Abstract
The genetic manipulation of marine double-stranded DNA (dsDNA) bacteriophage PM2 (Corticoviridae) has been limited so far. The isolation of an autonomously replicating DNA element of Pseudoalteromonas haloplanktis TAC125 and construction of a shuttle vector replicating in both Escherichia coli and Pseudoalteromonas enabled us to design a set of conjugative shuttle plasmids encoding tRNA suppressors for amber mutations. Using a host strain carrying a suppressor plasmid allows the introduction and analysis of nonsense mutations in PM2. Here, we describe the isolation and characterization of a suppressor-sensitive PM2 sus2 mutant deficient in the structural protein P10. To infect and replicate, PM2 delivers its 10-kbp genome across the cell envelopes of two gram-negative Pseudoalteromonas species. The events leading to the internalization of the circular supercoiled dsDNA are puzzling. In a poorly understood process that follows receptor recognition, the virion capsid disassembles and the internal membrane fuses with the host outer membrane. While beginning to unravel the mechanism of this process, we found that protein P10 plays an essential role in the host cell penetration.
The study of membrane-containing bacteriophages has produced notable knowledge of the assembly and structure of biological membranes and virus structures (18, 32). The availability of genetic tools has been valuable in the study of many bacterial viruses with a membrane (36, 37). The understanding of these sophisticated virus systems has relied on the isolation and analysis of phage mutants. A recent example of this is the X-ray crystallography structure of the membrane-containing phage PRD1, which was solved using a nonsense mutant particle, Sus539, lacking the receptor binding protein P2 (1, 8). Moreover, suppressor-sensitive mutants of PRD1 have been valuable in assigning functions to viral proteins. Recently, we initiated the structural characterization of bacteriophage PM2 as a new model system (2, 19; N. G. Abrescia, J. M. Grimes, H. M. Kivelä, R. E. Assenberg, G. E. Sutton, S. J. Butcher, J. K. H. Bamford, D. H. Bamford, and D. I. Stuart, submitted for publication). However, construction of PM2 mutants has been limited by the fact that there have been no host bacteria containing suppressor activity available for PM2.
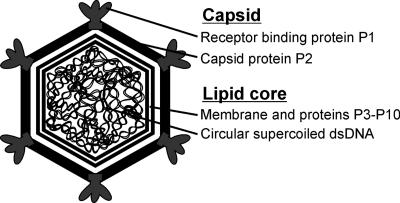
The double-stranded DNA (dsDNA) virus PM2 has at least two host bacteria: it replicates in the marine gram-negative bacterium Pseudoalteromonas espejiana BAL-31 and in Pseudoalteromonas sp. strain ER72M2 (14, 16, 25). PM2 exhibits distinctive features, such as a highly supercoiled, circular dsDNA genome (10 kbp) and a membrane residing under an icosahedral protein coat (for a review, see reference 3). PM2 is the type virus of the family Corticoviridae and still the only known isolate of this virus group. A schematic presentation of the PM2 virion organization is shown in Fig. 1. Recently, using a comparative genomic approach, putative corticoviral prophage elements were identified in chromosomes from aquatic bacteria of the phylum Proteobacteria (28). It appears that aquatic bacteria have been and still are in contact with numerous PM2-like viruses. The structure of the tailless phage PM2 has been studied using cryoelectron microscopy combined with three-dimensional image reconstruction (19) and, more recently, X-ray crystallography. The entire 45-MDa PM2 virion with the inner membrane and the circular DNA has been crystallized (Abrescia et al., submitted). The virion contains 10 virus-encoded protein species (Fig. 1). The internal membrane is covered by the protein capsid, composed of the major capsid protein and the receptor binding protein (24). The trimeric capsid protein P2, with a double β-barrel motif, is organized on an icosahedral pseudo-T=21 lattice (19; Abrescia et al., submitted). The P2 fold is similar to that observed in phage PRD1, the type virus of the family Tectiviridae, and a number of other large icosahedral dsDNA viruses, such as adenovirus, Paramecium bursaria chlorellavirus 1, and the archaeal virus Sulfolobus turreted icosahedral virus (5, 21, 38). At the vertices, the pentameric, trumpet-shaped P1 protein complexes build up the spike structures (19; Abrescia et al., submitted). The virus membrane is composed of phosphatidylglycerol (∼66%) and phosphatidylethanolamine (∼34%), deviating from the composition of the host's cytoplasmic membrane (phosphatidylglycerol, 25%; phosphatidylethanolamine, 75%) (31). The membrane lipids are selectively acquired from the host cytoplasmic membrane during virus assembly (13). Other structural proteins, P3 to P10, associate with the viral lipid bilayer, enclosing the genome and forming a lipid core particle (Fig. 1) (19, 24, 25).
FIG. 1.
Schematic organization of bacteriophage PM2. PM2 has an icosahedral protein coat consisting of 200 copies of capsid protein P2 trimers. The vertices contain the pentameric host recognition protein P1, forming the spikes. The capsid surrounds a lipid bilayer with associated viral proteins enclosing a highly supercoiled circular dsDNA genome (10,097 bp).
During the virus replication cycle, receptor binding on the host cell surface leads to genome entry. Bacteriophages have adopted various strategies to overcome the barrier of the bacterial cell envelope. Generally, the capsid of bacterial viruses remains outside the cell while the nucleic acid is delivered from the capsid via genome delivery machinery located at one of the vertices. This strategy is employed by all known tailed dsDNA phages and icosahedral single-stranded RNA phages, as well as dsDNA tectiviruses, e.g., PRD1 (for a review, see reference 40). In most membrane-containing viruses, viral and host membranes fuse during viral entry (for reviews, see references 9 and 22). The fusion event is triggered by receptor recognition or by lowering of pH in the endosome. The bacterial virus PRD1 also uses its internal membrane vesicle for genome delivery (17). The receptor recognition is a signal for conformational changes in the vertex structure, allowing the membrane to undergo a transformation from a spherical vesicle to a tubular structure, which protrudes though an opening of the vertex. Also in PM2, the internal membrane plays a key role in the viral DNA delivery, but the entry process leads to dissociation of the virion capsid, and the fusion-active membrane vesicle carrying the nucleic acid is exposed (23). Thus, the mechanism differs from the other described entry mechanism of dsDNA bacteriophages, resembling more closely that of the enveloped animal viruses.
PM2 infection starts with the specific recognition of the receptor by the distal C-terminal domain of the spike protein P1 (23). The virus receptor is still unidentified. The presence of calcium in the crystal structure of the receptor binding domain of P1 and its strong structural similarity to the family of bacterial calcium-dependent carbohydrate binding modules (CBM36) suggest a calcium-mediated carbohydrate binding activity for P1 (probably to the lipopolysaccharides) (Abrescia et al., submitted). The receptor binding is a signal that is transmitted to the capsid and leads to the release of the protein shell, which exposes the internal membrane surface. The evidence that the PM2 membrane fuses with the host cell has been obtained by measuring ion fluxes across the cell envelope during virus entry (23). Transient permeability of the host cell membrane to lipophilic compounds is induced, apparently due to the lack of lipopolysaccharides in the host outer membrane area contributed by the PM2 membrane. One peptidoglycan active protein has been shown to be associated with the lipid core (23, 44). The membrane-bound lytic enzyme presumably assists in the opening of the bacterial cell wall. Efflux of potassium ions from the cytoplasm concomitant with PM2 entry indicates that the cytoplasmic membrane is permeabilized due to the virus genome transfer via a channel (34).
