Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) are hypervariable loci widely distributed in prokaryotes that provide acquired immunity against foreign genetic elements. Here, we characterize a novel Streptococcus thermophilus locus, CRISPR3, and experimentally demonstrate its ability to integrate novel spacers in response to bacteriophage. Also, we analyze CRISPR diversity and activity across three distinct CRISPR loci in several S. thermophilus strains. We show that both CRISPR repeats and cas genes are locus specific and functionally coupled. A total of 124 strains were studied, and 109 unique spacer arrangements were observed across the three CRISPR loci. Overall, 3,626 spacers were analyzed, including 2,829 for CRISPR1 (782 unique), 173 for CRISPR2 (16 unique), and 624 for CRISPR3 (154 unique). Sequence analysis of the spacers revealed homology and identity to phage sequences (77%), plasmid sequences (16%), and S. thermophilus chromosomal sequences (7%). Polymorphisms were observed for the CRISPR repeats, CRISPR spacers, cas genes, CRISPR motif, locus architecture, and specific sequence content. Interestingly, CRISPR loci evolved both via polarized addition of novel spacers after exposure to foreign genetic elements and via internal deletion of spacers. We hypothesize that the level of diversity is correlated with relative CRISPR activity and propose that the activity is highest for CRISPR1, followed by CRISPR3, while CRISPR2 may be degenerate. Globally, the dynamic nature of CRISPR loci might prove valuable for typing and comparative analyses of strains and microbial populations. Also, CRISPRs provide critical insights into the relationships between prokaryotes and their environments, notably the coevolution of host and viral genomes.
The dairy industry relies heavily on the use of microbial starter culture systems. Among domesticated bacteria widely used in industrial applications, Streptococcus thermophilus is a key species involved in the acidification of milk and the development of texture in various fermented dairy products (3). Recent advances in genomics have provided novel insights into the many critical physiological functions carried out by S. thermophilus in fermentation processes (12). Specifically, unraveling the full genome sequences of three different strains has led to a better understanding of genes involved in acidification, texture development, and flavor enhancement (3, 12, 20). Also, comparative genomic analyses have identified specific loci involved in particular traits attributed to selected strains and pointed out differential content, notably with regard to exopolysaccharide synthesis and phage resistance (3, 12). Recent studies have also established a correlation between distinctive genomic content such as clustered regularly interspaced short palindromic repeats (CRISPR) and resistance to phages (2, 4).
CRISPRs are a peculiar family of DNA repeats widely distributed in Bacteria and Archaea (8, 9, 11, 13, 18). CRISPR loci usually consist of short and highly conserved DNA repeats, typically 21 to 48 bp, repeated up to 250 times (9). The repeated sequences, typically specific to a given CRISPR locus, are interspaced by variable sequences of constant and similar length, called spacers, usually 20 to 58 bp depending on the species or the CRISPR locus. Several distinct CRISPR loci can be located on a particular prokaryotic genome (18); for example, in Methanocaldococcus jannaschii, 18 distinct CRISPR loci have been identified on the chromosome, totaling almost 1% of the genome (5). In addition, cas (CRISPR-associated) genes are often present in the direct vicinity of CRISPR loci (8, 11, 14). Based on similarities between CRISPR spacers and phage or plasmid sequences (4, 21, 23), it was proposed in the literature that CRISPR and cas genes might be involved in conferring immunity to the host cell against foreign DNA (19, 21). In the S. thermophilus chromosome, two distinct CRISPR loci have been identified, namely, CRISPR1 and CRISPR2 (3, 4). Comparative analysis of CRISPR1 sequence between various S. thermophilus strains has revealed polymorphisms (4). In addition, it was recently reported that CRISPR provides acquired resistance against viruses in prokaryotes, notably in S. thermophilus (2). This is consistent with the putative CRISPR-cas immunity system based on RNA interference (RNAi) proposed by Makarova et al. (19), although the mechanism of action remains uncharacterized.
The correlation between CRISPR spacer content and phage susceptibility suggests that spacer content might provide a historical perspective of phage exposure. Since the CRISPR system is reactive to the environment, it might play a critical role in the adaptation of the host to its surroundings and explain the persistence of particular bacterial strains in ecosystems where phages are present. CRISPRs may also provide insight into the codirected genomic evolution of the phage and its host. Here, we provide a comparative analysis of CRISPR activity and diversity across a collection of S. thermophilus strains. We specifically analyzed the relationship between spacer hypervariability and CRISPR locus activity across three distinct CRISPR loci. In addition, we provide insight into the functional coupling between a given CRISPR repeat and the accompanying cas gene set. Finally, we investigated the various origins of CRISPR spacers and discuss the evolutionary modifications of CRISPR loci in direct response to genetic elements present in the environment, notably bacteriophages and plasmids.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, media, and growth conditions.
S. thermophilus strains and their derivatives were grown in M17 broth (Difco) supplemented with 0.5% (wt/vol) lactose and 0.5% (wt/vol) saccharose, followed by incubation at 42°C. For bacteriophage exposure and phage-resistant mutant selection, experiments were carried out according to protocols previously outlined (2, 7).
DNA sequencing of S. thermophilus CRISPR loci.
S. thermophilus genomic DNA was extracted and purified by using a DNeasy Blood & Tissue kit (Qiagen). Prior to sequencing, each CRISPR locus was amplified by PCR with LA-Taq DNA polymerase (Takara/Lonza). PCR amplification of the CRISPR1 locus was performed with the primers yc70 (5′-TGCTGAGACAACCTAGTCTCTC-3′) (4) and CR1-rev (5′-TAAACAGAGCCTCCCTATCC-3′). The CRISPR2 locus was amplified by using the primers CR2-fwd (5′-TTAGCCCCTACCATAGTGCTG-3′) and CR2-rev (5′-TTAGTCTAACACTTTCTGGAAGC-3′), whereas primers CR3-fwd (5′-CTGAGATTAATAGTGCGATTACG-3′) and CR3-rev (5′-GCTGGATATTCGTATAACATGTC-3′) were used for the amplification of CRISPR3. Each PCR product was then subjected to DNA sequencing from each end with the same primers on a CEQ8000 system (Beckman). For long PCR fragments, sequence walking was achieved using internal primers designed in spacer sequences.
