Abstract
Hydrogenases are enzymes involved in hydrogen metabolism, utilizing H2 as an electron source. [NiFe] hydrogenases are heterodimeric Fe-S proteins, with a large subunit containing the reaction center involving Fe and Ni metal ions and a small subunit containing one or more Fe-S clusters. Maturation of the [NiFe] hydrogenase involves assembly of nonproteinaceous ligands on the large subunit by accessory proteins encoded by the hyp operon. HypE is an essential accessory protein and participates in the synthesis of two cyano groups found in the large subunit. We report the crystal structure of Escherichia coli HypE at 2.0-Å resolution. HypE exhibits a fold similar to that of PurM and ThiL and forms dimers. The C-terminal catalytically essential Cys336 is internalized at the dimer interface between the N- and C-terminal domains. A mechanism for dehydration of the thiocarbamate to the thiocyanate is proposed, involving Asp83 and Glu272. The interactions of HypE and HypF were characterized in detail by surface plasmon resonance and isothermal titration calorimetry, revealing a Kd (dissociation constant) of ∼400 nM. The stoichiometry and molecular weights of the complex were verified by size exclusion chromatography and gel scanning densitometry. These experiments reveal that HypE and HypF associate to form a stoichiometric, hetero-oligomeric complex predominantly consisting of a [EF]2 heterotetramer which exists in a dynamic equilibrium with the EF heterodimer. The surface plasmon resonance results indicate that a conformational change occurs upon heterodimerization which facilitates formation of a productive complex as part of the carbamate transfer reaction.
Hydrogenases (H2ases) are key enzymes involved in hydrogen metabolism, catalyzing reversible redox reactions involving molecular hydrogen (for reviews see references 5, 7, 10, 12, 43, and 47). These enzymes are widely distributed in Archaea and Bacteria but are rarely found in Eukarya (12, 47). H2ases are grouped into three classes (5), the [NiFe] H2ase, the [FeFe] H2ase, and [Fe] H2ases such as the methylene-tetrahydromethanopterin dehydrogenases from methanogenic archaea (14, 16, 49). [NiFe] H2ases function as multisubunit complexes containing a minimum of two nonidentical subunits. Some of these enzymes are found associated with membranes while others are localized to the periplasm or cytoplasm. In contrast, [FeFe] H2ases are predominantly monomeric and contain only the catalytic subunit; some are known, however, to be dimeric, trimeric, or tetrameric enzymes (47). All three H2ase families are unrelated at the amino acid sequence level, however. [FeFe] as well as [NiFe] H2ases possess at least one [Fe-S] center that is involved in electron transport from the active site toward its redox partner (7).
Escherichia coli has four [NiFe] H2ase isozymes with different subunit compositions, all invariably containing a large subunit (∼60 kDa) and a small subunit (∼30 kDa) that form part of a larger complex that performs either evolution of H2 or its utilization as an electron source (47). The H2ase isozymes Hyd1 and Hyd2 are uptake H2ases that utilize molecular H2 to produce protons and electrons coupled to fumarate reduction. The isoenzyme Hyd3 is involved in reduction of protons to elemental hydrogen and is a component of formate-hydrogen lyase (37, 43). The fourth isozyme, Hyd4, has been proposed to be a part of a separate formate-hydrogen lyase system that seems to be a proton-translocation enzyme (2). Each H2ase operon encodes the large and the small subunit, redox carriers, membrane anchor proteins, and the accessory proteins required for H2ase maturation.
In H2ases, the large subunit contains the reaction center involving Fe and Ni metal ions, while the small subunit harbors Fe-S clusters that participate in an electron relay from the active site toward the redox partner (7, 47). In most [NiFe] H2ases, a single CO and two CN molecules act as nonprotein ligands to form a Fe(CN)2(CO) cluster in the large subunit (15, 30, 39). A model of the chemical reaction catalyzed by H2ase maturation proteins assumes that carbamoyl phosphate acts as the precursor for synthesis of both the CO and CN groups (36). However, recent studies show that carbamoyl phosphate leads only to the formation of CN but not CO (11, 24, 42). The source of CO is not completely resolved, although it may be derived from the acetate pool (42) or CO itself (24) and incorporated into the large subunit.
The accessory proteins required for maturation of the large subunit (HyaB, HybC, and HycE) of the E. coli H2ase are encoded by the hyp operon (hypABCDE) that is situated adjacent to the genes encoding Hyd3. The gene encoding HypF, another accessory protein, is situated on a separate operon (7). Among the accessory proteins participating in functional maturation of the H2ase large subunit, HypE is required by at least three isozymes to achieve full functionality (19). The route of CN incorporation into the large subunit involves interaction of HypE with several protein complexes (7, 40). Initially, HypF in the presence of ATP converts carbamoyl phosphate to AMP-OCONH2 (35). The carbamoyl moiety of AMP-OCONH2 is then transferred to the C-terminal cysteine of HypE as a thiocarbamate, and this reaction takes place while HypF and HypE are bound to each other (4). Subsequently, the thiocarbamate is dehydrated by HypE to thiocyanate in a reaction that is coupled with ATP hydrolysis (4, 40).
The extensive biochemical characterization of HypE from E. coli with respect to its function and interaction with other partners has led to a need to determine its three-dimensional structure. Here we report the crystal structure of HypE in its apoenzyme form. Very recently, the crystal structures of HypC, HypD, and HypE from Thermococcus kodakarensis KOD1 (48) and HypE from Desulfovibrio vulgaris (45) have been reported. Previously, the structure of HypB required for Ni insertion (13) has been determined. An acylphosphatase-like domain (residues 1 to 91, Protein Data Bank [PDB] identification [ID] 1GXU) of the 750-residue-long HypF has also been solved (41). Specific HypE residues involved in interactions with the carbamoyl group, Mg2+, and ATP are proposed based on structural comparisons with ThiL and PurM and sequence conservation of residues near the putative active site. A reaction mechanism based on the crystal structure is postulated for the enzymatic dehydration of the thiocarbamate to the thiocyanate. Moreover, as HypE exerts its function in part through binding to HypF, we have characterized in detail the stoichiometry and binding affinity of the HypE-HypF complex.
MATERIALS AND METHODS
Cloning and purification.
Escherichia coli O157:H7 genomic DNA (38) was used as the template to amplify the hypE gene (AAG57838) and hypF gene (AAG57819) by PCR using Pfu polymerase (Stratagene) and oligonucleotide primers (IDT, Coralville, IA). The hypE gene product, consisting of residues 15 to 336, starting from the second methionine (4), but which is fully active (results not shown), was cloned into a modified pET15b vector (GE Healthcare, Baie d'Urfe, Quebec, Canada) and expressed in E. coli BL21(DE3) as a fusion protein with a noncleavable N-terminal (His)6 tag. The hypF gene was cloned into a modified pET15b vector containing an N-terminal (His)6 tag followed by a tobacco etch virus (TEV) cleavage site and expressed in E. coli BL21(DE3). The hypE and hypF genes were also cloned into a modified pGEX-4T1 vector (GE Healthcare, Baie d'Urfe, Quebec, Canada) and expressed in E. coli BL21 as an N-terminal glutathione S-transferase (GST) fusion protein with a TEV protease cleavage site used to remove the tag. The E. coli methionine auxotroph strain DL41 was transformed by the plasmid for the production of selenomethionine-labeled protein (17).