Genetic manipulation of PM2 and its marine host bacteria belonging to the genus Pseudoalteromonas has been limited so far. By screening Antarctic bacteria for the presence of autonomously replicating extrachromosomal genetic elements, a cryptic plasmid, pMtBL, from a gram-negative bacterium, Pseudoalteromonas haloplanktis TAC125, was identified (46). The isolation of the autonomously replicating sequence from the plasmid and the development of a shuttle vector replicating in Escherichia coli and in Pseudoalteromonas (46) enabled us to set up genetic methods for PM2. In this study, we utilized synthetic suppressor tRNA genes in constructing a set of vectors and making them able to suppress amber mutations. We also describe a site-directed mutagenesis system for the PM2 virus. Using host bacteria carrying a suppressor plasmid, the mutant could be rescued.
MATERIALS AND METHODS
Bacteria and phage.
E. coli HMS174 (7) and strain HB101 (41) were used as cloning hosts, and S17-1(λpir) (43) was used as a donor for interspecies conjugation of the plasmids from E. coli to Pseudoalteromonas. E. coli cells were grown at 37°C in Luria-Bertani medium (41). Bacteriophage PM2 (14) was propagated on either Pseudoalteromonas sp. strain ER72M2 (25) or P. espejiana BAL-31 (13) at 28°C in SB broth (25).
Construction of the suppressor tRNA-carrying plasmids.
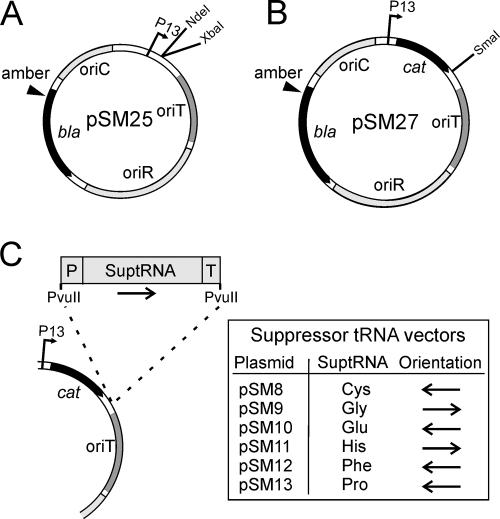
The plasmids and DNA constructions used in the study are listed in Table 1. DNA manipulations were done using standard molecular biology techniques (41). The shuttle vector cloneQ was transferred into ER72M2 and BAL-31 cells by transformation (47). Plasmid pPM13 is a shuttle vector replicating in both E. coli and P. haloplanktis TAC125 (12, 46, 48). It was used for expression of the E. coli-derived tRNA genes in the PM2 host bacteria. For selection of the suppressor activity, the codon for glutamine (Q37) of the ampicillin resistance gene (β-lactamase) of pPM13 was replaced by an amber stop codon (TAG) using the Quick Change site-directed mutagenesis method (Stratagene) and Pfu Ultra DNA polymerase (Stratagene). Specific complementary primers containing altered nucleotides were used to produce the amino acid change and to create a SpeI restriction site for screening. The mutated DNA was transformed into the HB101 strain, which contains a tRNA suppressor (supE44), and the clones were selected with ampicillin. The recovered plasmid was named pSM25 (Fig. 2A), and the mutation was verified by DNA sequencing. Another antibiotic resistance marker, the gene for chloramphenicol transacetylase, was cloned between the NdeI and XbaI restriction sites of pSM25. The gene was amplified by PCR using pSU18 as a template and specific primers carrying NdeI and XbaI restriction sites at the ends. The resulting construction was named pSM27 (Fig. 2B).
TABLE 1.
Plasmids used in the study
| Plasmid | Descriptiona | Selective markerb | Repliconc | Reference or source |
|---|---|---|---|---|
| pBR332 | Cloning vector | Tc, Ap | pMB1 | 41 |
| pSU18 | Cloning vector | Cm | p15A | 4 |
| pSU19 | Cloning vector | Cm | p15A | 4 |
| cloneQ | Shuttle vector | Ap | oriR, oriC | 46 |
| pPM13 | Shuttle vector | Ap | oriR, oriC | 12, 48 |
| pGF1 | pGFIB PvuIIΩsuptRNACys | Ap | pMB1 | Promega |
| pGF2 | pGFIB PvuIIΩsuptRNAGly | Ap | pMB1 | Promega |
| pGF3 | pGFIB PvuIIΩsuptRNAGlu | Ap | pMB1 | Promega |
| pGF4 | pGFIB PvuIIΩsuptRNAHis | Ap | pMB1 | Promega |
| pGF5 | pGFIB PvuIIΩsuptRNAPhe | Ap | pMB1 | Promega |
| pGF6 | pGFIB PvuIIΩsuptRNAPro | Ap | pMB1 | Promega |
| pHMK1 | pSU18Δ(BamHI-EcoRI)Ω(9877-2502) | Cm | p15A | This study |
| pHMK2 | pSU19Δ(EcoRI-XbaI)Ω(2502-4863) | Cm | p15A | This study |
| pHMK3 | pSU18Δ(XbaI-SacI)Ω(4863-7173) | Cm | p15A | This study |
| pHMK4 | pSU19Δ(SacI-BamHI)Ω(7173-9877) | Cm | p15A | This study |
| pJB140 | pBR322 PstIΩPM2 | Ap | pMB1 | This study |
| pSM8 | pSM27 SmaIΩsuptRNACys | Ap (amber), Cm | oriR, oriC | This study |
| pSM9 | pSM27 SmaIΩsuptRNAGly | Ap (amber), Cm | oriR, oriC | This study |
| pSM10 | pSM27 SmaIΩsuptRNAGlu | Ap (amber), Cm | oriR, oriC | This study |
| pSM11 | pSM27 SmaIΩsuptRNAHis | Ap (amber), Cm | oriR, oriC | This study |
| pSM12 | pSM27 SmaIΩsuptRNAPhe | Ap (amber), Cm | oriR, oriC | This study |
| pSM13 | pSM27 SmaIΩsuptRNAPro | Ap (amber), Cm | oriR, oriC | This study |
| pSM25 | pPM13 with amber mutation in bla (Q37) | Ap (amber) | oriR, oriC | This study |
| pSM27 | pSM25Δ(NdeI-XbaI)Ωcat | Ap (amber), Cm | oriR, oriC | This study |
| pSM76 | pSM124 with amber mutation in gene X (H47) | Cm | p15A | This study |
| pSM93 | pBR322 BamHIΩsus2 | Ap | pMB1 | This study |
| pSM96 | pBR322 BamHIΩPM2V1 | Ap | pMB1 | This study |
| pSM123 | pSU18Δ(BamHI-XbaI)Ω(9877-4863) | Cm | p15A | This study |
| pSM124 | pSU18Δ(XbaI-BamHI)Ω(4863-9877) | Cm | p15A | This study |
bla, β-lactamase; cat, chloramphenicol transacetylase. The numbers refer to the PM2 genome coordinates (GenBank accession no. AF155037); Ω, insertion; Δ, deletion.
Cm, chloramphenicol; Ap, ampicillin; Tc, tetracycline.
oriR, origin of replication in P. haloplanktis TAC125; oriC, pMB1, and p15A, origin of replication in E. coli.
FIG. 2.