Computational analyses.
Microbial genome sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov). Also, previously published S. thermophilus CRISPR sequences were retrieved from the National Center for Biotechnology Information, including entries from Bolotin et al. (4) and Barrangou et al. (2). In addition, the CRISPR database CRISPRdb (9) and the CRISPR identification application CRISPRFinder (10) were used to retrieve and find CRISPR repeats and spacer sequences, respectively. CRISPR spacers were visualized as color combinations, as previously described (2). For sequence similarity analyses, comparisons to public sequences were carried out using BLASTn (1) at the National Center for Biotechnology Information. Matches to sequences found within S. thermophilus CRISPR loci were ignored, notably for published CRISPR sequences (DQ072985 to DQ073008 and EF434458 to EF434504) and S. thermophilus chromosomal CRISPR loci (CP000023, CP000024, and CP000419). All matches with a bit score above 40.0 were retained, corresponding to 100% identity over at least 20 bp, and only the top hit annotation was considered for classification. Secondary structure predictions were obtained by using the Mfold 3.2 program (26). Multiple sequence alignments and phylogenetic analyses were carried out by using CLUSTAL X (15). Sequence consensus motifs were visualized by using WebLogo (6).
RESULTS
Identification of CRISPR3 in S. thermophilus.
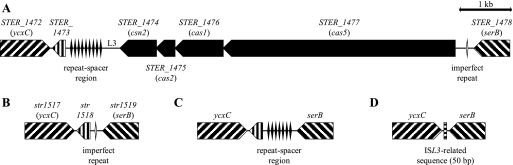
Two CRISPR loci, CRISPR1 and CRISPR2, have previously been described in S. thermophilus strains CNRZ1066 and LMG 18311 (3, 4). The genome sequence of strain LMD-9 was recently determined (20). We identified a new CRISPR locus, CRISPR3, located at base pair positions 1377794 to 1377229 on the negative strand of the LMD-9 genome, between open reading frames (ORFs) STER_1474 and STER_1473 (Fig. 1A). In this strain, CRISPR3 is composed of nine identical repeats of a nearly perfect 36-bp palindrome (5′-GTTTTAGAGCTGTGTTGTTTCGAATGGTTCCAAAAC-3′), interspaced by eight unique spacers of 30 or 32 bp. In addition, four CRISPR-associated (cas) genes—cas5, cas1, cas2, and csn2—are present in the vicinity of CRISPR3, corresponding to ORFs STER_1477 to STER_1474, respectively (Fig. 1A). Further, sequence analysis of the leader in CRISPR3 revealed that it is different from those typical of CRISPR1 and CRISPR2, with no particular sequence conservation observed, although leader sequence conservation has previously been described (13, 14, 18). Leader sequences are generally high-A/T, noncoding sequences, conserved within a CRISPR type, located on one side of the CRISPR repeat-spacer units (14, 18).
FIG. 1.
S. thermophilus CRISPR3 locus overview. (A) CRISPR3 locus in the LMD-9 genome; (B) CRISPR3 locus in the genome of CNRZ1066, also present in the LMG 18311 genome; (C) CRISPR3 locus in strain DGCC7984, without cas genes; (D) CRISPR3 locus in strain DGCC7857, without cas genes and a repeat-spacer region.
Surprisingly, the CRISPR3 locus is not present in the genomes of strains CNRZ1066 and LMG 18311, although the overall genome content is highly conserved between the three strains (3, 20). Nevertheless, one imperfect CRISPR3 repeat can be identified between ORFs str1518/stu1518 (hypothetical gene) and str1519/stu1519 (serB gene) in the genomes of CNRZ1066 and LMG 18311 (Fig. 1B). A degenerate repeat is also present in the genome sequence of LMD-9, between cas5 (STER_1477) and serB (STER_1478). Interestingly, LMD-9 genome sequences located upstream of CRISPR3 repeats and downstream of the imperfect repeat are highly similar to the sequences found in CNRZ1066 and LMG 18311 in the vicinity of this imperfect repeat, suggesting that a recombination event may have occurred between the terminal CRISPR3 repeat and the imperfect repeat close to serB, potentially leading to the deletion or insertion of a whole CRISPR3-cas segment. Analyses of similar loci in other S. thermophilus strains suggested that alternative recombination events may have also occurred in strain DGCC7984 between the imperfect repeat close to serB and another CRISPR3 repeat, leading to the presence at this chromosomal locus of a repeat-spacer region without cas genes (Fig. 1C). In S. thermophilus strain DGCC7857, the chromosomal region between serB and ycxC is even more reduced, since neither cas genes nor the repeat-spacer region nor str1518 is present (Fig. 1D). An additional 50-bp segment of ISL3-related sequence can be identified at the junction of ycxC and serB adjacent sequences (Fig. 1D).
Overall, we investigated the occurrence of CRISPR3 in 66 strains and found this locus in 53 of them (80%). Specifically, among the four different structures documented (Fig. 1), we identified structure A (Fig. 1A) in 52 strains and structure B (Fig. 1B) in 12 strains, whereas structures C (Fig. 1C) and D (Fig. 1D) were each found in only one strain.
Activity of CRISPR loci.
We propose that CRISPR activity be defined as the ability of a CRISPR locus to integrate novel repeat-spacer units in response to exposure to foreign genetic elements, such as bacteriophage, to provide resistance.