For expression of native protein E. coli was grown in 1 liter of Terrific broth containing 100 μg ml−1 of ampicillin for 2 h at 37°C followed by induction with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 h at room temperature. The same induction protocol was followed to express the selenomethionine-labeled HypE protein using LeMaster medium (17). Cells were harvested by centrifugation (4,000 × g, 4°C, 20 min) and resuspended in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 400 mM NaCl, 1% (vol/vol) Triton X-100, 5% (vol/vol) glycerol, 10 mM β-mercaptoethanol, 20 mM imidazole, 10 mM magnesium acetate, and a cocktail of protease inhibitors (Complete; Roche Diagnostics). Cells were disrupted by sonication for a total of 2 min with alternating cycles of 15 s on and 15 s off, and the lysate was clarified by centrifugation (150,000 × g, 45 min, 4°C). The protein supernatant was incubated with a 1-ml bed volume of nickel-nitrilotriacetic acid resin (Qiagen, Mississauga, Canada), preequilibrated with lysis buffer, for 1 h with gentle shaking. The beads were washed first with buffer containing 50 mM Tris, pH 7.5, 1 M NaCl, 5% (vol/vol) glycerol, 10 mM β-mercaptoethanol, 20 mM imidazole, 10 mM magnesium acetate followed by the same buffer containing 0.4 M NaCl and 40 mM imidazole. Elution of proteins was performed in the same buffer containing 250 mM imidazole. The purified protein was concentrated by ultrafiltration to 10 mg ml−1 in a final buffer consisting of 20 mM Tris-HCl, pH 7.5, 0.2 M NaCl, 5% (vol/vol) glycerol, 5 mM dithiothreitol (DTT), and 10 mM magnesium acetate. Magnesium acetate is included in all buffers during purification to stabilize the protein as its absence led to precipitation of the purified protein. Incorporation of selenium in HypE was confirmed by electrospray ionization-mass spectrometry performed with an Agilent 1100 Series LC/MSD (Agilent Technologies, Palo Alto, CA) by direct injection of the protein sample for ionization and monitoring in the positive mode using isocratic buffer conditions of 30% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid.
Cells expressing HypF were lysed in 10 mM HEPES, pH 7.5, 400 mM NaCl, 5% (vol/vol) glycerol, 0.1% (vol/vol) Triton X-100, and one tablet of protease inhibitor cocktail (Complete EDTA-free; Roche Diagnostics). Following sonication, the protein supernatant was clarified by centrifugation (15,000 × g, 45 min, 4°C), and the protein supernatant was incubated with 1 ml of glutathione Sepharose resin (GE Healthcare, Mississauga, Canada). Beads were washed with 10 column volumes of TEV cleavage buffer (10 mM HEPES, pH 7.5, 300 mM NaCl). Cleavage of GST-HypF while bound to glutathione Sepharose was performed with TEV protease (1:250 [wt/wt]) for 20 h at room temperature. Cleaved HypF protein was collected and loaded on a Hi-Load Superdex 200 16/60 column (GE Healthcare) equilibrated in the same buffer. Equimolar quantities of HypE and HypF were combined to form the complex and purified by size exclusion chromatography (SEC) on the Superdex 200 column. SEC protein calibration standards (Sigma Chemical Co.) were used to calibrate the Superdex 200 column. The HypE GST fusion protein was purified under conditions similar to those for GST-HypF, except that it was not cleaved using TEV protease.
Cross-linking and densitometry.
HypE and HypF and their cognate complex (pooled fractions from SEC) were cross-linked by mixing stock solutions of BS3 [10 mM bis(sulfosuccinimidyl) suberate; Pierce] or EDC (50 mM 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride; Pierce) in buffer with protein in buffer to give a final concentration of 1 mM BS3 or 5 mM EDC in 20 mM HEPES, pH 7.5, 100 to 200 mM NaCl with magnesium acetate concentrations from 0 to 10 mM. The final protein concentrations were between 1 and 2 mg ml−1. The protein-cross-linker mixtures were incubated at room temperature for 30 to 60 min, and the reactions were quenched by adding equal volumes of 2× sodium dodecyl sulfate (SDS) gel loading buffer. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), together with broad-range molecular weight markers (PageRuler Prestained Protein Ladder Plus; Fermentas) together with samples of HypE and HypF not incubated with cross-linking agents as controls.
The SEC fraction corresponding to the peak maximum was used to determine the stoichiometry of HypE and HypF in the complex. The protein peak fraction at a concentration of 1.5 mg ml−1 together with HypE or HypF protein standards (1 mg ml−1) was analyzed by SDS-PAGE. The standards for densitometry correspond to 16, 12, 8, 4, and 2 μg of protein. Gels were scanned using a Bio-Rad ChemiDoc XRS densitometer (Hercules, CA), and protein bands were quantified using the ImageJ software (NIH freeware; Bethesda, MD).
Isothermal titration calorimetry.
Isothermal titration calorimetry measurements were performed using a VP isothermal titration calorimeter (Microcal, Northampton, MA) at 15°C. HypE and HypF were dialyzed extensively against 20 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM magnesium acetate. The titrations involved a total of 29 10-μl injections of 400 μM HypE into a 1.5-ml solution of 40 μM HypF. Stirring speed was set to 310 rpm, and the time between injections was 350 s. The last 10 injections were averaged as heats of dilution of HypE. Titration curves were fit by a nonlinear least-squares method (Origin software; Microcal, Northampton, MA) using a one-site model. Three independent titrations gave similar thermodynamic and binding estimates.
SPR.
Surface plasmon resonance (SPR) analysis of the HypE-HypF interaction was performed using a Biacore 3000 (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). HypE-GST was captured by noncovalent immobilization to a 7,400-resonance-unit (RU) rabbit anti-GST antibody surface (a generous gift from Yves Fortin). The antibody was covalently immobilized to the surface using standard amine coupling methods with running buffer containing 10 mM HEPES, 10 mM magnesium acetate, 0.02% (vol/vol) Tween 20, 50 μM EDTA, pH 7.4. Briefly, a 35-μl injection of a mixture of 0.05 M N-hydroxysuccinimide and 0.2 M EDC was followed by the manual injection of antibody diluted 100-fold in 10 mM sodium acetate, pH 5, until 7,500 RU was immobilized. The remaining activated surface groups were inactivated by a 35-μl injection of 1 M ethanolamine. Similarly, a blank control surface was created by N-hydroxysuccinimide-EDC treatment followed immediately with ethanolamine.
For the varied contact time injections of His-HypF over HypE-GST, a method using Biacore MDL programming language was written to measure binding of 500 nM His-HypF with noncovalently captured HypE-GST for 60-, 150-, 375-, or 1,500-second contact times. Each cycle was analyzed at an individual contact time, and the order of contact times was selected at random. At a flow rate of 20 μl/min, 500 nM His-HypF was first injected using the KINJECT command with a 300-second dissociation over the anti-GST antibody surface. This was followed by a 10-μl injection of 200 nM GST-HypE solution diluted in running buffer using the INJECT command. Approximately 300 RU GST-HypE was captured, and this injection was immediately followed by 500 nM GST-HypE using an identical KINJECT command as the initial injection over the anti-GST surface alone. One 15-s pulse of 10 mM HCl followed His-HypF over the anti-GST antibody surface, and two 15-s pulses followed His-HypF over the captured GST-HypE surface to regenerate the anti-GST surface for the next cycle. The His-HypF over the anti-GST control injection sensogram was aligned to and subtracted from the His-HypF-captured GST-HypE interaction. The resulting sensogram was double referenced with the buffer injection controls of equal contact times. All of the sensograms were aligned and normalized to the start of the dissociation phase.
For a global fit analysis of the binding sensograms, a dilution series of His-HypF at 2- and 10-minute contact times over GST-HypE was generated. The sequence of injections over the blank and anti-GST antibody surfaces per cycle was performed as above, except that contact time for the His-HypF injections was either 2 min or 10 min. His-HypF samples were prepared by diluting the 2 μM stock solution twofold in running buffer. All of the His-HypF-GST-HypE binding sensograms were blank subtracted and double referenced as above. The resulting sensograms were normalized to account for slight variations in GST-HypE capture. Both the 2-min and 10-min sensogram dilution series were aligned with the start of the His-HypF injection, and binding kinetics were determined by globally fitting the sensograms to a conformational change model using BiaEvaluation software v3.2. Three independent runs were performed to obtain the average apparent kinetic and affinity values [Kd(eq) (dissociation constant)], defined as [ka1/kd1(1 + ka2/kd2)]−1 (26).
AUC.