Construction of the suppressor tRNA shuttle plasmids. (A) An amber stop codon (TAG) was introduced into the ampicillin resistance gene (bla) of the pPM13 shuttle vector to create the plasmid pSM25. P13, P. haloplanktis TAC125 P13 promoter; oriT, origin of conjugative transfer; oriR, origin of replication (P. haloplanktis); oriC, origin of replication (E. coli). (B) A selectable marker, the chloramphenicol resistance gene (cat) of pSU18, was amplified by PCR and cloned into the pSM25 plasmid between the NdeI and XbaI restriction sites to generate plasmid pSM27. (C) The PvuII fragments of amber suppressor plasmids (Interchange Amber Suppressor in vivo Mutagenesis System kit; Promega) carrying suppressor tRNA genes with a promoter and a transcription terminator were inserted into the SmaI restriction site of pSM27, generating the suppressor tRNA shuttle plasmids listed in the table. The arrows show the orientations of the cloned fragments. P, synthetic promoter derived from lipoprotein gene lpp; T, synthetic transcription terminator based on rRNA operon rrnC. See the text and Table 1 for details.
Suppressor tRNA genes were inserted into the SmaI site of pSM27. They were cut out with PvuII enzyme from amber suppressor plasmids (Interchange Amber Suppressor in vivo Mutagenesis System kit; Promega [26, 39]). The cloned PvuII fragments contained an E. coli lpp promoter, an rrnC transcription terminator, and one of the following amber suppressor tRNA genes: tRNACys, tRNAGly, tRNAGlu, tRNAHis, tRNAPhe, or tRNAPro, resulting in plasmids pSM8, pSM9, pSM10, pSM11, pSM12, and pSM13, respectively (Fig. 2C). The plasmids were transferred from E. coli S17-1(λpir) into Pseudoalteromonas sp. strain ER72M2 or P. espejiana BAL-31 cells by conjugation (45). The cells were grown on SB plates at 28°C; kanamycin was used for the counterselection of the donor bacterium, and chloramphenicol was used to select the plasmid-containing conjugants.
Cloning of the PM2 genome into a plasmid vector.
The PM2 DNA was isolated from the purified virus by sodium dodecyl sulfate (SDS) treatment, followed by three phenol extractions (35). It was linearized by cutting at a unique PstI site and ligated with a PstI-digested pBR332 vector. The construction was transformed into HB101 using tetracycline selection, and the resulting plasmid was named pJB140. The phage genome was digested from the vector by PstI endonuclease, ligated into a circular form, and transferred into ER72M2 cells by electroporation (30).
Site-directed mutagenesis of the PM2 genome.
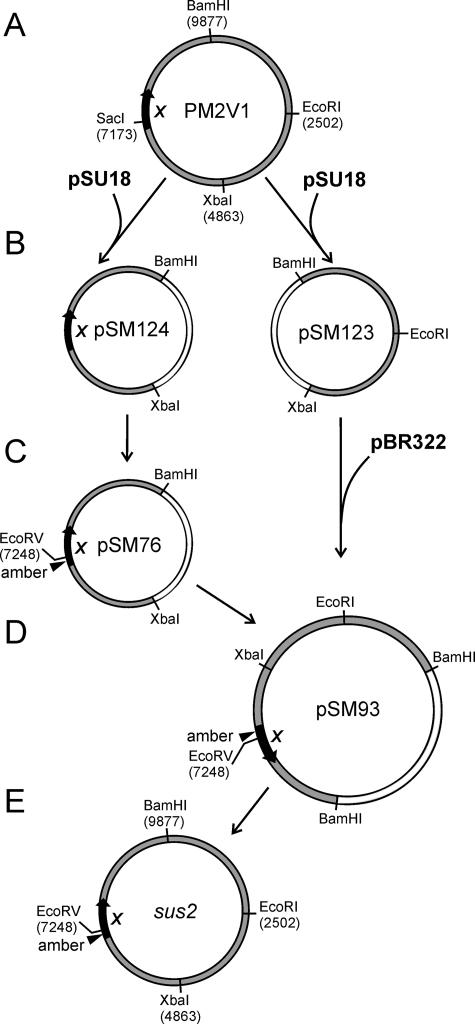
Four new unique restriction enzyme cleavage sites (EcoRI, XbaI, SacI, and BamHI at positions 2502, 4863, 7173, and 9877, respectively) were introduced to the PM2 genome by site-directed mutagenesis. The genome was amplified into four fragments, which were digested with the appropriate enzymes and ligated with a pSU18 or pSU19 vector. The resulting plasmids were named pHMK1 to pHMK4. The four genomic fragments were cut out from the plasmids, separated by agarose gel electrophoresis, mixed together, and ligated back to a circular form. The purified ligation mixture was transferred into ER72M2 by electroporation (30). Plaques were purified by three subsequent single-plaque purifications. The new cleavage sites in the genome were confirmed by digestions with the appropriate restriction enzymes. The altered phage genome was named PM2V1 (Fig. 3A).
FIG. 3.
Genetic engineering of the PM2 genome to construct the sus2 amber mutant. (A) Four unique restriction enzyme cleavage sites were introduced into the PM2 genome, resulting in PM2V1. The position of gene X is indicated. (B) PM2V1 was digested with BamHI and XbaI, and the two fragments were cloned into the corresponding sites of the pSU18 vector, generating plasmids pSM123 and pSM124. (C) pSM124 was used as a template to introduce a nonsense mutation in gene X (substituting a His47 codon for a TAG codon) and an EcoRV restriction cleavage site for screening, resulting in plasmid pSM76. (D) PM2-specific fragments were BamHI-XbaI double digested from the plasmids pSM123 and pSM76 and ligated with BamHI-digested pBR322 vector, generating plasmid pSM93. (E) The mutated genome amplified in E. coli was cut out with BamHI from pSM93 and ligated into circular form, resulting in the PM2 sus2 mutant. See the text and Table 1 for details. The numbers refer to the PM2 genome coordinates (GenBank accession no. AF155037).
PM2V1 was digested with BamHI and XbaI restriction enzymes and cloned as two separate fragments (5,059 bp and 5,020 bp) in pSU18 between BamHI and XbaI sites. The resulting plasmids were named pSM123 and pSM124, respectively (Fig. 3B). The PM2V1 fragments were cut out from the plasmids by BamHI and XbaI, isolated from the agarose gel, and ligated into BamHI-digested pBR322 as a three-fragment ligation. The resulting plasmid, pSM96, was amplified in E. coli HMS174 and selected with tetracycline. The PM2V1 genome was cut out from the plasmid (BamHI), ligated into a circular form, and transferred to ER72M2 cells by electroporation (30) to confirm the viability of the phage after DNA manipulations.
Plasmid pSM124 was used as a template to mutate the PM2 gene X. An amber mutation was introduced at position 7224 encoding a histidine residue (H47) of protein P10, and an EcoRV site was generated at position 7248 for screening of the mutation. The resulting plasmid, pSM76, was amplified in HMS174 (Fig. 3C). The PM2-specific fragment was cut out by BamHI and XbaI, isolated from the agarose gel, and ligated with the other half of the genome (a BamHI- and XbaI-digested wild-type [wt] PM2V1 fragment from pSM123) and the BamHI-digested pBR322. The resulting plasmid was named pSM93 (Fig. 3D). After amplification in HMS174, the plasmid was digested with BamHI to release the PM2V1 genome carrying the amber mutation in gene X. It was isolated from the agarose gel, ligated to a circular form, and transferred by electroporation into ER72M2 cells containing one of the suppressor plasmids.
Recovery of the mutant virus.
A single plaque was picked from the plate and resuspended in 500 μl of SB, diluted, and plated with soft agar on SB (chloramphenicol, 25 μg/ml) with the suppressor tRNA-containing host. The plates were grown overnight at 28°C. The soft agar from a semiconfluent plate was collected, 4 ml of SB (chloramphenicol, 25 μg/ml) was added, and the phage stock was grown for 1 h at 28°C. After centrifugation (in a microcentrifuge at 10,000 rpm for 10 min), the stock was plated on the suppressor tRNA-containing bacteria and wt host bacteria. Virus agar stocks were grown for 3 h, and they were prepared for each one-step growth of the virus.