CRISPR1 activity in S. thermophilus has been demonstrated previously (2). Here, we provide additional experimental data showing novel spacer acquisition after exposure to bacteriophage in four different strains, as shown on Fig. 2. Specifically, novel spacers are observed on lines 4 to 6 and lines 9 to 18 for strain DGCC7710, lines 24 to 26 for strain DGCC7778, lines 39 to 44 for strain LMD-9, lines 52 to 55 for strain SMQ-301, after exposure to a variety of bacteriophage.
FIG. 2.
Graphic representation of spacers across the three CRISPR loci for a variety of S. thermophilus strains. Repeats are not included; only spacers are represented. Each spacer is represented by a combination of one select character in a particular font color, on a particular background color. The color combination allows unique representation of a particular spacer, whereby squares with similar color schemes (combination of character color and background color) represent identical spacers, whereas different color combinations represent distinguishable spacers. Deleted spacers are represented by crossed squares. L1, L2, and L3, CRISPR leader sequences. Left, CRISPR1; center, CRISPR2; right, CRISPR3. Question marks and empty spaces indicate elements that were not sequenced.
We also investigated the ability of CRISPR3 to integrate novel spacers after phage challenge in phage-resistant mutants. We isolated bacteriophage-insensitive mutants (BIMs), derived from either DGCC7710 or LMD-9, after challenge with phage Φ858, Φ3821, or Φ4241 (Fig. 2). For LMD-9, two BIMs with one novel CRISPR3 spacer each were obtained after exposure to Φ4241 (see Fig. 2, lines 45 to 46). For DGCC7710, one BIM was obtained with two new CRISPR3 spacers after exposure to Φ3821 (see Fig. 2, line 29). Also, for DGCC7710, a mutant was obtained with one new CRISPR3 spacer after exposure to Φ858, derived from a strain (DGCC7710Φ858+S1S2ΔCRISPR1) where the whole CRISPR1 locus was deleted and replaced by a unique terminal repeat (2) (see Fig. 2, line 30). The addition of new spacers in response to phage infection was polarized toward the leader end of the CRISPR3 locus. This shows that CRISPR3, in addition to CRISPR1, has the ability to integrate novel spacers during the natural generation of phage-resistant mutants.
Overall, a total of 41 distinct CRISPR BIMs were isolated, including 37 for CRISPR1, none for CRISPR2, and 4 for CRISPR3. Accordingly, it appears that CRISPR1 has the highest ability to integrate novel spacers in response to phage exposure, followed by CRISPR3, whereas we have no evidence that CRISPR2 is active.
CRISPR repeats are locus specific.
Three distinct CRISPR loci have now been identified in S. thermophilus and may be simultaneously present on the chromosome, as is the case in LMD-9 (20). Usually, each CRISPR locus is defined by the sequence of the repeat. We define the typical repeat as the most frequent repeat within a CRISPR locus. The typical repeat sequences of the three S. thermophilus CRISPR loci are different (Table 1), although they all are 36 bp long. Sequence comparison between the three typical repeats showed that CRISPR1 and CRISPR2 share 30.6% identity, while CRISPR1 and CRISPR3 are 52.8% identical, and CRISPR2 and CRISPR3 share 47.2% nucleotides.
TABLE 1.
Analysis of CRISPR repeat sequences
| CRISPR | Type | Repeat sequence (5′-3′)a | No. of sequenced repeats | Frequency (%) |
|---|---|---|---|---|
| CRISPR1 | Typical repeat | GTTTTTGTACTCTCAAGATTTAAGTAACTGTACAAC | 2,820 | 99.7 |
| Repeat variants | GTTTTTGTACTCTCAAGATTTAAGTAACTGTGCAAC | 8 | 0.28 | |
| GTTTTTGTACTCTCAAGATTTAAGTAACCGTACAAC | 1 | 0.04 | ||
| Terminal repeat | GTTTTTGTACTCTCAAGATTTAAGTAACTGTACAGT | 107 | 87.0 | |
| GTTTTTGTACTCTCAAGATTTAAGTAGCTGTACAGT | 10 | 8.1 | ||
| GTTTTTGTATTCTCAAGATTTAAGTAACTGTACAGT | 6 | 4.9 | ||
| CRISPR2 | Typical repeat | GATATAAACCTAATTACCTCGAGAGGGGACGGAAAC | 129 | 74.6 |
| Repeat variant | GATATAAACCTAATTACCTCGAGAAGGGACGGAAAC | 44 | 25.4 | |
| Terminal repeat | GATATAAACCTAATTACCTCGAGAGGGGACTTTTTT | 59 | 100 | |
| CRISPR3 | Typical repeat | GTTTTAGAGCTGTGTTGTTTCGAATGGTTCCAAAAC | 670 | 99.8 |
| Repeat variant | TTTAACTCGCTGTGTTGTTTCGAATGGTTCCAAAAC | 1 | 0.15 | |
| Terminal repeat | Not different from the typical repeat |
Mutations as compared to the typical CRISPR repeat sequence are indicated by underscoring.
Sequence comparison within each CRISPR locus showed that, although the repeat sequence is usually highly conserved throughout the locus, polymorphisms can be observed, notably for the terminal repeat (Table 1). Specifically, sequence degeneracy is observed at the 3′ end of the terminal repeat. This observation is particularly important for the correct annotation and orientation of CRISPR loci, since the last spacer/repeat unit, including the terminal repeat, which is usually degenerate, is often missed, or the repeats are frequently annotated on the opposite DNA strand. The repeat orientation appears to be consistent with the orientation of adjacent cas genes. In addition, variations within the repeat sequence can be observed throughout a CRISPR locus. The ratios of atypical repeats were 0.3% for CRISPR1, 25% for CRISPR2, and 0.2% for CRISPR3 (Table 1), indicating that repeat sequence degeneracy was much higher for CRISPR2, while CRISPR1 and CRISPR3 seemed to possess highly conserved repeat sequences. Interestingly, for CRISPR3, no polymorphism was observed in the terminal repeat.