Fractions of the HypE-HypF complex following SEC were pooled from the corresponding peak and extensively dialyzed against the sample buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT). Analytical ultracentrifugation (AUC) analysis was performed at 13 μM, 9 μM, and 4 μM concentrations of the 1:1 E-F complex. A 13 μM solution of the complex corresponded to an A260 of 1.03. Sedimentation velocity data were collected using a Beckman XL-I analytical ultracentrifuge at 60,000 rpm at 20°C. Sedimentation velocity data were analyzed with the program SEDFIT (V. 8.5, http://www.analyticalultracentrifugation.com/sedfit.htm). Absorbance data were fit to the continuous c(S) model, allowing for the estimate of sedimentation coefficients and molecular weights.
Crystallization.
Initial crystallization conditions were obtained using sparse matrix screens from Hampton Research (Hampton Research, Aliso Viejo, CA). Optimization of initial hits for selenomethionine-labeled protein led to the best diffracting crystals, at 20°C, from microbatch conditions when 1 μl of protein in buffer (20 mM Tris-HCl [pH 7.5], 200 mM NaCl, 5% [vol/vol] glycerol, 5 mM DTT) was mixed with 1 μl of reservoir solution (1.28 M Na/K phosphate, pH 5.0). For native crystals, hanging drop vapor diffusion was employed and crystals were obtained by mixing 3 μl of protein (8 mg ml−1) in buffer (10 mM HEPES, pH 7.5, 5 mM magnesium acetate, 10 mM ATP) with 3 μl of reservoir solution (0.6 M Na/K phosphate, pH 5.0). Crystals grew to their final size in 3 to 4 days. Prior to data collection, crystals were briefly transferred to a cryoprotection solution consisting of reservoir solution supplemented with 20 to 25% (vol/vol) glycerol before flash cooling in the N2 cold stream at 100 K (Oxford Cryosystem, Oxford, United Kingdom). The crystals belong to space group C2 with unit cell dimensions a = 258.6 Å, b = 72.1 Å, c = 113.1 Å, and β = 90° (Z = 16) with four molecules in the asymmetric unit and Vm (Matthew's coefficient figure-of-merit) of 3.5 Å3 Da−1 (28), corresponding to a solvent content of 64%.
Data collection and refinement.
A data set from a selenomethionine-substituted HypE crystal was collected to 2.7-Å resolution at the Se peak energy wavelength 0.9792 Å using a Quantum-4 charge-coupled device detector (Area Detector Systems Corp., San Diego, CA) at beamline X12B at the National Synchrotron Light Source, Brookhaven National Laboratory. A 2.0-Å resolution data set from a native crystal cocrystallized with 10 mM ATP was subsequently collected at SGX CAT, Advanced Photon Source, and used for refinement. Data processing and scaling were performed using HKL2000 (34). Of the 20 expected Se sites, 16 were identified using SHELXD (44) with cross-correlation values for all and weak reflections of 34.7 and 20.1, respectively. These sites were refined using SHARP (6), resulting in phases having a FOM (figure-of-merit) of 0.65. The phases from SHARP were then utilized in solvent flattening and iterative model building and refinement using the RESOLVE autobuild procedure (46). The resulting model included 80% of the main chain and 46% of the side chain atoms. This model was completed manually using XtalView (29) and Coot (8) and refined to a final resolution of 2.0 Å using the program REFMAC5 (33). The final model has a Rwork of 0.176 and Rfree of 0.208 for all reflections. Noncrystallographic symmetry was not applied. The model lacks side chain density for residues 14 to 17 in all four subunits, the region adjacent to Cys193, and residues 34 to 36 in subunits A and D. A serine (residue 14) that is part of the linker between the (His)6 tag and the HypE sequence was found only in subunit D. Lys142 and Thr223 in all subunits were an outlier in the Ramachandran plot from the program PROCHECK (23); however, their electron density is well defined. No electron density consistent with bound nucleotide could be interpreted from these maps. Final data collection and refinement statistics are presented in Table 1.
TABLE 1.
X-ray crystallographic data
| Data set or parameter | Se peak | Apo-HypE |
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Unit cell | ||
| a (Å) | 258.6 | 254.6 |
| b (Å) | 72.1 | 72.3 |
| c (Å) | 113.1 | 112.6 |
| β (degrees) | 117.0 | 115.3 |
| Za | 16 | 16 |
| Resolution (Å) | 50-2.7 (2.80-2.70) | 50-2.0 (2.07-2.0) |
| Wavelength (Å) | 0.9792 | 0.97978 |
| Observed hkl | 154,890 | 457,738 |
| Unique hkl | 92,233b | 121,997 |
| Redundancy | 1.9 (1.8) | 3.8 (3.7) |
| Completeness | 92.3 (80.4) | 98.7 (98.7) |
| Rsymc | 0.051 (0.261) | 0.057 (0.204) |
| I/σ (I) | 21.9 (3.8) | 10.7 (3.8) |
| Wilson B (Å)b | 61.8 | 23.1 |
| Refinement | ||
| Resolution (Å) | 43.7-2.0 | |
| Rworkd (hkl) | 0.176 (117,379) | |
| Rfree (hkl) | 0.208 (6,153) | |
| Average B factors | ||
| Protein | 25.0 (9,862) | |
| Solvent | 30.0 (714) | |
| Ramachandran plot | ||
| Allowed (%) | 99.2 | |
| Generous (%) | 0.5 | |
| Disallowed (%) | 0.4 | |
| RMS, bonds (Å) | 0.008 | |
| RMS, angles (degrees) | 1.151 | |
| PDB code | 2RB9 |
Number of molecules in the unit cell.
Number of Bijovet pairs unmerged.
Rsym = (ΣIobs − Iavg)/ΣIavg.
Rwork = (ΣFobs − Fcalc)/ΣFobs.
Protein structure accession number.
The coordinates and structure factors have been deposited in the PDB under accession code 2RB9.
RESULTS AND DISCUSSION
The protein that we used for crystallization, structure determination, and biophysical characterization lacks the first 14 residues, as our construct starts at the second methionine. It was, therefore, necessary to confirm that this shortened enzyme possesses the expected catalytic activity. This was assessed in vivo by a complementation assay of the DHP-E strain containing the inactivated hypE gene (19). The DHP-E strain produces no H2 gas, as would be expected, because it cannot synthesize active H2ase 3, and consequently no active formate-hydrogen lyase complex is made. However, the mutant transformed with the vectors carrying either full-length hypE or the shortened version showed production of gas (data not shown), confirming that the short version of HypE is fully functional.
Monomer structure.
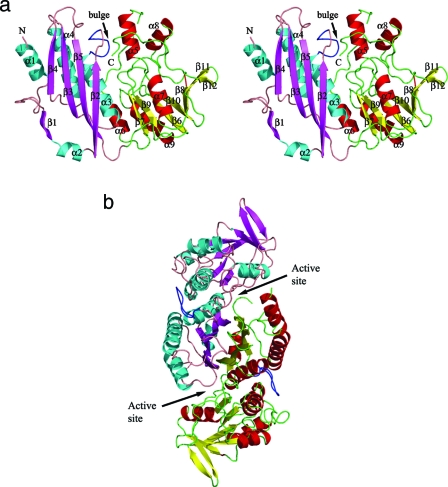
The recombinant protein corresponds to a shortened HypE construct starting from the second methionine (Met15 in the full-length structure). Four HypE molecules are present in the asymmetric unit. The monomers superimpose well on each other with a root mean square deviation (RMSD) of 0.33 to 0.43 Å for ∼313 Cα pairs. Each monomer consists of two domains—the N-terminal domain spanning residues 15 to 157 and the C-terminal domain comprised of residues 158 to 336 (Fig. 1a). The N-terminal domain is composed of a four-stranded mixed β-sheet (β2↑-β5↓-β3↑-β4↑) and three α-helices nearly parallel to the β-strands covering one side of the β-sheet. An additional extended strand, β1, and a short helix, α2, are involved in dimer formation (see below). The C-terminal domain contains a central, seven-stranded bent β-sheet assembled in the order β7↑-β9↓-β6↑-β10↓-β8↑-β11↑-β12↓. The helix α7 is positioned in the center of the concave side of this β-sheet; helices α6 and α9 are on the convex side of the β-sheet along strands β6, β7, and β9; and helices α5 and α8 form a minidomain on one side of the β-sheet (Fig. 1a). The N-terminal helix α1 (residues 15 to 28) is less well ordered and has higher temperature factors.