Growth and purification of virus particles.
Wt and mutant virus particles were produced by infecting ER72M2 cells (7 × 108 CFU/ml) with fresh virus agar stock using a multiplicity of infection (MOI) of 10. After cell lysis and treatment of the lysate with DNase I (60 μg/ml; 30 min; +28°C), particles were precipitated with polyethylene glycol 6000 and purified by rate zonal centrifugation in sucrose as described previously (24). The particles were collected by differential centrifugation.
To separate the capsid and the lipid core, the purified particles resuspended in 20 mM MOPS (morpholinopropanesulfonic acid), pH 7.2, 100 mM NaCl buffer were disrupted by freezing and thawing them three times (−20°C). The soluble capsid components were separated from the lipid cores by differential centrifugation (Beckman Airfuge, A110 rotor; 29 lb/in2; 20 min). The lipid cores were washed once with the buffer and collected (as described above). The protein compositions were analyzed by SDS-polyacrylamide gel electrophoresis using 14% polyacrylamide-Tricine-SDS gels (42). Protein concentrations were determined with Coomassie brilliant blue by the Bradford assay using bovine serum albumin as a standard (6).
Phage adsorption assay.
The phage adsorption was assayed in SB broth at 28°C by infecting ER72M2 cells (3 × 108 CFU/ml) at an MOI of ∼0.3 (23). The adsorption mixture was incubated with shaking for 10 min. After the removal of the cells by centrifugation, the number of nonadsorbed particles was determined from the supernatant by plaque assay. To test Sus2 receptor binding, ER72M2 cells were first incubated with cell lysate containing Sus2 particles for 15 min to saturate the receptors and then infected with wt virus (MOI, ∼0.3). After a 10-min adsorption period, the number of nonadsorbed wt particles was determined by plaque assay. MOIs for Sus2 were calculated by assuming that the Sus2 lysate contained an amount of particles corresponding to that of the wt lysate (at least 2 × 1011 PFU/ml).
Measurements of ion fluxes.
Ion fluxes were measured as described previously (10, 23) with minor modifications. Briefly, ER72M2 cells were grown to a density of ∼6 × 108 CFU/ml, collected by centrifugation, and resuspended in a buffer (50 mM Tris-HCl, pH 8.0, 450 mM NaCl) to an ∼1:65 proportion of the original volume. The cells were diluted with the same buffer supplemented with 10 mM CaCl2 to the original cell density, incubated for 7 minutes with intensive aeration at 28°C, and infected with purified wt or Sus2 particles using an MOI of ∼20. Sus2 particles were added corresponding to the amount of wt PM2 (measured as 5 × 1012 PFU/mg of protein). The concentrations of tetraphenylphosphonium (TPP+) and K+ in the medium were measured in 5-ml thermostatic vessels with selective electrodes. The intracellular K+ content and the nonspecific binding of TPP+ were determined by adding gramicidin D (GD) (5 μg/ml) and polymyxin B sulfate (25 μg/ml). The characteristics of the TPP+-selective electrode have been described previously (10). The K+-selective electrode (Orion model 93-19) was from Thermo Inc. The Ag/AgCl reference electrodes (Thermo Inc.; Orion model 9001) were indirectly connected to the measuring vessels through agar salt bridges.
RESULTS
The replicon of the plasmid from the Antarctic gram-negative bacterium P. haloplanktis TAC125 is functional in the PM2 host bacteria BAL-31 and ER72M2.
No plasmids replicating in P. espejiana BAL-31 or Pseudoalteromonas sp. strain ER72M2 had been previously identified. By testing the sensitivities of the species to common antibiotics, it was revealed that BAL-31 and ER72M2 were resistant to kanamycin (25 to 50 μg/ml) but sensitive to ampicillin (10 to 150 μg/ml), chloramphenicol (15 to 25 μg/ml), and tetracycline (10 to 20 μg/ml). The isolation of the autonomously replicating element of the multicopy plasmid pMtBL from the phychrophilic gram-negative P. haloplanktis TAC125 strain and development of a shuttle vector system (46) widened the options for setting up genetic methods to manipulate PM2. The constructed shuttle vector, cloneQ, carrying the autonomously replicating element from P. haloplanktis, the origin of replication from E. coli, and the origin of conjugative transfer (oriT) was able to replicate in two cold-adapted gram-negative bacterial species, as well as in E. coli (46). To determine whether it could replicate in PM2 host bacteria, cloneQ was transferred into ER72M2 and BAL-31 cells by transformation. The cells were grown in SB medium supplemented with ampicillin. Several ampicillin-resistant colonies appeared. CloneQ was isolated from recombinant strains using a standard plasmid isolation alkali-lysis method. Later, it was demonstrated that cloneQ and its derivatives could be transferred from E. coli S17-1(λpir) to ER72M2 and BAL-31 by conjugation. The presence of the vector in the host bacteria did not interfere with virus replication. The sensitivity of P. haloplanktis TAC125 cells to the phage was also tested using a plaque assay. It appeared that PM2 did not form plaques on P. haloplanktis TAC125.
The shuttle vector-encoded amber suppressor tRNA genes are active in E. coli.
In an attempt to produce PM2 viruses carrying a nonsense mutation in an essential gene, our first trial was to construct host bacteria with suppressor activity based on a plasmid replicon. A cloneQ derivative shuttle vector, pPM13, replicates in E. coli, ER72M2, and BAL-31 (Table 1) (12, 48). To screen the suppressor activity, we first constructed E. coli strains carrying a plasmid with both a suppressor tRNA gene and a nonsense mutation in a selectable marker. Using site-directed mutagenesis, an amber stop codon (TAG) was introduced into the pPM13 shuttle vector. The amber codon replaced the codon for glutamine (Q37) in the ampicillin resistance gene (bla) (Fig. 2A). The resulting plasmid, assigned the name pSM25, was transformed into the suppressor strain HB101 (supE44) and strain HMS174, which has no suppressor activity. The clones were selected with ampicillin. Only the colonies of HB101 survived, which indicates that the altered selectable marker conferred bacterial resistance to ampicillin only upon suppression of the amber stop codon. Another selectable marker was introduced into the plasmid pSM25 by amplifying the gene for chloramphenicol transacetylase (cat) of plasmid pSU18 and ligating it between NdeI and XbaI restriction sites. The resulting plasmid, pSM27, was transformed into HB101, and the growth of the recombinant colonies was selected for on chloramphenicol-containing plates (Table 1 and Fig. 2B). The cat gene in pSM27 was also functional in PM2 host bacteria, ER72M2 and BAL-31. However, when selected with ampicillin, the growth of ER72M2 and BAL-31 was inhibited, which indicates that they are not naturally suppressor strains.
Synthetic amber suppressor tRNA genes (suptRNA genes from the Interchange Amber Suppressor in vivo Mutagenesis System kit; Promega) (26, 39) were utilized for the construction of suppressor vectors replicating in both Pseudoalteromonas and E. coli. The tRNA genes are transcribed constitutively from the llp promoter included in the DNA construction. Six different suptRNA genes derived from E. coli (PvuII fragments from the plasmids in the Interchange kit) were cloned into the SmaI site of pSM27. The resulting suptRNA shuttle vectors, pSM8 to pSM13, contained the amber suppressor tRNA genes for tRNACys, tRNAGly, tRNAGlu, tRNAHis, tRNAPhe, or tRNAPro, respectively (Table 1 and Fig. 2C). The constructs were amplified one at a time in the strain HB101 (supE) containing suppressor activity and transferred into the nonsuppressor strain HMS174 to test the functionality of the constructions in E. coli. Ampicillin-resistant colonies of HMS174 appeared, showing that the plasmid-encoded amber suptRNA genes were active in E. coli due to the production of ampicillin-hydrolyzing β-lactamase. Sequencing of the resulting plasmids confirmed the orientation of the cloned fragments. The PvuII fragments of the amber suppressor tRNAs for cysteine, glutamic acid, phenylalanine, and proline were oriented in the opposite direction to the chloramphenicol transacetylase gene. Histidine- and glycine-inserting suptRNA genes were transcribed in the same direction as the cat gene (Fig. 2C).