CRISPR repeats may form secondary structures.
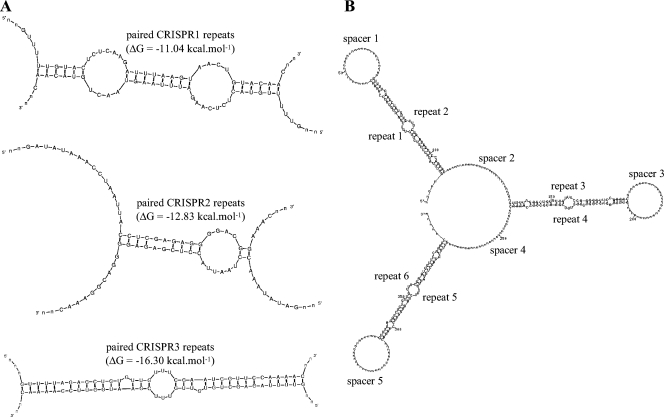
Previous studies have suggested that CRISPR repeats may form stable hairpin-like secondary structures due to their partially palindromic nature (13, 16). Moreover, CRISPR loci appear to be transcribed as a single-stranded RNA molecule starting from the CRISPR leader (18, 22, 25). Accordingly, as transcription is progressing, two consecutive single-stranded CRISPR repeats may conceivably be able to interact and form secondary structures by pairing head to foot, giving rise to an even more stable structured RNA.
We determined the putative secondary structures of the three S. thermophilus CRISPR repeats by using the Mfold program (26), as shown in Fig. 3A. These hypothetical structures seem relatively stable (ΔG < −10 kcal mol−1), notably for CRISPR3 paired repeats. In the case of CRISPR3, the pairing of two repeats is nearly perfect and does not, overall, require shifting one repeat compared to the other. In contrast, the pairing of two CRISPR1 repeats results in four unpaired nucleotides at the 5′ end and one unpaired nucleotide at the 3′ end. The pairing of two CRISPR2 repeats appears to be more questionable since the default parameters of the Mfold program did not result in any secondary structure; to obtain the structure shown on Fig. 3A, the pairing of eight nucleotides of the major stem had to be forced. As previously discussed, repeat sequence is subject to discrete variations, which might slightly affect the stability of such putative structures.
FIG. 3.
Putative secondary structures of the three S. thermophilus CRISPR repeats. Putative structures were predicted by using the Mfold program (26). (A) Putative structures of paired CRISPR repeats for the three loci; (B) putative structure obtained by pairing of six consecutive CRISPR3 repeats.
Interestingly, assuming transcription of the whole CRISPR locus as a single-stranded RNA molecule, and if such a pairing between consecutive repeats would occur, every-other spacer would be constrained as a loop, whereas the remaining spacers would remain unconstrained, alternatively (Fig. 3B).
cas genes are locus specific.
The three S. thermophilus CRISPR loci are associated with cas genes. For CRISPR1 and CRISPR3, the architecture is seemingly conserved, with four cas genes located upstream of the repeat-spacer region (Fig. 4). In contrast, the content and organization for CRISPR2 is different, with hypothetical cas genes on both sides of the repeat-spacer region. Notably, in the genome of CNRZ1066, several CRISPR2 elements appear to be deleted, including the latter portion of the leader, the complete repeat-spacer region, as well as cas6, csm1, and most of csm2.
FIG. 4.
Overview of the three S. thermophilus CRISPR loci in the LMD-9 genome. The cas genes are shown in black. Numbers within the genes indicate the genomic ORF number. Numbers on the gray shading indicate percent identity (top) and percent similarity (bottom) between homologous Cas protein sequences. Other Cas protein sequences do not share significant similarity.
Although gene organization is similar between CRISPR1 and CRISPR3, sequence similarity is low, even at the protein level, as shown on Fig. 4. Interestingly, cas1 seems to be the only gene conserved between the three loci, with 22% similarity at the protein level between CRISPR1 and CRISPR2. The relatedness of the three cas systems is similar to that of the three CRISPR repeats, whereby CRISPR1 and CRISPR3 are more closely related, and CRISPR2 is more distant.
Coupling CRISPR repeats/cas genes.
Since we observed a correlation between the relatedness of cas genes and CRISPR repeats within S. thermophilus, we investigated whether there was a relationship between CRISPR repeats and cas genes across bacterial species. Across a variety of species, the clustering of the typical CRISPR repeats was similar to that of the Cas proteins (Fig. 5), which is consistent with previous observations by Kunin et al. (16), showing a correspondence between CAS subtypes and repeat clusters in prokaryotes. Specifically, CRISPR1 appeared to be present only in a few streptococcal species such as S. thermophilus, Streptococcus vestibularis, and Streptococcus suis. CRISPR3 seemed to be present across most Streptococcus species (including S. agalactiae, S. mutans, S. pyogenes, and S. thermophilus) and also in other genera, including Listeria and Enterococcus. CRISPR2, which is only present in one other Streptococcus species, namely, S. sanguinis, is also present in several mycobacteria (4) and Staphylococcus epidermidis. Additional information regarding the presence of the three S. thermophilus CRISPR loci in various genera and species is available (see Table S1 in the supplemental material).
FIG. 5.