FIG. 1.
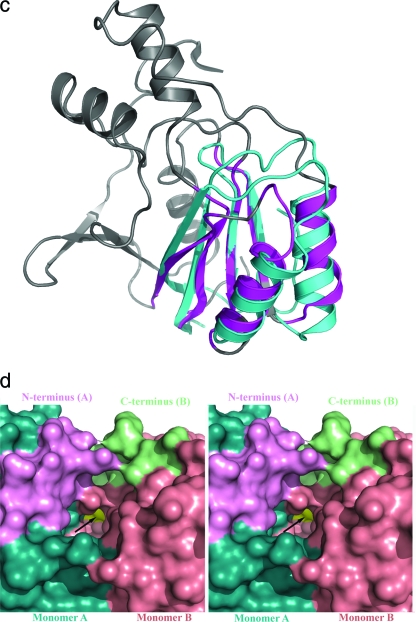
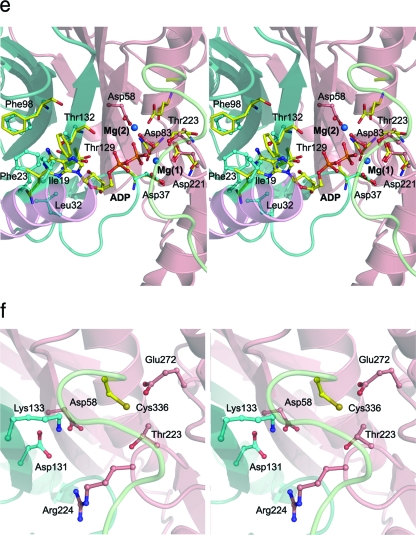
Cartoon representation of HypE. (a) Stereo view of the monomer. The domains are individually colored (N terminal: helices, cyan; strands, magenta; loops, salmon; C terminal: helices, red; strands, yellow; loops, green). The “bulge” corresponding to residues 64 to 71 is colored blue. This figure and subsequent figures were prepared using PyMol (www.pymol.org). (b) HypE dimer viewed along the twofold axis colored as described above. The active sites are indicated with arrows. (c) Superposition of the C-terminal domain of HypE (gray) with the N-terminal domain of HypF (cyan) shown in a cartoon representation. The parts of HypE that share the fold with HypF are colored in magenta. (d) Surface representation of the active site of HypE formed at the interface of monomers A (cyan) and B (salmon). The N-terminal helix (pink) of monomer A and the C-terminal loop (light green) of monomer B are situated proximal to each other and contribute toward the active site (shown by arrow). (e) Stereo view of the active site of HypE with modeled ADP positioned based on the superposition of HypE with PurL. One monomer is depicted in cartoon format while the second monomer is shown in surface representation. (f) Blowup of the putative carboxamide binding site situated near the C-terminal cysteine (Cys336).
Dimer structure.
The HypE structure observed in the crystal is a dimer. There are two such dimers in the asymmetric unit, which can be superimposed with an RMSD of ∼0.6 Å for 622 Cα pairs. Gel filtration studies carried out either in the presence or in the absence of 5 mM DTT at a protein concentration similar to that used for crystallization showed that HypE in both cases elutes as a dimer (data not shown), suggesting that a disulfide linkage through the C-terminal cysteines is not a prerequisite for dimer formation, contrary to previous suggestions (4, 40). The crystal structure supports the above conclusion, as the C-terminal cysteines of the two monomers do not form a disulfide bridge and are located ∼25 Å apart.
The dimer has an elongated shape with overall dimensions of ∼95 × 48 × 40 Å3. The two monomers associate through interactions predominantly involving their N-terminal domains and, in particular, back-to-back stacking of their β-sheets (Fig. 1b). The β1 strand from one monomer associates in an antiparallel fashion with the β2 strand of the second monomer, providing the fifth strand in the β-sheet of the N-terminal domain. The two N-terminal domains therefore form an αββα layered structure. The interacting surfaces of the β-sheets have two hydrophobic patches, one centered at Met150 and the other centered at Phe144. The hydrogen bonding interactions between the monomers are found only at the edges of the β-sheets. A deep, cone-shaped cavity is formed between the N-terminal domain of one monomer and the C-terminal domain of the second monomer in the vicinity of the N-terminal helix. This putative active site cavity is occupied by the C terminus of the second monomer, with the terminal Cys336 residing at the very bottom of this cavity (Fig. 1d).
Sequence and structural comparisons.
The crystal structure of E. coli HypE is similar to that of the “inward” conformation of the enzyme from T. kodakarensis (48), showing an RMSD upon superposition of 1.1 Å for 262 Cα pairs for the monomers, reflecting the 38.5% sequence identity between the two proteins. Superposing the dimers shows an RMSD of 1.3 Å for 478 Cα pairs, indicating that the mode of dimerization is the same for the two enzymes. As our crystallization conditions also included ATP at a concentration of 10 mM, our results are consistent in that the C-terminal ends of the molecules adopt the same, inward conformation (48). The most significant differences are in the conformation of the N-terminal segment. In the E. coli HypE the first 40 residues are well ordered and begin with an α-helix which packs along the β-sheet of the N-terminal domain next and parallel to helix α3. The T. kodakarensis structure corresponds to the long form (15 additional residues on the N terminus), and the N terminus is poorly ordered. The well-ordered region starts at Asp43, and the residues Asp43 to Phe49 form an additional, first β-strand of the main β-sheet (48), on the side opposite where the corresponding segment is placed in the E. coli enzyme. In the dimer, this position is occupied by the N terminus of the other subunits. The most recent structure of the HypE from D. vulgaris shows the same conformation of the N terminus as in the E. coli enzyme described here. Therefore, in the T. kodakarensis HypE there is a swapping of the N-terminal segment between the two subunits of the dimer, or the connectivity has been modeled incorrectly due to poor electron density in this region. Other differences between the HypE proteins from different organisms are in a few connecting loops between secondary structure elements.
Within the PFAM database (9) HypE is clustered together with a large pool of aminoimidazole ribonucleotide synthase (AIRS)-related protein sequences comprising the PF00586 and PF02769 families corresponding to the N- and C-terminal HypE domains, respectively. These sequences correspond to the IPR00078 and IPR010918 families in the InterPro database (32). There are over 160 HypE-like sequences that contain both domains (InterPro, IPR11854). Among these proteins several have known three-dimensional structures, including E. coli AIRS (PurM, EC 6.3.3.1) and Thermotoga maritima phosphoribosyl formylglycinamide synthase (FGAM/PurL, EC 6.3.5.3), both of which are involved in de novo purine biosynthesis, as well as thiamine-monophosphate kinase (ThiL, EC 2.7.4.16). HypE shares low overall sequence identity (∼19%) with ThiL from Aquifex aeolicus and ∼15% with PurM from E. coli. Interestingly, all the above enzymes perform ATP-dependent processing of their substrates (1, 25).
Sequence analysis of HypE and HypE-related proteins shows conservation of many residues throughout the entire sequence. The C-terminal sequence motif 333PR(I/V)C336, which includes the catalytically important Cys336 (40), and two other sequence motifs, 130GDTKV134 and 220RDX(T/S)RGG226, are conserved in all HypE sequences. Additionally, the conserved signature motif DX4GAXP, involved in ATP binding in PurM/PurL and ThiL (1), is also found conserved in HypE (83DX4GAXP91) and related sequences. Interestingly, the first aspartate in the ATP binding motif is involved in metal binding (1), and therefore, the presence of this signature motif in HypE suggests metal-mediated ATP binding, most likely of a Mg2+ ion. These conserved motifs cluster together within the HypE dimer and form the substrate binding site near the dimer interface.