Plasmid-encoded synthetic E. coli-derived suppressor tRNA genes are active in pseudoalteromonads.
After finding that suptRNA shuttle vector constructs were functional in E. coli, the constructs were tested in Pseudoalteromonas strains. Since ER72M2 and BAL-31 do not carry indigenous suppressors, ampicillin-resistant colonies could appear only if the suptRNA genes in the plasmid replicon could suppress the nonsense mutation of the bla gene. The suppressor plasmids pSM9 to pSM13 were transferred from strain S17-1(λpir) into ER72M2 and BAL-31 cells by conjugation. The conjugants were selected with chloramphenicol (25 μg/ml) and kanamycin (50 μg/ml). To test the tRNA-mediated suppression, the strains were grown on SB plates supplemented with chloramphenicol (25 μg/ml) and ampicillin (50 μg/ml). Plasmids pSM9 (suptRNAGly), pSM11 (suptRNAHis), pSM12 (suptRNAPhe), and pSM13 (suptRNAPro) were functional in ER72M2 (Table 2). In BAL-31, pSM8, encoding the suptRNA for cysteine, was also active. In both cells, plasmid-encoded suptRNAGlu (pSM10) did not suppress the stop codon in the ampicillin resistance gene. We compared the nonsense suppressor efficiencies by growing the clones with increasing amounts of ampicillin (Table 2). The plasmid-encoded suppressor tRNAs for glycine, histidine, phenylalanine, and proline restored the activity of β-lactamase, which hydrolyzed ampicillin even at 800 μg/ml (Table 2). All ER72M2 and BAL-31 strains containing suppressor activity were sensitive to PM2 at the same level as the wt host strain, as determined by plaque assay.
TABLE 2.
Abilities of ER72M2 and BAL-31 suppressor tRNA strains to suppress the amber mutation (Gln37amber) in the ampicillin resistance genea
| Plasmid | OD600
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ER72M2 with ampicillin (μg/ml)
|
BAL-31 with ampicillin (μg/ml)
|
|||||||
| 50 | 200 | 400 | 800 | 50 | 200 | 400 | 800 | |
| pSM8 (suptRNACys) | 2.0 | 2.2 | 1.6 | 1.8 | ||||
| pSM9 (suptRNAGly) | 2.7 | 2.7 | 2.6 | 2.6 | 2.4 | 2.4 | 2.2 | 1.4 |
| pSM10 (suptRNAGlu) | ||||||||
| pSM11 (suptRNAHis) | 3.4 | 3.0 | 2.6 | 2.7 | 4.0 | 3.9 | 3.8 | 3.6 |
| pSM12 (suptRNAPhe) | 3.6 | 3.3 | 3.1 | 3.4 | 4.4 | 4.0 | 3.6 | 3.0 |
| pSM13 (suptRNAPro) | 2.7 | 2.6 | 2.3 | 2.4 | 3.4 | 3.4 | 3.3 | 3.2 |
The strains harboring one of the plasmids were grown in liquid SB medium supplemented with 25 μg/ml of chloramphenicol and the indicated amount of ampicillin for 24 h at +28°C. The optical density (OD) of the cultures at saturation was recorded at 600 nm.
PM2 genome can be manipulated in E. coli.
To test whether the PM2 genome can be manipulated in vitro by DNA techniques, amplified in E. coli, and transferred back to the host bacterium, we cloned the phage genome into the pBR322 vector. After isolation from E. coli strain HB101, the PM2 genome was digested from the construction, ligated into a circular form, and transferred by electroporation into ER72M2, which resulted in plaques, showing that the virus was viable. The tight supercoiled structure of the circular PM2 genomic DNA (15) is not a prerequisite for electroporation and transfection.
Construction of a PM2 nonsense mutant.
To exploit the usefulness of the standard recombinant DNA techniques in E. coli and to manipulate the genome more easily, we inserted four unique restriction enzyme cleavage sites into the PM2 genome by site-directed mutagenesis. The genome was amplified in four fragments. The new cleavage sites were designed for endonucleases EcoRI, XbaI, SacI, and BamHI cutting the genome at positions 2502, 4863, 7173, and, 9877, respectively. The circular PM2 genome was used as a template, and specific oligonucleotides with the desired restriction enzyme sites were used as primers. Introduction of new restriction enzyme recognition sites in the PM2 genome resulted in four amino acid changes in the protein-coding regions of gene IX (V84L), gene X (V34L), and gene XVII (L34I and I35L). The fragments were cloned in pSU18 or pSU19 vectors for amplification (Table 1, plasmids pHMK1 to pHMK4). The phage-specific fragments from pHMK1 to pHMK4 were isolated and ligated back into circular form. The reconstructed genomes were transferred into ER72M2 by electroporation. The modified phage was designated PM2V1 (Fig. 3A). The titer of PM2V1 and the one-step growth curve of PM2V1 infection were the same as those of the wt phage (data not shown).
For site-directed mutagenesis, the PM2V1 genome was cloned as two separate fragments into the (BamHI-XbaI and XbaI-BamHI) pSU18 vector, creating plasmids pSM123 and pSM124 (Fig. 3B and Table 1). Plasmid pSM124 was used as a template, and an amber mutation was generated in gene X at the position 7224 by PCR using oligonucleotides with the desired mutation. The resulting plasmid, pSM76, had an amber stop codon, TAG, replacing the histidine residue (H47) in the beginning of the coding region of protein P10 (Fig. 3C). In order to construct and amplify the entire mutated virus genome in E. coli, the virus-specific fragments from the plasmids pSM123 and pSM76 were cut, isolated, and cloned together with the BamHI-digested pBR322 vector, resulting in the construct pSM93 (Fig. 3D and Table 1). The digestion of the pSM93 plasmid with BamHI restriction enzyme released the full-length mutated virus genome, which was circularized into its native form (Fig. 3E) to be tested in suppressor Pseudoalteromonas host cells.
The amber mutation H47 in PM2 gene X can be suppressed by suptRNAPro and suptRNAHis.
To recover nonsense mutant phages, the mutated genome was transferred by electroporation into ER72M2 hosts expressing plasmid-encoded tRNA suppressors. The resulting plaques were picked up and tested directly by plating them on the corresponding suppressor host and wt host. One turbid plaque smaller than those of the wt virus was detected on the suppressor strain ER72M2(pSM13). The plaque was purified by three subsequent single plaque isolations using the suppressor strain. This PM2 nonsense mutant having a mutation in the gene X was assigned as sus2.
The abilities of the different plasmid-encoded suppressor tRNAs to suppress the nonsense mutation in sus2 were tested by plating the mutant virus on different suppressor hosts. The mutant phenotype of sus2 could be rescued when the suptRNAPro or suptRNAHis gene was provided in trans in a plasmid replicon (pSM13 or pSM11, respectively). The determined titers of the mutant virus grown as agar stocks were ∼5 × 1011 and ∼1 × 1010 PFU/ml on the suppressor ER72M2(pSM13) and ER72M2(pSM11) strains, respectively, and ∼5 × 103 PFU/ml on the wt host.