Coclustering of CRISPR repeats and Cas sequences. CRISPR repeats and Cas sequences were aligned by using CLUSTAL X (15). (A) Analysis of various CRISPR repeats; (B) analysis of concatenated Cas sequences; (C) analysis of Cas1 sequences. Several sequences were retrieved from the CRISPRdb database (9) or found by using CRISPRFinder (10). Lin, Listeria innocua Clip11262 (AL592022); Mbo, Mycobacterium bovis BCG Pasteur 1173P2 (AM408590); Mtu, Mycobacterium tuberculosis F11 (CP000717); Sag, Streptococcus agalactiae A909 (CP000114); Sep, Staphylococcus epidermidis RP62A (CP000029); Smu, Streptococcus mutans UA159 (AE014133); Spy, Streptococcus pyogenes MGAS5005 (CP000017); Ssa, Streptococcus sanguinis SK36 (CP000387); Streptococcus suis 89/1591 (AAFA00000000); Sth, Streptococcus thermophilus LMD-9 (CP000419). CR1, CRISPR1; CR2, CRISPR2; CR3, CRISPR3; CRa and CRb, two CRISPR loci in S. pyogenes.
Comparative analysis of the trees obtained revealed similar clustering patterns (Fig. 5), with different clusters for each CRISPR locus, namely, CRISPR1, CRISPR2, and CRISPR3. Sequence alignments are provided in supplementary material (see Fig. S1 in the supplemental material). CRISPR1 consistently clusters with S. suis, while CRISPR2 consistently clusters with S. sanguinis and CRISPR3 consistently clusters with S. mutans, S. agalactiae, and S. pyogenes. Although the trees were based on widely different element sizes (the repeat size varied between 32 and 37 bp, while the cas tree was generated using concatenated Cas protein sequences, which varied between 1,628 and 3,029 amino acids), the congruence between them is relatively high. This observation suggests a potential coevolution of the cas genes and the CRISPR repeats, perhaps indicating a functional link between the two.
Spacer diversity in S. thermophilus CRISPR loci.
We investigated CRISPR spacer diversity across a variety of S. thermophilus strains (Fig. 2). A total of 124 strains were partially analyzed (Table 2). For CRISPR1, all 124 tested strains contained this locus, indicating that it is likely ubiquitous in S. thermophilus. A total of 105 unique spacer arrangements were found, with an average number of 23 spacers per locus, and a spacer number between 2 and 51. For CRISPR2, 65 strains were analyzed, and among the 59 strains containing a CRISPR2 locus (91%), only seven unique spacer arrangements were observed, with an average number of three spacers per locus, and a spacer number between zero and eight. For CRISPR3, 66 strains were analyzed, and among 53 strains containing a CRISPR3 repeat-spacer region (80%), 20 unique spacer arrangements were observed, with an average number of 13 spacers per locus, and a spacer number between 0 and 29. Although differences were observed at the three CRISPR loci, the highest degree of polymorphism occurred at the CRISPR1 locus, followed by CRISPR3. Specifically, the relative proportions of unique arrangements were 84.7, 11.2, and 37.7% for CRISPR1, CRISPR2, and CRISPR3, respectively. Further, all strains showing differences at the CRISPR2 locus also had different CRISPR1 spacer combinations. Similarly, all strains showing differences at the CRISPR3 locus had different CRISPR1 locus, except for the four phage-resistant CRISPR3 mutants mentioned above. Overall, CRISPR1 showed the most diversity among the three CRISPR loci, followed by CRISPR3.
TABLE 2.
CRISPR diversity in S. thermophilus
| Parameter | CRISPR1 | CRISPR2 | CRISPR3 |
|---|---|---|---|
| No. of strains analyzed | 124 | 65 | 66 |
| No. of strains with locus (%) | 124 (100) | 59 (90.8) | 53 (80.3) |
| No. of unique arrangements (%) | 105 (84.7) | 7 (11.9) | 20 (37.7) |
| Avg no. of spacers per locus ± SD | 22.9 ± 10.4 | 2.9 ± 1.1 | 13.2 ± 5.2 |
| Minimum no. of spacers per locus | 2 | 0 | 0 |
| Maximum no. of spacers per locus | 51 | 8 | 29 |
| Total no. of spacers | 2,829 | 173 | 624 |
| No. of unique spacers (%) | 782 (27.6) | 16 (9.2) | 154 (24.7) |
The identification of arrays of common consecutive spacers allowed the grouping of different S. thermophilus strains into clusters (Fig. 2). In general, identical spacers between different strains or subclusters occur more frequently at the 3′ end (the trailer end) of the CRISPR1 locus. In contrast, for strains clusters sharing most of their spacers, hypervariability is more frequently visible at the 5′ end (the leader end) of the locus. Accordingly, the spacers located at the trailer end of the CRISPR loci can be used to anchor clusters of strains. In some cases, the spacers located at the trailer end of the loci are actually common to divergent strain groups that may be derived from a common ancestor. For instance, strains shown on lines 1 to 29 and lines 31 to 36 (Fig. 2) may be two subgroups derived from a common ancestor. Also, strains shown on lines 37 to 50 and lines 54 to 57 are likely derived from a parental strain and probably diverged after the integration of the 10 spacers they have in common. Similarly, strains shown on lines 74 to 80 and lines 82 to 87 may also be two subgroups derived from a common parental strain and likely have diverged after the integration of the first five spacers they share. Accordingly, comparison of CRISPR spacer arrangements between different strains, within and across the three CRISPR loci clearly provides insight into the relatedness of various S. thermophilus strains.
Within a particular CRISPR locus, beside the diversity derived from the presence of additional spacers, we also observed probable deletions of select spacers. For spacer additions, it was previously reported that integration of novel spacers primarily occurs at the leader end of CRISPR1 (2, 23), which is consistent with the diversity observed at the leader end of the CRISPR1 locus in several clusters (Fig. 2), specifically for strains shown on lines 1 to 29, 37 to 57, and 69 to 73. Also, a similar pattern of polarized spacer acquisition after bacteriophage challenge was observed for CRISPR3 in BIMs derived from LMD-9 and DGCC7710 (see the strains on lines 29 and 30 and lines 45 and 46 in Fig. 2). In addition to the polarized integration of novel spacers, differences were also observed in several strains that appeared to be missing consecutive internal spacers. For most of the strain clusters shown on Fig. 2, it appears that deletions occurred, resulting in the loss of internal spacers, more frequently toward the trailer end of the loci. Interestingly, while both polarized addition of new spacers and deletion of vestigial spacers were observed, it was recently reported that internal addition of novel spacers can occur simultaneously with internal deletion of vestigial spacers (7).