Despite low sequence identity, the fold of HypE is similar to that of PurM (PDB ID 1CLI [25]), PurL (PDB ID 2HRU [31]), and ThiL (PDB ID 1VQV [S. Eswaramoorthy and S. Swaminathan, unpublished data]). Structural superposition of HypE on PurM and ThiL suggests that the chain assignment of the N-terminal regions (residues 1 to 32) of both subunits in ThiL should be swapped to obtain the correct dimer. This misassignment was likely the result of disorder in the segment consisting of residues 33 to 37 in both subunits and their close proximity in the dimeric structure. PurL (PDB ID 1T3T [1]), though being a monomer, possesses an internal fold repeat that makes this molecule resemble the HypE dimer. Although the dimers of HypE, ThiL, and PurM are rather similar, the relative orientations of the monomers within these dimers differ somewhat, leading to differences in the size of the individual active site pockets.
Rather unexpectedly, the first six structural elements (β7-β9-β6-β10-α6-α9) of the C-terminal domain of HypE exhibit fold similarity with the N-terminal acylphosphatase domain (residues 1 to 91) of HypF (1GXU [41], another accessory protein involved in H2ase maturation), with an RMSD of 1.4 Å for 42 Cα atom pairs (2.4 Å for 55 pairs) (Fig. 1c).
Among this group of enzymes, a structural feature observed only in HypE is a bulging loop between strand β2 and helix α3 (residues 65 to 72, Fig. 1c). This structural element displaces the adjacent segment corresponding to residues 195 to 201 (an α-helix in PurM and ThiL) and aids in the formation of a cavity suitable for burying the C-terminal Cys336, which plays a pivotal role in accepting the carbamoyl moiety from HypF (4). This bulge in HypE is caused by two prolines in this sequence (64PXXXP68), in which the first proline is conserved in all HypE sequences. The corresponding structural element that connects the β-strand and α-helix in PurM and ThiL is a loop that varies in the number of residues between the proteins. In PurM, an additional α-helix is present in this loop segment. Another difference is that the C terminus in HypE is buried at the dimer interface.
Active site. (i) ATP binding site.
Biochemical evidence shows that HypE catalyzes a bisubstrate reaction involving carboxamide and ATP (4, 40). Despite many attempts, we were unable to trap ATP or its nonhydrolyzable analogs in the crystal structure of HypE, similarly to unsuccessful trials reported for PurM and ThiL. However, the structure of D. vulgaris HypE was determined in complex with ATP and confirmed the predicted location of its binding site. Also, the recent crystal structures of PurL have revealed a molecule of bound ADP (1) or ATP (31). All of these structures show a common ATP binding location.
The ATP binding pocket in E. coli HypE forms a deep cavity, very similar to that in HypE from D. vulgaris (HypEDv). In PurM and PurL the presence of a β-hairpin between helix α1 and strand β3 creates a suitable cavity for proper positioning of the adenine ring between the N-terminal β-sheet and helix α1. However, the absence of this β-hairpin in HypE and ThiL shifts the N-terminal helix by 2 to 4 Å relative to the α1 helix in PurM and PurL and places the helix much closer to the N-terminal β-sheet, while still maintaining the adenine binding site. The residues that were identified in HypEDv as interacting with the adenine base are conserved in the E. coli enzyme, namely, Ile19, Phe23, and Leu32. The requirement of Mg2+ ions for efficient catalysis (4) supports their binding to the enzyme. We and others have not found Mg2+ ions in the apo structure of HypE, PurM (25), or PurL (1, 31). However, Mg2+ ions are present in the enzyme-ATP complex, indicating that the binding of ATP occurs with the participation of Mg2+. Two such ions coordinated by the phosphate groups of ATP and protein side chains were observed in HypEDv (45) and in PurL complexes (31). The side chains that coordinate Mg2+ ions are structurally conserved in HypE and PurL. In E. coli HypE, one Mg2+ ion would be coordinated by the side chains of Asp37 (Asp39HypEDv), Asp83 (Asp79HypEDv), and Asp221 (Asp218HypEDv) (Fig. 1e). The second Mg2+ ion would be coordinated by the side chains of Asp58 (Asp54HypEDv) and Asp131 (Asp127HypEDv). Previous studies of HypE showed that Asp83 (4) is essential for ATPase activity, indicating that the Mg2+ is necessary for ATP hydrolysis.
(ii) Putative carboxamide binding site.
Based on the observation that the C-terminal thiocarbamate is the substrate that undergoes dehydration to produce thiocyanate (4, 40), the cavity situated adjacent to Cys336 is identified as the putative carboxamide binding site (Fig. 1f). The residues situated in the vicinity of Cys336 that could participate in ligand interactions are Asp58, Asp131, Lys133, Thr223, Arg224, and Glu272 and are highly conserved in HypE-related sequences. Of these, Asp58, Lys133, Thr223, and Glu272 are predicted to be within hydrogen bonding distance of the carbamate when bound to Cys336. We also expect small reorientation of the side chain of Cys336 upon covalent attachment of the carboxamide group to accommodate the substrate in the active site pocket.
Reaction mechanism.
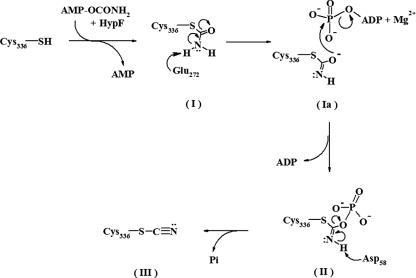
The dehydration reaction proposed for the catalytic conversion of the thiocarbamate to the thiocyanate by HypE involves two steps (40). A unique feature of HypE among AIRS family enzymes is that both the substrate and the product remain covalently attached to the C-terminal cysteine residue. In the first step, phosphotransfer from the Mg2+-ATP complex leads to phosphorylation of the substrate's amide oxygen and formation of a phosphoryl anhydride intermediate (Fig. 2). Subsequently, proton abstraction from the N atom of the anhydride intermediate by a base generates the final product containing the CN group. The structure of HypE allows us to interpret the original proposal at the molecular level.
FIG. 2.
Proposed reaction mechanism for HypE. The reaction intermediates that are formed starting from free cysteine (Cys336) are the thiocarbamate after reaction with carbamylated AMP in the presence of HypE before (I) and after (Ia) activation, thiocarbamic phosphoryl anhydride (or iminophosphate intermediate) (II), and thiocyanate (III)—the final product. Glu272 is proposed to act in the deprotonation of the amino nitrogen of the substrate (I), and Asp58 is proposed to act in the deprotonation of the imino nitrogen (II).
The reaction catalyzed by HypE has features in common with those of PurM and PurL, namely, the ATP-dependent dehydration of an amide to generate a phosphoryl anhydride intermediate (1, 25). Our proposed mechanism for HypE is shown in Fig. 2. The amide group of the substrate (I, Fig. 2), covalently attached to the S atom of Cys336, is a neutral ligand. Therefore, proton abstraction by a base is necessary to activate the amide oxygen for in-line attack on the γ-phosphate of ATP. The modeled position of the thiocarbamoyl nitrogen atom is equidistant between the carboxylate groups of Asp58 and Glu272. One likely residue that could function as a base is Glu272, which is within a suitable distance of the putative position of the carbamoyl moiety and is strictly conserved in HypE-related sequences. An equivalent glutamate, Glu269, was suggested for the role of a general base in HypEDv. The attack of the amide oxygen of the thiocarbamate (Ia, Fig. 2) on the γ-phosphate of ATP results in the formation of a thiocarbamic phosphoanhydride intermediate (II, Fig. 2), possibly stabilized by Thr223 and Lys133. A related substrate-derived carbamic-phosphoryl anhydride reaction intermediate has been trapped crystallographically for dethiobiotin synthetase (21). The mechanisms employed by various enzymes for the transfer of the terminal phosphoryl group from a Mg2+-nucleotide complex to the substrate involving the formation of a pentavalent transition-state intermediate are well documented (27). In such transfers, the geometric alignment of the γ-phosphate for in-line transfer to the oxygen of the substrate is through specific metal coordination. Based on the structural comparison of HypE with other structurally related proteins as well as the available biochemical data for HypE (4), we propose that Asp83, through coordination of the metal, plays a role in anchoring the ATP and aiding in proper orientation of the γ-phosphate for in-line attack by the amide oxygen of the thiocarbamate.
