.
Experiments with E. coli O157:H7 Sakai carrying an affinity-tagged RNAP β′ subunit. E. coli O157:H7 Sakai and a derivative carrying a chromosomal rpoC::PrA fusion were grown in MSM. (A) SDS-PAGE analysis of total cell extracts. Only the segment of the gel with the β′ subunit is shown. Lanes 1 and 3 show the tagged strain and lanes 2 and 4 show the wild-type strain. The proteins in lanes 1 and 2 were stained with Coomassie blue. Lanes 3 and 4 are from a Western blot probed with antibody specific for protein A. (B) Growth curve experiment comparing wild-type growth (dashed line) with the tagged strain (solid line). (C) SDS-PAGE analysis of proteins coisolated with RNAP:4PrA. Proteins were stained with Coomassie blue. Core RNAP subunits are indicated.
RNAP was affinity isolated from cells grown to mid-exponential phase in three different media: minimal salts supplemented with 0.4% glucose; minimal salts supplemented with 0.4% glucose and subjected to an acid shock to mimic the human gastric acid barrier; and DMEM, which contains sodium bicarbonate, a key compound in the lower intestinal tract known to promote O157:H7 Sakai colonization (1). We also examined whether the growth phase affected the binding of proteins to RNAP by affinity isolating RNAP from cells grown to stationary phase in minimal salts supplemented with 0.4% glucose. Importantly, prior to being affinity isolated, the samples were incubated with DNase I and RNase A to prevent DNA binding proteins (e.g., transcription factors) and RNA-associated proteins (e.g., ribosomes) from being coisolated with RNAP. Protein complexes isolated in each growth condition were analyzed by SDS-PAGE. Figure 1C shows a typical gel lane with the locations of the major RNAP subunits. The entire gel lane for each affinity isolation was sliced, and the proteins were analyzed after being digested with trypsin.
Table 1 lists the proteins that were identified from each of the four growth conditions. In total, 29 proteins were identified. As expected, these include the β′:4PrA fusion protein (RpoC) along with the other three components of the core RNAP enzyme complex, β (RpoB), α (RpoA), and ω (RpoZ). Four of the seven sigma factors (14) were identified: σ70 (RpoD), the most-abundant sigma factor; σ38 (RpoS), the stationary-phase sigma factor, σ54 (RpoN), the sigma factor that controls expression of nitrogen-related genes, and σ24 (RpoE), a sigma factor that drives transcription of genes required under heat shock conditions. All four sigma factors were identified in each of the exponential-phase affinity isolates, whereas only σ70 and σ38 were coisolated in stationary-phase isolates. Two proteins, RapA, which is involved in the recycling of RNAP (23), and NusG (15), which is involved in transcription antitermination, were coisolated in all growth conditions. Of the remaining proteins, nine have previously been reported to associate with RNAP in E. coli K-12 (3, 5, 7). DnaK, NusA, YegD, TufA, DnaJ, GreB, YacL, and CedA were coisolated in one or more of the growth conditions, whereas Crl was isolated only in DMEM. The other 10 proteins that coisolated with RNAP are GadB, AtpD, OmpC, OmpA, RfaD, YgfB, OmpX, Dps, YgaU, and ElaB. None of these identified proteins were found to be as abundant as the core RNAP subunits when viewed on the SDS-PAGE gel (Fig. 1C), suggesting that none of the proteins are associated with all of the RNAP enzymes within the cell.
TABLE 1.