A total of 3,626 spacers (2,829 for CRISPR1, 173 for CRISPR2, and 624 for CRISPR3) were analyzed (Fig. 2 and Table 2), including previously described S. thermophilus CRISPR spacers (2-4, 20). Globally, 952 unique spacers were identified (26%), including 782, 16, and 154 spacers for CRISPR1, CRISPR2, and CRISPR3, respectively. Accordingly, most of the unique spacers are present in CRISPR1 (82%), while very few are found in CRISPR2 (<2%), which further indicates that CRISPR1 showed the most diversity among the three CRISPR loci, followed by CRISPR3 (16%). Overall, the degree of polymorphism was highest for CRISPR1, in terms of unique spacers, unique spacer combinations, and average spacer content.
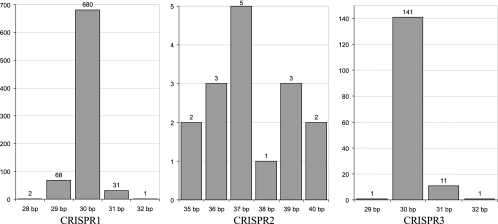
In addition, spacer polymorphisms were also observed with regard to spacer size. Specifically, analysis of the spacer size distribution indicated that variability was lower for CRISPR1 and CRISPR3 than for CRISPR2 (Fig. 6). The typical spacer size was 30 bp with ranges of 28 to 32 bp for CRISPR1 and 29 to 32 bp for CRISPR3, whereas the typical spacer size was 37 bp with a range of 35 to 40 bp for CRISPR2. In addition, the proportions of spacers of typical size were 87% (680 of 782), 31% (5 of 16), and 92% (141 of 154), for CRISPR1, CRISPR2, and CRISPR3, respectively.
FIG. 6.
CRISPR spacer size variability. The x axis represents the size of a CRISPR spacer, in nucleotides. The y axis represents the number of CRISPR spacer sequences of a given size. (A) CRISPR1 spacers; (B) CRISPR2 spacers; (C) CRISPR3 spacers.
Analysis of CRISPR spacer sequences.
We analyzed the CRISPR spacer sequences and investigated similarity and identity to phage, plasmid, and bacterial sequences (see Table S2 in the supplemental material). A total of 952 unique spacer sequences were analyzed, including 782, 16, and 154 spacers for CRISPR1, CRISPR2, and CRISPR3, respectively. Overall, among the 500 spacers with matches above the selected cutoff (see Materials and Methods), 384 (77%) showed similarity to viral sequences, whereas 80 (16%) showed similarity to plasmid sequences, and 36 (7%) showed similarity to chromosomal sequences (Table 3). Among CRISPR spacers homologous to viral sequences, the large majority (97% [374 of 384]) showed similarity to S. thermophilus phage sequences, including 147 spacers (39%) showing 100% identity. For CRISPR spacers showing similarities with plasmid sequences, nearly all of them (96% [77 of 80]) showed similarity to S. thermophilus plasmid sequences, including 41 spacers (51%) showing 100% identity. In contrast, only four CRISPR spacers (11% [4 of 36]) showed identity to S. thermophilus chromosomal sequences. Spacers identical to known sequences are particularly insightful, since it was previously shown that 100% identity between spacer and proto-spacer sequences is required to provide immunity (2, 7). Nevertheless, other similarities provide insight into the probable origin of the spacers.
TABLE 3.
S. thermophilus CRISPR spacer sequence matches
| Parameter | Viruses
|
Plasmids
|
Chromosomes
|
|||
|---|---|---|---|---|---|---|
| S. thermophilus | Other | S. thermophilus | Other | S. thermophilus | Other | |
| Identity | 147 | 0 | 41 | 1 | 4 | 0 |
| Similarity | 227 | 10 | 36 | 2 | 0 | 32 |
| Total | 374 | 10 | 77 | 3 | 4 | 32 |
Several matches were found across particular S. thermophilus phage genomes (see Table S2 in the supplemental material), notably phages 2972, DT1, 7201, Sfi11, Sfi19, and Sfi21 (17). As previously reported, no particular phage genome module seemed to be targeted, and similarities were observed across the genome, on both strands, with a bias toward the leading strand (2, 7, 17). Also, we investigated the occurrence of the spacer sequences in bacterial sequences, outside of CRISPR loci. Interestingly, the spacers showed identity to sequences found in 27 different S. thermophilus plasmids, notably several matches in pSt08, pt38, pSt106, pSt1, and pND103. In some cases, spacers showed identity to genes involved in plasmid replication, notably repA and repS, as is the case for spacer 059_1_01 (see Table S2 in the supplemental material), which showed identity to sequences found in pND103, pSt1, pt38, pER36, pER35, pER16, and pJ34. Interestingly, most of these plasmids, which are found in a variety of S. thermophilus strains, belong to the same family, namely, the pC19/pUB1104 rolling-circle family (24).
Further, three CRISPR1 spacers and one CRISPR3 spacer showed 100% identity to S. thermophilus chromosomal sequences, notably in dtpT (encoding a di- or tripeptide proton symporter), rexA (encoding an ATP-dependent exonuclease), and str_0775 (phage-associated DNA primase), as well as a match in an intergenic region between STER_0810 (hypothetical protein) and STER_0811 (transposase).