The mechanism described for PurL and dethiobiotin synthetase suggests that nucleophilic attack on the phosphoanhydride intermediate is required to complete the dehydration reaction. The requirement of a base for the dephosphorylation step is consistent with the expected reaction chemistry of HypE (40). The crystal structure of HypE shows the presence of Asp58 and Lys133 in the putative substrate binding site, either of which could carry out the role of the second base. Modeling shows that the anhydride intermediate (II, Fig. 2) can be positioned at the HypE active site such that the imino group is proximal to Asp58 with the phosphate group near Lys133, Arg224, and Thr223 allowing for charge stabilization. This suggests the possibility that Asp58 participates as the second base in the overall reaction, abstracting the proton from the imino nitrogen and leading to the formation of thiocyanate (III, Fig. 2) with concomitant release of phosphate. This aspartate is conserved in the HypE sequences from various organisms. All of the putative active site residues identified in E. coli HypE are structurally identical in HypE from T. kodakarensis (48) and D. vulgaris (45).
HypE-HypF interaction.
HypE is one of the essential accessory proteins required for the synthesis of mature H2ase and is involved in interactions with multiple partners. It has been established that HypE exerts its activity through forming a complex with HypF (4, 20) and, subsequently, through interactions with HypC-HypD (3, 20). Using a variety of methods, we have investigated the binding affinity and stoichiometry of the HypE-HypF complex.
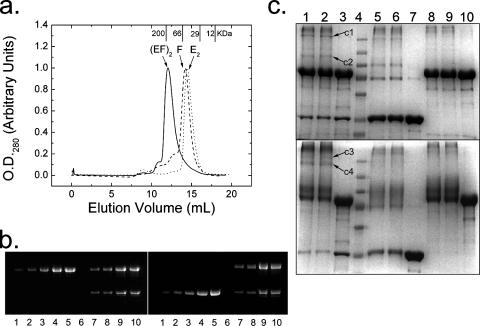
SEC (Fig. 3a) of HypE (35 kDa) and HypF (80 kDa) suggests that a complex between these two proteins is formed. There is a clear shift to a lower elution volume for the mixture of the two proteins compared to that of either the dimeric HypE (E2) or monomeric HypF (F) alone. The presence of magnesium acetate (10 mM), ATP (1 mM), carbamoyl phosphate (1 mM), or the nonhydrolyzable ATP analog ATP-β-γ-methylene triphosphate (1 mM) did not affect the volume of elution by SEC either of HypE or HypF alone or of the complex. Analysis of HypE-HypF mixtures by SEC in which one protein or the other is added in molar excess reveals the expected additional peak in the chromatogram corresponding to the protein in excess (result not shown). The calculated molecular weights from a calibrated SEC column based on the elution volumes of the major peaks (Table 2) suggest that HypE and HypF form an [EF]2 complex under these conditions. This result is supported by densitometry of Coomassie blue-stained SDS-polyacrylamide gels (Fig. 3b) of fractions from the major elution peak of the complex from SEC, confirming an equimolar stoichiometry (average ratio is 1.17 ± 0.11 HypE:HypF) of the two proteins. Cross-linking of the complex using EDC or BS3, a cross-linker with ∼3-Å or ∼20-Å spacing between the connected groups, respectively, also alludes to the formation of [EF]2 (Fig. 3c). When cross-linking the E and F complex with EDC, the appearance of bands c1 and c2 suggests the formation of [EF]2 and EF, respectively. Cross-linking with a longer spacer (BS3) shows bands c3 and c4 near the 250-kDa marker protein, suggestive of differently cross-linked forms of [EF]2. The absence of the EF band in BS3-cross-linked products may be due to the relatively longer length of the linker and the dynamic nature of the EF-[EF]2 equilibrium.
FIG. 3.
Characterization of HypE-HypF interactions. (a) SEC: elution profiles of HypE (dots) and HypF (dashes) and an equal mixture of HypE and HypF by mass (solid line). Calculated molecular weights averaged from at least three runs (Table 2) confirm a monomeric HypF, a dimeric HypE, and a 2:2 complex of HypE and HypF. The elution volumes of four molecular mass standards are indicated on the top x axis. (b) Quantitation of HypE and HypF from the major elution peak of the HypE-HypF complex (solid line, marked by an arrow) following SEC. Lanes 1 to 5 of the SDS gel are increasing amounts of the HypF (left panel) and HypE (right panel) standards, lane 6 is empty, and lanes 7 to 10 are different amounts of the HypE and HypF complex from the peak fraction. Estimates from two independent measurements gave an average HypE/HypF ratio of 1.17 ± 0.11. (c) Cross-linking by BS3 (top gel) and EDC (bottom gel) analyzed by SDS-PAGE: lanes 1 and 2 are cross-linked HypE and HypF, lane 3 is HypE and HypF, lane 4 is a broad-range molecular mass marker (from top, 250, 130, 100, 70, 55, 35, and 27 kDa), lanes 5 and 6 are cross-linked HypE, lane 7 is HypE alone, lanes 8 and 9 are cross-linked HypF, and lane 10 is HypF alone. Bands c1 to c4 correspond to cross-linked HypE and HypF complexes.
TABLE 2.
Apparent molecular masses of HypE, HypF, and their complex estimated from gel filtration chromatography and dynamic light scattering
| Protein(s) | SEC
|
Dynamic light scattering
|
||
|---|---|---|---|---|
| Concn (mg/ml) | Molecular mass (kDa) | Concn (mg/ml) | Molecular mass (kDa) | |
| HypE | 4-10 | 64 ± 2 | 2-5 | 70 ± 6 |
| HypF | 5-10 | 72 ± 4 | 2-5 | 81 ± 5 |
| HypE-HypF | 5-10 | 212 ± 9 | 3-8 | 201 ± 19 |
| 1 | 96 | |||
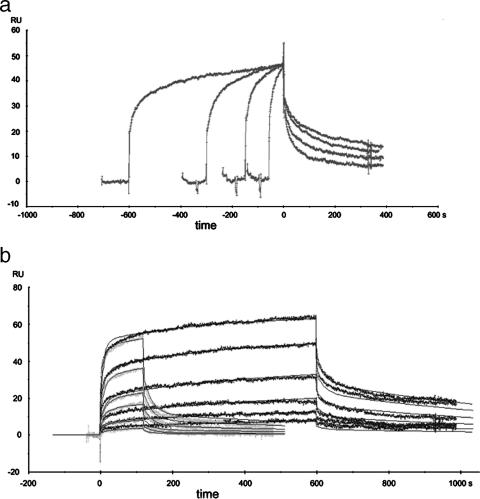
SPR was used to further investigate the association between HypE and HypF. A series of injections of His-tagged HypF protein over the α-GST sensor surface saturated with GST-HypE dimer revealed sensograms showing a kinetic response varying with the contact time (from 60 s to 1,500 s) (Fig. 4). These data could not be satisfactorily fit to a 1:1 Langmuir binding model. These experiments showed that the longer the association time, the slower the dissociation of HypF from the GST-HypE surface, a characteristic of linked reactions (22). These data offer evidence of a conformational change accompanying binding of HypF to HypE, an interpretation supported by similar SPR results with other proteins (18, 26) and noted also in T. kodakarensis HypE in the presence or absence of ATP (48). Global fitting of these data gave an apparent Kd(eq) of 464 ± 94 nM. Separate SEC experiments showed that GST-HypE behaved as an apparent dimer and that it was capable of forming an apparent heterotetramer with (His)6-HypF (results not shown).
FIG. 4.
SPR analysis of HypF binding to GST-HypE. (a) Varied contact times (60, 150, 375, or 1,500 s) of 500 nM His-HypF over approximately 300 RU HypE-GST noncovalently captured over a 7,400-RU rabbit anti-GST antibody surface. The binding sensograms are normalized to the start of the dissociation phase to highlight the change of kinetic values with time. (b) Global fit of 2-min (light gray) and 10-min (dark gray) injections of HypF-His (2 μM to 62.5 nM) over 300 RU HypE-GST noncovalently captured using a 7,400-RU rabbit anti-GST antibody surface. The sensograms were blank subtracted, double referenced, and normalized to account for slight variations in HypE-GST capture. Evaluation of the curves was performed using a global fit of both the 2- and 10-min injections using a conformational change model in the BiaEvaluation software v3.2. The modeled curves are shown as the black lines and fit to the experimental data with χ2 < 2 in this example. Three independent runs were performed and resulted in a Kd(eq) of 464 ± 94 nM.