Proteins coisolated with O157:H7 Sakai RNAP under different growth conditions
| Protein | Molecular mass (kDa) | Role | Exponential phase
|
Stationary-phase minimal media | ||
|---|---|---|---|---|---|---|
| Minimal media | Acid challenge | DMEM | ||||
| RpoC | 155.16 | RNAP β′ subunit | X | X | X | X |
| RpoB | 150.63 | RNAP β subunit | X | X | X | X |
| RapA | 109.77 | RNAP recycling | X | X | X | X |
| RpoD | 70.26 | Primary σ factor | X | X | X | X |
| DnaK | 69.12 | Chaperone | X | X | X | X |
| NusA | 54.87 | Transcription antitermination | X | X | ||
| RpoN | 53.99 | Nitrogen regulating σ factor | X | X | X | |
| GadB | 52.66 | Glutamate decarboxylase B | X | |||
| AtpD | 50.33 | ATP synthase β subunit | X | |||
| YegD | 49.37 | DnaK homologue | X | X | X | |
| TufA | 43.28 | Elongation factor Tu | X | X | X | |
| DnaJ | 41.10 | Chaperone with DnaK | X | X | ||
| OmpC | 40.37 | Outer membrane porin C | X | |||
| RpoS | 37.97 | Stationary-phase σ factor | X | X | X | X |
| OmpA | 37.20 | Outer membrane porin A | X | |||
| RpoA | 36.51 | RNAP α subunit | X | X | X | X |
| RfaD | 34.89 | l-Glycero-d-mannoheptose epimerase | X | X | ||
| RpoE | 21.70 | Heat shock σ factor | X | X | X | |
| NusG | 21.53 | Transcription antitermination | X | X | X | X |
| YgfB | 21.23 | Unknown | X | X | X | |
| OmpX | 18.60 | Outer membrane porin X | X | |||
| GreB | 18.53 | Transcription elongation factor | X | X | X | |
| Dps | 18.70 | Iron binding DNA binding protein | X | X | ||
| YgaU | 16.06 | Unknown | X | |||
| Crl | 15.66 | Holoenzyme formation | X | |||
| YacL | 13.94 | RNAP association | X | X | X | X |
| ElaB | 11.31 | Unknown | X | |||
| RpoZ | 10.24 | RNAP ω subunit | X | X | X | X |
| CedA | 10.20 | Cell division modulator | X | X | X | X |
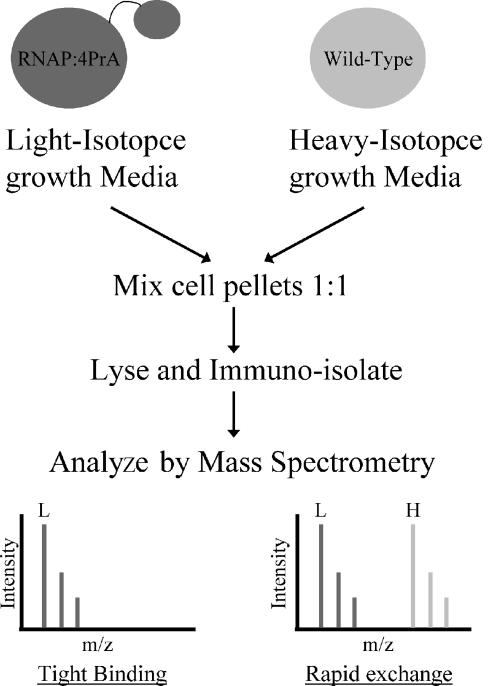
I-DIRT analysis of the RNAP complex from exponential- and stationary-phase cultures.
In order to distinguish RNAP binding proteins that bind tightly to the complex within the cell from those that are nonspecific contaminants, we utilized the I-DIRT technique, which is outlined in Fig. 2 (24). Cells expressing the 4PrA-tagged RNAP were grown in light-isotope media, whereas the parent strain was grown in heavy-isotope media. Cells from each were mixed and lysed, and after affinity isolation of the RNAP enzyme complex, copurified proteins were separated by SDS-PAGE and analyzed by mass spectrometry. Specifically bound proteins that remained tightly associated with the RNAP complex (i.e., those having a slow exchange rate) consisted of 100% light-isotope proteins, whereas proteins that interacted with RNAP only after cell lysis and during the affinity isolation procedure had an equal chance of being represented by light- and heavy-isotope proteins. There is an intermediate possibility for specific but relatively fast-exchanging proteins; these would contain somewhere between 50% light (for fast-exchanging proteins) and 100% light (for slow-exchanging proteins) isotopes.
FIG. 2.