Interestingly, a number of similarities were observed with sequences found in closely related genera, including Lactococcus, Lactobacillus, and Staphylococcus (see Table S2 in the supplemental material). For phage sequences, similarities were found in Staphylococcus aureus phage Twort and Lactobacillus plantarum phage LP65. For plasmid sequences, similarity was found in Streptococcus pneumoniae pSpnP1, and identity was actually observed in the Lactococcus lactis plasmid pCIS3, within hsdS, which encodes a type I restriction-modification system. For chromosomal sequences, homologies were identified with Lactococcus lactis subsp. cremoris MG1363 and SK11, Lactobacillus reuteri F275, and Streptococcus pyogenes MGAS10394. Interestingly, in L. lactis subsp. cremoris, homology was found in hsdR, a gene also encoding a type I restriction-modification system.
DISCUSSION
In addition to the two CRISPR loci previously described in S. thermophilus (3), we report here the identification of CRISPR3 in the LMD-9 genome (20). Interestingly, this particular CRISPR locus is not ubiquitous in S. thermophilus genomes. Between the three CRISPR loci present in S. thermophilus genomes, diversity is observed at many levels, including (i) the typical CRISPR repeat sequence; (ii) the cas gene content, organization, and sequence; (iii) locus architecture and content; and (iv) spacer content, arrangement, and sequence. Diversity was observed across the three CRISPR loci between 124 different S. thermophilus strains. Specifically, CRISPR1 was ubiquitous, whereas CRISPR2 was present in 59 of 65 strains, and CRISPR3 was present in 53 of 66 strains. A total of 49 strains (39.5%) carried all three loci.
Comparative genome analysis of CRISPR content in streptococci and various bacterial genera and species indicates that the three S. thermophilus CRISPR loci are distributed differently. Notably, CRISPR1 is present in only a few streptococci, whereas CRISPR3 can be found in most Streptococcus species. The distribution of these three CRISPR loci suggests that CRISPR1 may have recently become more specific to a few streptococcal species, whereas CRISPR3 is more widespread across streptococci, and CRISPR2 may be a vestige of a gram-positive ancestor. This is consistent with the absence of CRISPR2 and/or CRISPR3 in various S. thermophilus strains. In fact, detailed sequence analysis of distinct CRISPR3 locus architectures in various S. thermophilus strains suggests that deletions may have occurred via homologous recombination events involving CRISPR3 repeats, likely including the degenerate repeat in the vicinity of serB (Fig. 1).
When equivalent CRISPR loci between strains are compared, a high degree of polymorphism is observed for spacer content and sequences. Specifically, 105 of 124, 7 of 59, and 20 of 53 unique spacer arrangements were observed for CRISPR1, CRISPR2, and CRISPR3, respectively. This indicates that the overall CRISPR content was unique in most strains. Perhaps the polymorphisms observed in the spacer contents of the three CRISPR loci across different S. thermophilus strains are an indicator of the activity of the locus, whereby spacer hypervariability is directly correlated with historical phage exposure. Arguably, the degree of spacer polymorphism, in terms of both total number of unique spacers and total number of unique spacer arrangements, for a given CRISPR locus, could be directly correlated with its activity. Consequently, we propose that in S. thermophilus CRISPR1 is the most active locus, followed by CRISPR3. This is supported by several observations: (i) repeat degeneracy seems to correlate with relative activity, whereby the most degenerate repeats are found in the least active locus, namely, CRISPR2; (ii) spacer size is more highly conserved in the most active loci, namely, CRISPR1 and CRISPR3, and least conserved in the least active locus, namely, CRISPR2; (iii) the average and maximum numbers of spacers are highest for CRISPR1 and lowest for CRISPR2; and (iv) the number of CRISPR BIMs obtained is higher for CRISPR1 than CRISPR3.
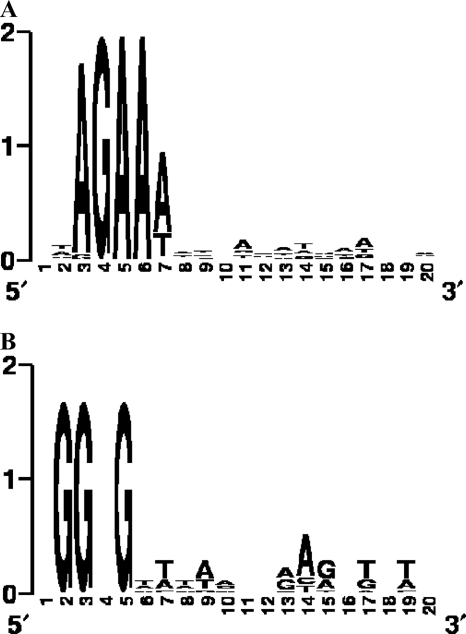
Previous data have suggested that the enzymatic machinery of a specific locus cannot be effective in conjunction with the CRISPR genetic content of another (2). Specifically, when cas genes are inactivated in a particular CRISPR locus, the ability of this locus to provide resistance and integrate novel spacers is lost, despite the concurrent presence of other CRISPR loci and cas genes elsewhere in the chromosome (2). Here, we provide data indicating that each CAS system may be directly linked to a particular CRISPR repeat sequence, which is consistent with the observed comparable clustering of CRISPR repeats and Cas sequences (Fig. 5), as previously suggested by Kunin et al. (16). Further studies investigating the mechanism of action of CRISPRs are currently under way and might provide insights into the roles of the various cas genes and the functional link between specific Cas proteins and a particular CRISPR repeat. Among Cas proteins, some are likely involved in the addition of novel repeat-spacer units, via a molecular interaction with CRISPR repeats. Other Cas proteins are likely involved in the spacer-encoded resistance, which may be mediated via a RNAi-like mechanism (19). These Cas proteins probably include at least one nuclease which might recognize and digest a specific target sequence. This is supported by the recent discovery of a highly conserved motif, which we propose to name CRISPR motif, immediately downstream of the proto-spacers found in phage sequences (7). For CRISPR1, the AGAAW CRISPR motif located two nucleotides downstream of the proto-spacer might serve as a recognition site for a CRISPR1-specific Cas nuclease (Fig. 7). A different CRISPR motif was also identified for CRISPR3 (Fig. 7), GGNG, located one nucleotide downstream of the proto-spacer, which suggests again that each CRISPR locus has a unique CRISPR motif which may serve as a sequence recognition pattern, specific to a particular Cas enzymatic machinery. Further, CRISPR motifs may serve as additional elements to define a particular CRISPR/Cas system.