Isothermal titration calorimetry was utilized to further probe the interaction between these two proteins. Titrating HypF at a concentration of 40 μM at 15°C with HypE showed an isotherm depicting a single binding event with derived thermodynamic and binding parameter estimates as follows: Kd, 0.223 ± 0.032 μM; n, 0.990 ± 0.0058; ΔH, −1,364 ± 20.7 cal/mol; ΔS, 25.7 cal/mol K. Thermodynamic estimates show that this interaction is exothermic and is both entropically and enthalpically favored. The stoichiometry, as represented by the n value of ∼1, suggests that the interaction of HypE and HypF is equimolar. The estimated Kd of ∼220 nM is also in reasonable agreement with that obtained by SPR and indicates an interaction of medium affinity.
The dependence of heterodimer-heterotetramer (EF↔ [EF]2) equilibrium on protein complex concentration was verified by dynamic light scattering and AUC sedimentation velocity studies. At the lower protein concentration (1 mg ml−1), the complex is predominantly EF (96 kDa), while at higher concentrations of 3 to 8 mg ml−1, it is likely [EF]2 (201 kDa). An AUC sedimentation velocity study of the HypE-HypF complex from the main SEC fraction further substantiates these observations. The analysis (Table 3) shows the presence of several species. At equimolar concentrations of HypE-HypF increasing from 0.5 to 1.5 mg ml−1, the predominant species is EF, with an increase in the relative abundance of [EF]2 with increasing concentration of the two proteins. Consistently, neither AUC nor SEC showed the presence of an EEF species. Overall, these results suggest a strong interaction between HypE and HypF with a concentration-dependent 2[EF]↔ [EF]2 equilibrium. This conclusion is also consistent with the HypE crystal structure in which the putative active site, including Cys336, is buried at the domain interface of two HypE subunits (Fig. 1d), suggesting that the transfer of the CN moiety from HypE to HypC may require a monomeric form of HypE with its C terminus accessible to the recipient protein. Further studies will be necessary to identify specific HypE and HypF residues involved in complex formation and in transfer of the carbamoyl group.
TABLE 3.
Relative abundances of various species from AUC sedimentation velocity
| Peak no. | Molecular mass (kDa)
|
Species | Relative abundance (%) at concn of HypE-HypF complex in AUC cell (mg/ml):
|
|||
|---|---|---|---|---|---|---|
| Theoretical | Estimated | 1.5 | 0.9 | 0.5 | ||
| 1 | 35.1 | 46 ± 5 | E | 5 | 9 | 9 |
| 2 | 81.9, 70.2 | 77 ± 6 | F or E2 | 11 | 9 | 20 |
| 3 | 117 | 110 ± 7 | EF | 72 | 70 | 52 |
| 4 | 163.8, 140.4 | 169 ± 8 | F2 or E4 | 6 | 8 | 15 |
| 5 | 234 | 215 ± 19 | [EF]2 | 6 | 5 | 3 |
In the current model, the transfer of the carbamoyl group from HypF to the C-terminal Cys336 of HypE occurs first, followed by the ATP-dependent dehydration of the thiocarbamate to thiocyanate (4, 40). Synthesis of the CN group is dependent on the formation of a HypE-HypF complex. Similarly, the formation of the HypE-HypC-HypD complex is essential for the transfer of the CN group to Fe, and subsequently, to the large subunit of the H2ase complex (3, 42). In both cases, Cys336HypE has to interact both with the active sites of HypF and with the HypC-HypD complex for accepting the substrate (carbamoyl group) or delivering the product (transfer of CN to Fe), respectively. In the crystal structure of apo-HypE, the C-terminal Cys336 is situated within a deep cleft, buried between two domains at the dimer interface. This arrangement of the C terminus completes the active site of HypE for the dehydration reaction. To access the active site on HypF or on HypC-HypD, the C terminus of HypE must become accessible to the partnering protein(s). While an ATP-dependent conformational change in HypE has been described (45, 48), further conformational changes in HypE, HypF, or both proteins could result upon their interaction. Dissociation of the HypE dimer as it interacts with HypF would also allow interaction between the active site around Cys336 and the carbamoyl moiety donated by HypF.
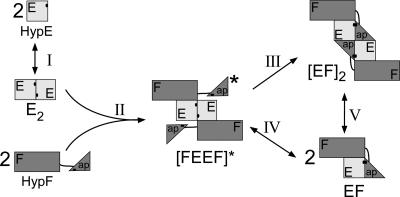
Combining all the data presented above, we propose a model of HypE-HypF complex formation shown in Fig. 5. HypE exists predominantly in a dimeric form at higher concentrations but in monomer-dimer equilibrium at lower concentrations. At the concentrations used in this study two molecules of HypF bind to the EE dimer to form [FEEF]*, followed by conformational rearrangement to a stable [EF]2 form. At lower concentrations there is an equilibrium between the [FEEF]* and/or [EF]2 and a heterodimer EF. The absence of a detectable EEF species in the SEC studies, where HypE is in molar excess, supports this proposition.
FIG. 5.
Model for HypE-HypF interaction. The HypE monomer-dimer E2 equilibrium is represented by reaction I. E2 and F interact to form an unstable tetrameric intermediate [FEEF]* (reaction II). [FEEF]* rearranges to form a stable tetramer [EF]2 (reaction III), which allows proximity of the putative active sites of E and F. Alternatively, the E dimer in [FEEF]* would dissociate to form EF (reaction IV). The protein concentration-dependent heterodimer [EF]-heterotetramer [EF]2 equilibrium is represented by reaction V. “ap” refers to the acylphosphatase domain of HypF.
Acknowledgments
X-ray diffraction data for this study were measured at beamlines X12B and X29 of the National Synchrotron Light Source and SGX CAT at the Advance Photon Source. Their financial support comes principally from the Office of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and the National Center for Research Resources of the National Institutes of Health.
We thank Alexei Soares, Dieter Schneider (beamline X12B), Howard Robinson (beamline X29), and Tongpil Min for assistance in data collection; Jean-Claude Lavoie for AUC analysis; and Holger Lindner and Traian Sulea for helpful discussions. AUC data analysis was made possible using UltraScan, DEMELER, and The University of Texas Health Science Center at San Antonio. We thank John Wagner for cloning.
This research was funded by a grant from the Canadian Institutes of Health Research, GSP-48370, to M.C.