I-DIRT procedure for analysis of a protein complex. Cells from a strain containing an affinity-tagged protein were grown in light-isotope media, whereas wild-type cells were grown in heavy-isotope media [(15NH4)2SO4]. Equal amounts of the cells were mixed, and the tagged protein complex was affinity isolated. Proteins within the complex were identified by mass spectrometry. Tight binding proteins were identified as mostly isotopically light, and rapidly exchanging proteins were identified as an equal mixture of light and heavy isotopes.
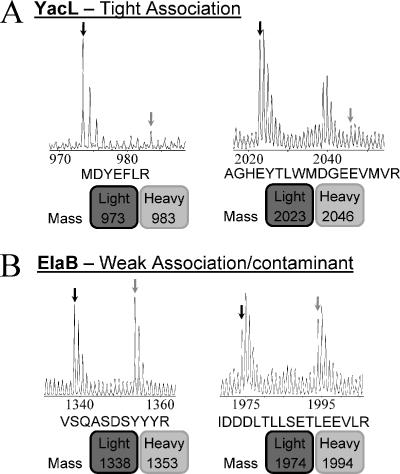
Figure 3 shows an example of the mass spectrum of two identified peptides from YacL and two from ElaB. Figure 3A shows the spectrum of a singly protonated YacL peptide with a mass-to-charge ratio (m/z) of 973 Da. The mass of the corresponding heavy-isotope peptide is dependent on the number of nitrogen (15N) atoms within the peptide. The calculated m/z for this peptide, which contains 10 nitrogen atoms, is 983 Da—i.e., 10 Da heavier than the light-isotope-containing peptide. No peak at this mass is discernible above the chemical noise in the mass spectrum, indicating that YacL represents a tight, specific interaction. This is also the case for the YacL peptide, with a m/z of 2,023 Da, confirming that YacL is a tight binding RNAP-associated protein. In contrast, the peptides corresponding to ElaB are present in both the light- and heavy-isotope forms, with a light/heavy ratio of approximately 1:1. Hence, ElaB is either a contaminant or involved in rapid exchange (Fig. 3B).
FIG. 3.
Examples of mass spectra for I-DIRT analysis. The figure shows single-stage mass spectra for the RNAP tightly associated protein YacL (A) and the contaminant ElaB (B). The peaks corresponding to the masses of the light-isotope peptides are indicated by black arrows and to those of the heavy-isotope peptides by light-gray arrows. The amino acid sequences of the light-isotope peptides are shown, and the masses of the light- and heavy-isotope peptides are indicated.
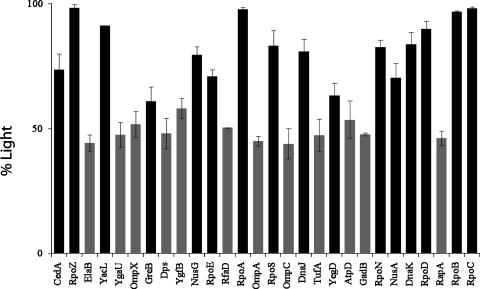
The isotopic ratios of at least three peptides for each of the proteins that were identified in the exponential phase or stationary phase were calculated, and the mean values for these ratios are shown in Fig. 4. Taking into account the statistical variance of the results, a cutoff of 60% light was applied, whereby those proteins consisting of >60% light isotopes were deemed to be bona fide interactors with various degrees of rates of exchange, whereas those under 60% were deemed to be either very rapidly exchanging proteins or contaminants. As expected, the composition of identified peptides for the four subunits of the RNAP core enzyme, β′ (RpoC), β (RpoB), α (RpoA), and ω (RpoZ), was almost 100% light isotope. All of the identified sigma subunits, RpoE, RpoS, RpoN, and RpoD, were identified as tightly associated proteins, along with the two transcription antitermination proteins NusA and NusG and the transcription elongation factor GreB. The chaperone complex DnaK/DnaJ and the predicted DnaK homologue, YegD, were also in this group, along with CedA and YacL. The remaining 12 proteins that were coisolated with RNAP all consist of heavy- and light-isotope peptides with a ratio approaching 1:1, indicating that these proteins are either very rapidly exchanging proteins or contaminants.