FIG. 7.
CRISPR motifs identified in the vicinity of the CRISPR proto-spacers. (A) Motif identified in the vicinity of CRISPR1 proto-spacers in the genome of the phage used in the challenge; (B) motif identified in the vicinity of CRISPR3 proto-spacers in the genome of the phage used in the challenge. Conserved sequence motifs were visualized by using WebLogo (6).
We have shown that two distinct CRISPR loci, namely, CRISPR1 and CRISPR3 have the ability to evolve directly in response to phages by the polarized addition of new spacers derived from viral genomic sequences. Accordingly, CRISPR spacers provide a historical perspective of phage exposure, whereby spacers present in the vicinity of the leader were relatively recently added, whereas distal spacers likely originated from previous events.
In addition to CRISPR variability due to the acquisition of novel spacers in response to phages, primarily at the leader end, we noticed that modifications can occur throughout the CRISPR locus, as seen in DGCC7710Φ2972+S15 (7), where a deletion occurred concomitantly with the insertion of a new spacer at the leader end (Fig. 2). Specifically, most of the variability observed at the trailer end of the locus seems to occur via deletion (Fig. 2), arguably resulting in the preferential deletion of older spacers, which are likely less valuable for the bacterium in its current environment. This phenomenon is probably due to homologous recombination events occurring between CRISPR direct repeats. On the other hand, spacers recently acquired may be more valuable and thus more likely to be retained in the current environment. In some instances, peculiar spacers seem to be retained between seemingly distant strains, perhaps indicating that they provide a critical function (Fig. 2), such as targeting a conserved phage sequence. Altogether, CRISPR loci seem to evolve both through additions and deletions of repeat-spacer units.
Similarities between CRISPR spacers and phage or plasmid sequences have been documented previously (2, 4, 21, 23). Although the majority of CRISPR spacers shows homology to phage (77%) and plasmid (16%) sequences, we identified four CRISPR spacers that are 100% identical to S. thermophilus chromosomal gene sequences, including dtpT and rexA. This might indicate that the CRISPR/Cas system, in addition to providing resistance against foreign genetic elements such as plasmids and phages, may also serve as a microbial regulatory system involved in the control of mRNA transcripts levels for genes encoded on the chromosome, perhaps using a system based on RNAi, as previously suggested (19).
Overall, the dynamic nature of CRISPR loci is potentially valuable for typing and comparative analyses of strains and microbial populations. Given that some loci are relatively active while others bear lower levels of polymorphism, the potential of a given CRISPR locus for typing and epidemiological studies has to be assessed on a case-by-case basis. Since CRISPRs are widely distributed in Bacteria and Archaea and actively involved in an adaptive immune system against foreign genetic elements, as well as intrinsic chromosomal elements, they provide critical insights into the relationships between prokaryotes and their environments, notably the coevolution of host and viral genomes.
Supplementary Material
Acknowledgments
This study was supported by funding from Danisco A/S. S.M. acknowledges support from the Natural Sciences and Engineering Research Council of Canada through its Discovery Program.
We thank Cécile Vos of Danisco Innovation for technical assistance.
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou, R., C. Fremaux, P. Boyaval, M. Richards, H. Deveau, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyrpides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 221554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 1512551-2561. [DOI] [PubMed] [Google Scholar]
- 5.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H. P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 2731058-1073. [DOI] [PubMed] [Google Scholar]
- 6.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonté, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 1390-1400. [DOI] [PMC free article] [PubMed]
- 8.Godde, J. S., and A. Bickerton. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62718-729. [DOI] [PubMed] [Google Scholar]
- 9.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 8172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haft, D. H., J. Selengut, E. F. Mongodin, and K. E. Nelson. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1474-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. D. Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29435-463. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, R., J. D. A. Van Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 623-33. [DOI] [PubMed] [Google Scholar]
- 14.Jansen, R., J. D. A. Van Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 431565-1575. [DOI] [PubMed] [Google Scholar]
- 15.Jeanmougin, F., J. D. Thompson, T. J. Gibson, M. Gouy, and D. G. Higgins. 1998. Multiple sequence alignment with CLUSTAL X. Trends Biochem. Sci. 23403-405. [DOI] [PubMed] [Google Scholar]
- 16.Kunin, V., R. Sorek, and P. Hugenholtz. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lévesque, C., M. Duplessis, J. Labonté, S. Labrie, C. Fremaux, D. Tremblay, and S. Moineau. 2005. Genomic organization and molecular analysis of the virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 714057-4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillestøl, R., P. Redder, R. Garrett, and K. Brügger. 2006. A putative viral defense mechanism in archaeal cells. Archaea 259-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova, K. S., N. V. Grishin, S. A. Shabalina, Y. I. Wolf, and E. V. Koonin. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mojica, F. J., C. Diez-Villasenor, J. Garcia-Martinez, and E. Soria. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60174-182. [DOI] [PubMed] [Google Scholar]
- 22.Peng, X., K. Brügger, B. Shen, L. Chen, Q. She, and R. A. Garrett. 2003. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 1852410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151653-663. [DOI] [PubMed] [Google Scholar]
- 24.Turgeon, N., M. Frenette, and S. Moineau. 2004. Characterization of a theta replicating plasmid from Streptococcus thermophilus. Plasmid 5124-36. [DOI] [PubMed] [Google Scholar]
- 25.Viswanathan, P., K. Murphy, B. Julien, A. G. Garza, and L. Kroos. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 1893738-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.