This is NRC publication no. 49530.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Anand, R., A. A. Hoskins, J. Stubbe, and S. E. Ealick. 2004. Domain organization of Salmonella typhimurium formylglycinamide ribonucleotide amidotransferase revealed by X-ray crystallography. Biochemistry 4310328-10342. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., B. C. Berks, J. McClay, A. Ambler, M. A. Quail, P. Golby, and J. R. Guest. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 1433633-3647. [DOI] [PubMed] [Google Scholar]
- 3.Blokesch, M., S. P. Albracht, B. F. Matzanke, N. M. Drapal, A. Jacobi, and A. Bock. 2004. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J. Mol. Biol. 344155-167. [DOI] [PubMed] [Google Scholar]
- 4.Blokesch, M., A. Paschos, A. Bauer, S. Reissmann, N. Drapal, and A. Bock. 2004. Analysis of the transcarbamoylation-dehydration reaction catalyzed by the hydrogenase maturation proteins HypF and HypE. Eur. J. Biochem. 2713428-3436. [DOI] [PubMed] [Google Scholar]
- 5.Bock, A., P. W. King, M. Blokesch, and M. C. Posewitz. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 511-71. [DOI] [PubMed] [Google Scholar]
- 6.Bricogne, G., C. Vonrhein, C. Flensburg, M. Schiltz, and W. Paciorek. 2003. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 592023-2030. [DOI] [PubMed] [Google Scholar]
- 7.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9228-237. [DOI] [PubMed] [Google Scholar]
- 8.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 602126-2132. [DOI] [PubMed] [Google Scholar]
- 9.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontecilla-Camps, J. C., M. Frey, E. Garcin, C. Hatchikian, Y. Montet, C. Piras, X. Vernede, and A. Volbeda. 1997. Hydrogenase: a hydrogen-metabolizing enzyme. What do the crystal structures tell us about its mode of action? Biochimie 79661-666. [DOI] [PubMed] [Google Scholar]
- 11.Forzi, L., P. Hellwig, R. K. Thauer, and R. G. Sawers. 2007. The CO and CN(-) ligands to the active site Fe in [NiFe]-hydrogenase of Escherichia coli have different metabolic origins. FEBS Lett. 5813317-3321. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich, B., and E. Schwartz. 1993. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu. Rev. Microbiol. 47351-383. [DOI] [PubMed] [Google Scholar]
- 13.Gasper, R., A. Scrima, and A. Wittinghofer. 2006. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 28127492-27502. [DOI] [PubMed] [Google Scholar]
- 14.Hagemeier, C. H., L. Chistoserdova, M. E. Lidstrom, R. K. Thauer, and J. A. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 2673762-3769. [DOI] [PubMed] [Google Scholar]
- 15.Happe, R. P., W. Roseboom, A. J. Pierik, S. P. Albracht, and K. A. Bagley. 1997. Biological activation of hydrogen. Nature 385126. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann, G. C., A. R. Klein, M. Linder, and R. K. Thauer. 1996. Purification, properties and primary structure of H2-forming N5, N10-methylenetetrahydromethanopterin dehydrogenase from Methanococcus thermolithotrophicus. Arch. Microbiol. 165187-193. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 91665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentschke, M., C. Schulze, U. Susens, and U. Borgmeyer. 2003. Characterization of calmodulin binding to the orphan nuclear receptor Errgamma. Biol. Chem. 384473-482. [DOI] [PubMed] [Google Scholar]
- 19.Jacobi, A., R. Rossmann, and A. Bock. 1992. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch. Microbiol. 158444-451. [DOI] [PubMed] [Google Scholar]
- 20.Jones, A. K., O. Lenz, A. Strack, T. Buhrke, and B. Friedrich. 2004. NiFe hydrogenase active site biosynthesis: identification of Hyp protein complexes in Ralstonia eutropha. Biochemistry 4313467-13477. [DOI] [PubMed] [Google Scholar]
- 21.Kack, H., K. J. Gibson, Y. Lindqvist, and G. Schneider. 1998. Snapshot of a phosphorylated substrate intermediate by kinetic crystallography. Proc. Natl. Acad. Sci. USA 955495-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson, R., A. Michaelsson, and L. Mattsson. 1991. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J. Immunol. Methods 145229-240. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 24.Lenz, O., I. Zebger, J. Hamann, P. Hildebrandt, and B. Friedrich. 2007. Carbamoylphosphate serves as the source of CN(-), but not of the intrinsic CO in the active site of the regulatory [NiFe]-hydrogenase from Ralstonia eutropha. FEBS Lett. 5813322-3326. [DOI] [PubMed] [Google Scholar]
- 25.Li, C., T. J. Kappock, J. Stubbe, T. M. Weaver, and S. E. Ealick. 1999. X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution. Structure 71155-1166. [DOI] [PubMed] [Google Scholar]
- 26.Lipschultz, C. A., Y. Li, and S. Smith-Gill. 2000. Experimental design for analysis of complex kinetics using surface plasmon resonance. Methods 20310-318. [DOI] [PubMed] [Google Scholar]
- 27.Matte, A., L. W. Tari, and L. T. Delbaere. 1998. How do kinases transfer phosphoryl groups? Structure 6413-419. [DOI] [PubMed] [Google Scholar]
- 28.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33491-497. [DOI] [PubMed] [Google Scholar]
- 29.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125156-165. [DOI] [PubMed] [Google Scholar]
- 30.Menon, N. K., J. Robbins, H. D. Peck, Jr., C. Y. Chatelus, E. S. Choi, and A. E. Przybyla. 1990. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J. Bacteriol. 1721969-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morar, M., R. Anand, A. A. Hoskins, J. Stubbe, and S. E. Ealick. 2006. Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima describe a novel ATP binding protein superfamily. Biochemistry 4514880-14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bradley, P. Bork, P. Bucher, L. Cerutti, R. Copley, E. Courcelle, U. Das, R. Durbin, W. Fleischmann, J. Gough, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, J. McDowall, A. Mitchell, A. N. Nikolskaya, S. Orchard, M. Pagni, C. P. Ponting, E. Quevillon, J. Selengut, C. J. Sigrist, V. Silventoinen, D. J. Studholme, R. Vaughan, and C. H. Wu. 2005. InterPro, progress and status in 2005. Nucleic Acids Res. 33D201-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 35.Paschos, A., A. Bauer, A. Zimmermann, E. Zehelein, and A. Bock. 2002. HypF, a carbamoyl phosphate-converting enzyme involved in [NiFe] hydrogenase maturation. J. Biol. Chem. 27749945-49951. [DOI] [PubMed] [Google Scholar]
- 36.Paschos, A., R. S. Glass, and A. Bock. 2001. Carbamoylphosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett. 4889-12. [DOI] [PubMed] [Google Scholar]
- 37.Peck, H. D., Jr., and H. Gest. 1957. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J. Bacteriol. 73706-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna, N. T., G. I. Plunkett, F. R. Blattner, B. Mau, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 39.Pierik, A. J., W. Roseboom, R. P. Happe, K. A. Bagley, and S. P. Albracht. 1999. Carbon monoxide and cyanide as intrinsic ligands to iron in the active site of [NiFe]-hydrogenases. NiFe(CN)2CO, biology's way to activate H2. J. Biol. Chem. 2743331-3337. [DOI] [PubMed] [Google Scholar]
- 40.Reissmann, S., E. Hochleitner, H. Wang, A. Paschos, F. Lottspeich, R. S. Glass, and A. Bock. 2003. Taming of a poison: biosynthesis of the NiFe-hydrogenase cyanide ligands. Science 2991067-1070. [DOI] [PubMed] [Google Scholar]
- 41.Rosano, C., S. Zuccotti, M. Bucciantini, M. Stefani, G. Ramponi, and M. Bolognesi. 2002. Crystal structure and anion binding in the prokaryotic hydrogenase maturation factor HypF acylphosphatase-like domain. J. Mol. Biol. 321785-796. [DOI] [PubMed] [Google Scholar]
- 42.Roseboom, W., M. Blokesch, A. Bock, and S. P. Albracht. 2005. The biosynthetic routes for carbon monoxide and cyanide in the Ni-Fe active site of hydrogenases are different. FEBS Lett. 579469-472. [DOI] [PubMed] [Google Scholar]
- 43.Sawers, G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie van Leeuwenhoek 6657-88. [DOI] [PubMed] [Google Scholar]
- 44.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 581772-1779. [DOI] [PubMed] [Google Scholar]
- 45.Shomura, Y., H. Komori, N. Miyabe, M. Tomiyama, N. Shibata, and Y. Higuchi. 2007. Crystal structures of hydrogenase maturation protein HypE in the Apo and ATP-bound forms. J. Mol. Biol. 3721045-1054. [DOI] [PubMed] [Google Scholar]
- 46.Terwilliger, T. C. 2003. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 5945-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25455-501. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, S., R. Matsumi, T. Arai, H. Atomi, T. Imanaka, and K. Miki. 2007. Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling. Mol. Cell 2729-40. [DOI] [PubMed] [Google Scholar]
- 49.Zirngibl, C., W. Van Dongen, B. Schworer, R. Von Bunau, M. Richter, A. Klein, and R. K. Thauer. 1992. H2-forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron-sulfur clusters in methanogenic archaea. Eur. J. Biochem. 208511-520. [DOI] [PubMed] [Google Scholar]