Protein P10 is not crucial for particle formation.
To analyze the phenotypic defects resulting from the mutation in gene X, ER72M2 cells were infected using an MOI of 10 with Sus2 grown on a suppressor strain, ER72M2(pSM13). The timing of the host cell lysis in the Sus2 infection followed that of the wt (data not shown). The released particles were purified by rate zonal sucrose centrifugation. After centrifugation through a 5 to 20% sucrose gradient, one light-scattering zone was detected similar to that seen in the wt particle purification. However, the infectivity of the mutant particles was reduced by ∼5 orders of magnitude. The specific infectivity of the purified mutant particles was ∼3.2 × 107 PFU/mg of protein. By comparing the number to the specific infectivity of the purified wt particles (∼2.8 × 1012 PFU/mg of protein), we can conclude that the Sus2 particles were noninfectious.
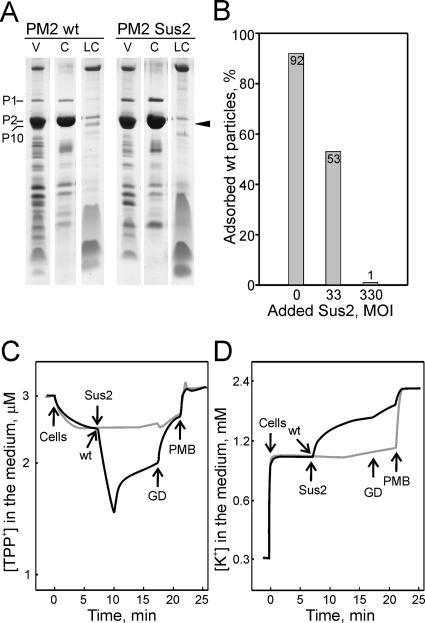
The protein compositions of the purified mutant and wt particles were determined by SDS-polyacrylamide gel electrophoresis (PAGE), followed by Coomassie blue staining. The protein P10 (29 kDa) is associated with the lipid core particle (Fig. 1), and its mobility in SDS-PAGE is the same as that of the abundant capsid protein P2 (30.2 kDa). To visualize protein P10, the phage membrane was separated from the capsid components. The purified mutant particles were disrupted by freezing and thawing them and by addition of EGTA, after which the lipid cores were separated from the soluble capsomers by differential centrifugation. Both phages showed the normal protein composition, except that the lipid core-associated protein P10 was absent from the mutant Sus2 particles (Fig. 4A). This shows that protein P10 is not essential for PM2 particle formation.
FIG. 4.
Phenotype of the PM2 sus2 mutant. (A) Analysis of the protein content of purified wt PM2 and mutant Sus2 virus particles by Tricine-SDS-PAGE and Coomassie blue staining. V, purified virus particles; C, released capsid components; LC, lipid core. The position of P10 is depicted with an arrowhead. The positions of PM2 proteins are indicated. (B) Phage adsorption to ER72M2 cells in the presence of Sus2 particles. The percentages of adsorbed wt particles after a 10-min adsorption period are shown. (C and D) Wt PM2 and mutant Sus2 induced changes in the envelope permeability of ER72M2 cells to TPP+ (C) and K+ (D) ions. The measurements were carried out at 28°C with a cell concentration of ∼6 × 108 CFU/ml. ER72M2 cells were infected with either wt (black curve) or Sus2 (gray curve) virus particles using an MOI of ∼20. GD and polymyxin B sulfate (PMB) were added to a final concentration of 5 μg/ml and 25 μg/ml, respectively.
Protein P10 is needed for host cell penetration but not for receptor recognition.
Our results showed that PM2 assembles normally in the absence of protein P10, but the resulting particles are noninfectious, which suggests that P10 plays a role in the infection process. Binding of the virions was studied by determining the number of nonadsorbed particles scored by plaque assay. When ER72M2 cells were infected with wt particles using a low MOI (0.3), at 10 min postinfection, over 90% of the virions were adsorbed to the host cell surface. Since Sus2 particles cannot produce progeny viruses on a solid medium, the number of nonadsorbed particles cannot be determined by titration assay. When the adsorption assay was used, the wt phage binding was challenged by Sus2 particles. Cells were first mixed with Sus2 particles (particle extract) or with an identical volume of medium (control) prior to the infection with wt PM2 (MOI = 0.3). If Sus2 particles bound to the host receptor, the number of adsorbed wt PM2 would be lower than in the control infection. Indeed, addition of Sus2 particles (MOI, ∼300) completely inhibited the adsorption of wt particles (Fig. 4B). Half of the particles were adsorbed in the presence of the Sus2 particle extract using an MOI of ∼30. Since Sus2 particles had an inhibitory effect on wt virus receptor binding, it is clear that protein P10 does not interfere with the initial step of infection, which is the recognition of the receptor.
Since the receptor recognition of the P10-deficient particles was unaltered, the following entry steps are most probably compromised. After binding to the receptor, the phage genome has to pass the cell envelope to reach the cytoplasm. In many cases, this event is associated with alterations in the membrane potential (ΔΨ) and efflux of potassium ions (34). The distribution of TPP+ between the cell cytosol and the surrounding medium is membrane voltage (ΔΨ) dependent and is therefore used to follow the voltage of the cytoplasmic membrane, while the efflux of intracellular K+ signifies the increase in the cytoplasmic membrane permeability. On the other hand, the low permeability of the outer membrane, due to the lipopolysaccharide layer, to lipophilic molecules, such as TPP+ and the ionophoric antibiotic GD, enabled us to follow the virus entry-specific effects on the outer membrane during virus penetration. For instance, a fusion between the phage membrane and the host outer membrane significantly increases the permeability of the latter to lipophilic cations and bulky ionophoric antibiotics (11, 23, 29). By measuring the ion fluxes during PM2 phage infection it has been shown that wt particles induce transient permeabilization of the outer membrane, which can be interpreted as a fusion event (23).
The effects of Sus2 particles on the envelope permeability of ER72M2 cells were studied by monitoring TPP+ accumulation in the cell cytoplasm and measuring the efflux of intracellular potassium ions. When ER72M2 cells were infected with wt PM2, an immediate drop in the extracellular TPP+ concentration was observed (Fig. 4C). This signified the increase of permeability in the outer membrane due to the fusion of the viral membrane with the outer membrane of the host, as proposed previously (23, 29). The fast accumulation of TPP+ was followed by the leakage of intracellular TPP+ into the medium, which indicated a decrease in ΔΨ. This is consistent with the efflux of intracellular K+ (Fig. 4D), which started shortly after infection with the wt PM2. However, when Sus2 particles were added to the cell suspension, no such effects on the cell envelope permeability were observed (Fig. 4C and D), which resembles the pattern of TPP+ and K+ fluxes in noninfected ER72M2 cells (reference 23 and data not shown). Furthermore, unlike the wt PM2, Sus2 particles were not able to increase outer membrane permeability for the translocation of GD (Fig. 4C and D and data not shown).
DISCUSSION
We applied well-established molecular-cloning techniques for E. coli to the analysis of corticovirus PM2 infecting marine Pseudoalteromonas to generate a system to construct mutant viruses. We describe a convenient system for inserting nonsense mutations into the genome of the dsDNA virus PM2, using site-directed mutagenesis. In addition to the characterization of the mutant phenotype, we present here the rescue of the mutant by plasmid-encoded synthetic amber suppressor tRNA. The development of methods for genetic analysis of bacteriophage PM2 is crucial for increasing our understanding of the structural characteristics of the family Corticoviridae (Fig. 1 is a schematic presentation of the virion organization). PM2 is a model system well-suited to the analysis of membrane-aided packaging and release of circular supercoiled dsDNA. Without methods for analyzing the functions of the virion components and for identifying which genes are involved in different stages of the virus replication cycle, the structural observations will remain static. Genetic engineering of the viral genome, producing mutants with different phenotypes, is the most direct method for addressing these questions.