FIG. 4.
I-DIRT analysis of RNAP-associated proteins. The figure illustrates the percentage of light isotopes in each component of the RNAP affinity isolate. Results for the tight binding proteins, consisting of more than 60% light-isotope peptides, are shown as black bars. Results for the rapidly exchanging or contaminating proteins are shown as light gray bars.
DISCUSSION
Using the I-DIRT technique, we identified 12 proteins that are tightly associated with the RNAP enzyme from E. coli O157:H7 Sakai during different stages of growth. These are RpoD, DnaK, NusA, RpoN, YegD, DnaJ, RpoS, RpoE, NusG, GreB, YacL, and CedA. These proteins also coisolated with RNAP purified from cells grown in virulence-inducing DMEM, and all but two (NusA and DnaJ) of these proteins were coisolated with RNAP in acid-shocked cells. We also identified 12 proteins that were categorized by I-DIRT as weak associates or contaminants: RapA, GadB, AtpD, TufA, OmpC, OmpA, RfaD, YgfB, Dps, OmpX, YgaU, and ElaB. It is important to note that some low-abundance proteins, or proteins that interact weakly with RNAP, may not have been detected in this study.
A key conclusion from this work is that none of the RNAP binding proteins are Sakai specific, despite the fact that the genome of O157:H7 Sakai codes for more than 1,600 proteins not present in the K-12 laboratory strain. This is surprising, given that around 40 of these proteins are predicted transcription regulatory proteins and more than 750 are of unknown function. Instead, every protein that we determined to tightly associate with RNAP has previously been shown to interact with RNAP in the E. coli K-12 laboratory strain (3, 5). Our results suggest that RNAP from O157:H7 Sakai is not tightly associated with any Sakai-specific proteins.
In addition to tightly associated proteins, we identified several proteins associated with RNAP that are categorized as contaminants or weak specifically associated proteins. Although most of these are likely to be true contaminants, two, RapA and TuFA, are known to form bona fide interactions with RNAP (18, 25). Presumably these proteins bind specifically, but weakly, to RNAP, with fast exchange rates. Note, however, that our observation of the association of these proteins with RNAP means that the rate of exchange is sufficiently slow or that the proteins are sufficiently abundant so as not to lose the protein during the affinity isolation procedure. There is also the possibility that the interaction is inhibited within the cell and forms only upon cell lysis, when RNAP is released from the DNA binding and is no longer involved in transcription.
Finally, the known RNAP-associated protein, Crl, was coisolated with RNAP only from cells grown in DMEM medium. Crl is involved in assisting sigma factor binding to core enzyme, particularly alternate sigma factors such as σ38 (7, 26). The DMEM medium contains sodium bicarbonate, which is an abundant compound in the lower intestine where the bacteria colonize. Since the environment in the gut is hostile, transcription directed by other sigma factors may play a role in its survival, suggesting a possible role for Crl in the programming of RNAP to express particular genes during colonization.
Acknowledgments
This work was funded by a Wellcome Trust program grant (S.J.W.B.), grants RR00862 and RR022220 from the National Institutes of Health (B.T.C.), and an EMBO short-term fellowship to D.J.L. Thanks also to Seth Darst for hospitality at Rockefeller University, New York, and additional funding (NIH grant GM61898).