The genetic engineering of Pseudoalteromonas has been primitive due to the lack of tools commonly used in modern molecular microbiology. Most probably due to unique replication properties, the only extrachromosomal replicon that has been shown to replicate in P. espejiana BAL-31 and Pseudoalteromonas sp. strain ER72M2 is the phage PM2 genome. No indigenous plasmids are known. The recent development of the shuttle vector system in the cold-adapted gram-negative bacterium P. haloplanktis TAC125 (46) gave us the opportunity to use bacterial genetics in PM2 host bacteria. It revealed that the shuttle vector cloneQ and its derivatives also replicate in both PM2 host species, in addition to the reported psychrophilic species of Pseudoalteromonas, Psychrobacter, and E. coli (46). No natural suppressor host bacteria are available for PM2. Therefore, we constructed strains with suppressor activity by genetics, exploiting the shuttle vector replicon. We refined the shuttle vector for the rescue of virus nonsense mutants with the help of synthetic suppressor tRNA genes. Moreover, the selectable marker bla gene encoding β-lactamase was mutated by inserting a nonsense mutation (Fig. 2). Employing the bla nonsense mutation for detection of suppressor activity, the constructed plasmids were first tested in E. coli and then in the Pseudoalteromonas strains BAL-31 and ER72M2, which showed that the constructs were active. Suppression of the amber stop codon in the bla gene by the suppressor strains resulted in the production of full-length β-lactamase, actively hydrolyzing ampicillin. The plasmids conferred resistance to ampicillin even with as high a concentration as 800 μg/ml (Table 2).
To demonstrate that Pseudoalteromonas suppressor strains can be utilized for rescuing viral mutants, an amber mutation was introduced into the PM2 genome (Fig. 3). The viral sus2 mutant generated by introducing a site-specific in-frame amber mutation into gene X (substituting a His47 codon for a TAG codon) could be rescued when the suptRNAPro or suptRNAHis gene was provided in trans in a plasmid replicon with a significant difference in plating efficiency on the suppressor strains (∼1 × 1010 to 5 × 1011 PFU/ml) and on the wt host (∼5 × 103 PFU/ml). In general, the efficiency of suppression is site specific, resulting in different levels of suppression that were detected by using different synthetic suppressor tRNAs (Table 2). The amber codon in bla replacing the glutamine tolerated substitution with all tested amino acids (Cys, Gly, Glu, His, Phe, and Pro) in E. coli. Negatively charged glutamic acid was not tolerated in that position in BAL-31 and ER72M2. Also, cysteine-inserting suptRNA did not restore the activity of β-lactamase in ER72M2. In these cases, however, ampicillin resistance, via suppression, was the selective pressure.
Our previous results suggested that the infection mechanism of PM2 is different from that described for other dsDNA bacteriophages (23). Despite considerable understanding of the general details of linear dsDNA delivery (for reviews, see references 20 and 33), we lack fundamental insights into the steps of a circular dsDNA genome penetration across the bacterial envelope. Here, we describe a new component, protein P10, essential for the phage DNA delivery. The absence of protein P10 did not affect DNA packaging or particle assembly. The replication cycle of sus2 was quite similar to that of the wt infection. Nonetheless, the mutation resulted in the production of noninfectious virus particles. The receptor binding domain of PM2 has been functionally defined as the distal carboxy-terminal domain of spike protein P1 (19). This fragment competed the binding of virions on the host cell surface consistently (Abrescia et al., submitted). The abundant receptor is uncharacterized, but recently, the X-ray structure of the receptor binding domain and its similarity to a marine bacterial carbohydrate binding protein, Aga16B-CBM6, provided evidence that the cell surface sugar moieties might serve as a receptor for PM2 (Abrescia et al., submitted). The complete competition of wt PM2 receptor binding by Sus2 particles demonstrated that the lack of protein P10 does not interfere with receptor binding (Fig. 4B).
Receptor binding is thought to trigger conformational changes within the P1 spike complex that are transmitted to the capsid. This leads to the uncoating of the virion and the exposure of the lipid core. Interactions between the membrane and the capsid are coordinated through 120 dimers of protein P3 and 60 molecules of protein P6 (Abrescia et al., submitted). The calcium ions found in the base of coat protein P2 trimers play a role in the stability of these interactions, since upon calcium depletion, the metastable capsid disassembles, exposing the hydrophobic lipid core (24; Abrescia et al., submitted). This mimics the events triggered by receptor recognition. Contrary to the characteristics of a successful phage entry injecting a linear dsDNA genome, after PM2 DNA delivery, no empty capsids are seen on the infected cell surfaces (23). It has been suggested that the lipid core fuses with the host outer membrane and mediates the translocation of the circular genome into the host cytoplasm by a mechanism reminiscent of those of certain animal viruses (23). The conformational change in the virus structure and disassembly promotes the fusion activity exposure.
We analyzed the Sus2 particle entry by evaluating the permeability of the host envelope. In the case of wt PM2, fusion with the host outer membrane results in a transient permeability to lipophilic ions, followed by events on the cytoplasmic membrane seen as a decrease in the membrane potential (23). The Sus2 particles did not induce an efflux of intracellular potassium ions or accumulation of lipophilic TPP+ indicator ions in the cell cytoplasm, which suggests that the outer membrane remained intact (Fig. 4C and D). Mutant Sus2 appeared to have normal receptor binding but no detectable membrane fusion activity. Every enveloped virus fuses its membrane with a host cell membrane. Typically, viral surface transmembrane glycoproteins mediate fusion. Common to all fusion mechanisms is the fact that in response to an activating trigger, the fusion protein converts to an extended structure, exposing its fusion peptide, which mediates insertion into the target membrane. Protein P10 is invisible in the electron density map of the crystallized virion (Abrescia et al., submitted). Thus, it is not icosahedrally organized. However, biochemical analyses of the virion dissociation products have shown that protein P10 is associated with the viral membrane (19). Also, the amino acid sequence of P10 shows characteristics of an integral membrane protein. The predicted transmembrane region is in its carboxy terminus (TMHMM prediction) (27). Therefore, the amino terminus would face outward on the lipid core surface. These results suggest that protein P10 acts early in the fusion pathway, perhaps involving the initial interaction of the virus membrane with the target membrane.
Suppression strains often grow slowly because of misreading elsewhere. Additionally, nonsense mutations might cause polar effects on downstream genes. Gene X is immediately upstream of gene I, encoding the receptor binding protein P1. According to Coomassie blue-stained SDS-PAGE gels, protein P1 was incorporated into the Sus2 particles in normal amounts (Fig. 4A). This indicates that the amber codon in gene X has no polar effects on downstream genes, showing that the entry-incompetent phenotype of sus2 is not caused by the lack of the receptor binding protein P1 or other gene products expressed downstream of gene X, proteins P5 and P6.
In conclusion, we have established a shuttle vector-based system for rescuing PM2 nonsense mutants in marine Pseudoalteromonas by making use of a set of synthetic suppressor tRNAs. The versatility of the system was demonstrated by the virus sus2 nonsense mutant with a specific amber mutation in the X gene. The results confirmed the essential role of the lipid core-associated protein P10 in virus entry. Together, these findings extended the results of earlier studies of PM2 DNA delivery, but clearly, many questions will need to be addressed concerning the mechanism of circular dsDNA genome delivery and penetration across the gram-negative cell envelope. However, considering that PM2 infection is supposed to depend on unidentified viral proteins, several candidates for promoting DNA delivery are present that associate with the viral membrane (Fig. 1). Further studies will reveal to what extent one can utilize the potential of generating virus mutants.