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Abe, H., I. Tatsuno, T. Tobe, A. Okutani, and C. Sasakawa. 2002. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 703500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitchison, J. D., G. Blobel, and M. P. Rout. 1995. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 1311659-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arifuzzaman, M., M. Maeda, A. Itoh, K. Nishikata, C. Takita, R. Saito, T. Ara, K. Nakahigashi, H. C. Huang, A. Hirai, K. Tsuzuki, S. Nakamura, M. Altaf-Ul-Amin, T. Oshima, T. Baba, N. Yamamoto, T. Kawamura, T. Ioka-Nakamichi, M. Kitagawa, M. Tomita, S. Kanaya, C. Wada, and H. Mori. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess, R. R., A. A. Travers, J. J. Dunn, and E. K. Bautz. 1969. Factor stimulating transcription by RNA polymerase. Nature 22143-46. [DOI] [PubMed] [Google Scholar]
- 5.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433531-537. [DOI] [PubMed] [Google Scholar]
- 6.Cristea, I. M., R. Williams, B. T. Chait, and M. P. Rout. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 41933-1941. [DOI] [PubMed] [Google Scholar]
- 7.Gaal, T., M. J. Mandel, T. J. Silhavy, and R. L. Gourse. 2006. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 1887966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 9.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311153-163. [DOI] [PubMed] [Google Scholar]
- 10.Ishihama, A. 1999. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4135-143. [DOI] [PubMed] [Google Scholar]
- 11.Krutchinsky, A. N., M. Kalkum, and B. T. Chait. 2001. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal. Chem. 735066-5077. [DOI] [PubMed] [Google Scholar]
- 12.Krutchinsky, A. N., W. Zhang, and B. T. Chait. 2000. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J. Am. Soc. Mass. Spectrom. 11493-504. [DOI] [PubMed] [Google Scholar]
- 13.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1774097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 283497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason, S. W., and J. Greenblatt. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 51504-1512. [DOI] [PubMed] [Google Scholar]
- 16.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 3521207-1212. [DOI] [PubMed] [Google Scholar]
- 17.Mooney, R. A., and R. Landick. 2003. Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev. 172839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzzin, O., E. A. Campbell, L. Xia, E. Severinova, S. A. Darst, and K. Severinov. 1998. Disruption of Escherichia coli hepA, an RNA polymerase-associated protein, causes UV sensitivity. J. Biol. Chem. 27315157-15161. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Rueda, E., and J. Collado-Vides. 2000. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 281838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pope, N. R., and J. A. Cole. 1982. Generation of a membrane potential by one of two independent pathways for nitrite reduction by Escherichia coli. J. Gen. Microbiol. 128219-222. [DOI] [PubMed] [Google Scholar]
- 21.Ray, P. E., and X. H. Liu. 2001. Pathogenesis of Shiga toxin-induced hemolytic uremic syndrome. Pediatr. Nephrol. 16823-839. [DOI] [PubMed] [Google Scholar]
- 22.Reznikoff, W. S., D. A. Siegele, D. W. Cowing, and C. A. Gross. 1985. The regulation of transcription initiation in bacteria. Annu. Rev. Genet. 19355-387. [DOI] [PubMed] [Google Scholar]
- 23.Sukhodolets, M. V., and D. J. Jin. 1998. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 2737018-7023. [DOI] [PubMed] [Google Scholar]
- 24.Tackett, A. J., J. A. DeGrasse, M. D. Sekedat, M. Oeffinger, M. P. Rout, and B. T. Chait. 2005. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 41752-1756. [DOI] [PubMed] [Google Scholar]
- 25.Trigwell, S., and R. E. Glass. 1998. Function in vivo of separate segments of the beta subunit of Escherichia coli RNA polymerase. Genes Cells 3635-647. [DOI] [PubMed] [Google Scholar]
- 26.Typas, A., C. Barembruch, A. Possling, and R. Hengge. 2007. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 261569-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westblade, L. F., L. Minakhin, K. Kuznedelov, A. J. Tackett, E. J. Chang, R. A. Mooney, I. Vvedenskaya, Q. Wang, D. Fenyö, M. P. Rout, R. Landick, B. T. Chait, K. Severinov, and S. A. Darst. Rapid isolation and identification of bacteriophage T4-encoded modifications of Escherichia coli RNA polymerase: a generic method to study bacteriophage/host interactions. J. Proteome Res., in press. [DOI] [PMC free article] [PubMed]
- 29.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 722482-2489. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., C. Laing, M. Steele, K. Ziebell, R. Johnson, A. K. Benson, E. Taboada, and V. P. Gannon. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8121. [DOI] [PMC free article] [PubMed] [Google Scholar]