Acknowledgments
This work was supported by the Finnish Centre of Excellence Program of the Academy of Finland (1213467, 2006 to 2011) and by Academy of Finland Research Grant 1213132 (to J.K.H.B).
Gennaro Marino and Dennis Bamford are thanked for fruitful discussions throughout the project. We thank P. Papponen for his skillful technical assistance and R. Hankkio for cloning.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 43268-74. [DOI] [PubMed] [Google Scholar]
- 2.Abrescia, N. G., H. M. Kivelä, J. M. Grimes, J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2005. Preliminary crystallographic analysis of the major capsid protein P2 of the lipid-containing bacteriophage PM2. Acta Crystallogr. F 61762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford, D. H., and J. K. H. Bamford. 2006. Lipid-containing bacteriophage PM2, the type organism of Corticoviridae, p. 171-174. In R. Calender (ed.), The bacteriophages. Oxford University Press, Oxford, United Kingdom.
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 10275-78. [DOI] [PubMed] [Google Scholar]
- 5.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16673-685. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 752276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockburn, J. J., N. G. Abrescia, J. M. Grimes, G. C. Sutton, J. M. Diprose, J. M. Benevides, G. J. Thomas, Jr., J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432122-125. [DOI] [PubMed] [Google Scholar]
- 9.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4309-319. [DOI] [PubMed] [Google Scholar]
- 10.Daugelavièius, R., J. K. H. Bamford, and D. H. Bamford. 1997. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 1795203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugelavičius, R., V. Cvirkaite, A. Gaidelyte, E. Bakiene, R. Gabrenaite-Verkhovskaya, and D. H. Bamford. 2005. Penetration of enveloped double-stranded RNA bacteriophages phi13 and phi6 into Pseudomonas syringae cells. J. Virol. 795017-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duilio, A., S. Madonna, M. L. Tutino, M. Pirozzi, G. Sannia, and G. Marino. 2004. Promoters from a cold-adapted bacterium: definition of a consensus motif and molecular characterization of UP regulative elements. Extremophiles 8125-132. [DOI] [PubMed] [Google Scholar]
- 13.Espejo, R. T., and E. S. Canelo. 1968. Properties and characterization of the host bacterium of bacteriophage PM2. J. Bacteriol. 951887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espejo, R. T., and E. S. Canelo. 1968. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34738-747. [DOI] [PubMed] [Google Scholar]
- 15.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1969. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc. Natl. Acad. Sci. USA 631164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45755-761. [DOI] [PubMed] [Google Scholar]
- 17.Grahn, A., M., R. Daugelavièius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 461199-1209. [DOI] [PubMed] [Google Scholar]
- 18.Huiskonen, J. T., and S. J. Butcher. 2007. Membrane-containing viruses with icosahedrally symmetric capsids. Curr. Opin. Struct. Biol. 17229-236. [DOI] [PubMed] [Google Scholar]
- 19.Huiskonen, J. T., H. M. Kivelä, D. H. Bamford, and S. J. Butcher. 2004. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 11850-856. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. E., and W. Chiu. 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17237-243. [DOI] [PubMed] [Google Scholar]
- 21.Khayat, R., L. Tang, E. T. Larson, C. M. Lawrence, M. Young, and J. E. Johnson. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 10218944-18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kielian, M., and F. A. Rey. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 467-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kivelä, H. M., R. Daugelavièius, R. H. Hankkio, J. K. Bamford, and D. H. Bamford. 2004. Penetration of membrane-containing double-stranded-DNA bacteriophage PM2 into Pseudoalteromonas hosts. J. Bacteriol. 1865342-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kivelä, H. M., N. Kalkkinen, and D. H. Bamford. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 768169-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262364-374. [DOI] [PubMed] [Google Scholar]
- 26.Kleina, L. G., J. M. Masson, J. Normanly, J. Abelson, and J. H. Miller. 1990. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J. Mol. Biol. 213705-717. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 28.Krupoviè, M., and D. H. Bamford. 2007. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genom. 8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupoviè, M., R. Daugelavièius, and D. H. Bamford. 2007. A novel lysis system in PM2, a lipid-containing marine double-stranded DNA bacteriophage. Mol. Microbiol. 641635-1648. [DOI] [PubMed] [Google Scholar]
- 30.Krupoviè, M., H. Vilen, J. K. H. Bamford, H. M. Kivelä, J. M. Aalto, H. Savilahti, and D. H. Bamford. 2006. Genome characterization of lipid-containing marine bacteriophage PM2 by transposon insertion mutagenesis. J. Virol. 809270-9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurinavièius, S., D. H. Bamford, and P. Somerharju. 2007. Transbilayer distribution of phospholipids in bacteriophage membranes. Biochim. Biophys. Acta 17682568-2577. [DOI] [PubMed] [Google Scholar]
- 32.Laurinmäki, P. A., J. T. Huiskonen, D. H. Bamford, and S. J. Butcher. 2005. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 131819-1828. [DOI] [PubMed] [Google Scholar]
- 33.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154283-287. [DOI] [PubMed] [Google Scholar]
- 34.Letellier, L., L. Plancon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150499-505. [DOI] [PubMed] [Google Scholar]
- 35.Männistö, R. H., H. M. Kivelä, L. Paulin, D. H. Bamford, and J. K. H. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262355-363. [DOI] [PubMed] [Google Scholar]
- 36.Mindich, L., D. Bamford, C. Goldthwaite, M. Laverty, and G. Mackenzie. 1982. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J. Virol. 441013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mindich, L., J. Cohen, and M. Weisburd. 1976. Isolation of nonsense suppressor mutants in Pseudomonas. J. Bacteriol. 126177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 9914758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Normanly, J., L. G. Kleina, J. M. Masson, J. Abelson, and J. H. Miller. 1990. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 213719-726. [DOI] [PubMed] [Google Scholar]
- 40.Poranen, M. M., R. Daugelavièius, and D. H. Bamford. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56521-538. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 43.Tascon, R. I., E. F. Rodriguez-Ferri, C. B. Gutierrez-Martin, I. Rodriguez-Barbosa, P. Berche, and J. A. Vazquez-Boland. 1993. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. J. Bacteriol. 1755717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukagoshi, N., R. Schäfer, and R. M. Franklin. 1977. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur. J. Biochem. 77585-588. [DOI] [PubMed] [Google Scholar]
- 45.Tutino, M. L., A. Duilio, M. A. Moretti, G. Sannia, and G. Marino. 2000. A rolling-circle plasmid from Psychrobacter sp. TA144: evidence for a novel rep subfamily. Biochem. Biophys. Res. Commun. 274488-495. [DOI] [PubMed] [Google Scholar]
- 46.Tutino, M. L., A. Duilio, R. Parrilli, E. Remaut, G. Sannia, and G. Marino. 2001. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles 5257-264. [DOI] [PubMed] [Google Scholar]
- 47.van der Schans, G. P., J. P. Weyermans, and J. F. Bleichrodt. 1971. Infection of spheroplasts of Pseudomonas with DNA of bacteriophage PM2. Mol. Gen. Genet. 110263-271. [DOI] [PubMed] [Google Scholar]
- 48.Vigentini, I., A. Merico, M. L. Tutino, C. Compagno, and G. Marino. 2006. Optimization of recombinant human nerve growth factor production in the psychrophilic Pseudoalteromonas haloplanktis. J. Biotechnol. 127141-150. [DOI] [PubMed] [Google Scholar]