Abstract
The mobile element IS256 causes phase variation of biofilm formation in Staphylococcus epidermidis by insertion and precise excision from the icaADBC operon. Precise excision, i.e., removal of the target site duplications (TSDs) and restoration of the original DNA sequence, occurs rarely but independently of functional transposase. Instead, the integrity of the TSDs is crucial for precise excision. Excision increased significantly when the TSDs were brought into closer spatial proximity, suggesting that excision is a host-driven process that might involve most likely illegitimate recombination.
IS256 is an insertion sequence (IS) element occurring in the genomes of staphylococci and enterococci, either as an integral part of the composite transposon Tn4001 or in multiple free copies (3, 6, 17). Recent studies indicate that IS256 is particularly common in the genomes of multiresistant, biofilm-forming Staphylococcus epidermidis isolates resident in the hospital environment (13). S. epidermidis biofilm formation is mediated mainly by a polysaccharide intercellular adhesin (PIA) (16), whose synthesis enzymes are encoded by the icaADBC operon (11). We demonstrated earlier that ica operon expression and subsequent biofilm formation are subject to phenotypic variation (18). In approximately 30% of spontaneous biofilm-negative variants, the switch from on to off in biofilm formation is caused by insertion of IS256 into icaA or icaC (19). Typically, IS256 creates 8-bp target site duplications (TSDs) on either end upon active transposition. Reversion from off to on was detected after repeated passages of S. epidermidis ica::IS256 insertion mutants (19). Interestingly, restoration of PIA production was accompanied by precise excision of IS256, including the initially duplicated target site, from the ica sequence (19). While the insertion frequency of IS256 into the ica operon is about 10−6 per cell generation, the excision frequency was reported to be considerably lower, at <10−8 per cell generation, indicating that restoration of the PIA-mediated biofilm by IS excision must be a rare event (19). However, the molecular mechanism of precise excision—complete elimination of IS256, including the duplicated target site, and restoration of the original ica sequence—has not been elucidated so far.
Precise excision of IS256 from the icaC gene does not require a functional IS256 transposase.
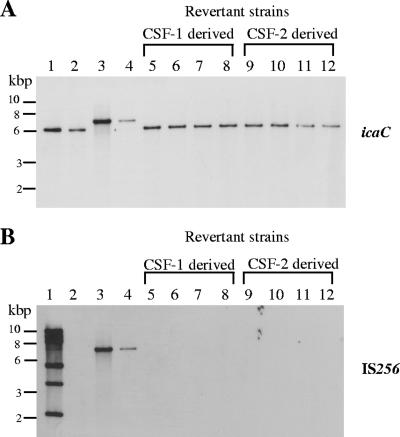
To test whether precise excision of IS256 from the ica operon is a transposase-driven process, we constructed two mutants of the biofilm-positive, IS256-negative strain S. epidermidis CSF41498 (Table 1). The first mutant, S. epidermidis CSF41498-1, carried a chromosomal copy of a previously described icaC::IS256 insertion (14), including an 8-bp TSD (GGTGGTTA), while the second, S. epidermidis CSF41498-2, carried an icaC::IS256 insertion with a 20-bp deletion within the tnp gene, rendering the transposase inactive (Table 1) (14). Both mutants exhibited a biofilm-negative phenotype. Since it was known from previous experiments that precise excision is a rare event (19), biofilm-positive revertants were enriched by serial passages in tissue culture flasks as described previously (19), with the following modification: at each passage step (every 24 h), the medium was discarded and flasks were washed with phosphate-buffered saline before new Trypticase soy broth was added. After 7 to 14 days, the adhering bacterial cells were scratched from the bottom and appropriate dilutions were plated onto Congo red agar (CRA). In all of three independent experiments, revertants could be detected as rough and black colonies on CRA, for both CSF41498-1 and CSF41498-2. Further analyses of these revertants by PCR using icaC-specific primers and by nucleotide sequencing revealed that the majority of them still carried the IS256 insertion (12), while others had lost the IS element completely and restored the original icaC sequence. IS256- and icaC-specific Southern hybridizations of eight members of the latter group demonstrated that the removal of the element was not accompanied by new IS256 insertions in the S. epidermidis chromosome (Fig. 1A and B). These results suggest that precise IS256 excision is not associated with transposition but is mediated by a transposase-independent mechanism.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| S. epidermidis strains | ||
| RP62A | ica positive, IS256 positive | ATCC 35984 |
| CSF41498 | ica positive, IS256-negative biofilm former | 8 |
| CSF41498-1 | icaC::IS256 insertion (at nt 600) | This study |
| CSF41498-2 | icaC::IS256 Δtnp insertion | This study |
| CSF41498-3 | icaC::IS256 insertion, 3-bp deletion of right TSD | This study |
| CSF41498-4 | icaC::IS256 Δtnp insertion, 3-bp deletion of right TSD | This study |
| Plasmids | ||
| pBT2 | E. coli-Staphylococcus shuttle vector with a thermosensitive origin of replication for gram-positive bacteria | 4 |
| pTH1 | pBT2 derivative carrying a spc::IS256 insertion at nt 63, including an 8-bp TSD (AGATCTAT) | This study |
| pTH2 | pTH1 derivative with a truncated IS256 copy of 166 bp consisting of IRL, IRR, and intact 8-bp TSDs | This study |
| pTH3 | pTH1 derivative with 2-bp deletion in the right-hand TSD | This study |
| pTH4 | pTH1 derivative with 4-bp deletion in the right-hand TSD | This study |
| pTH5 | pTH1 derivative with 6-bp deletion in the right-hand TSD | This study |
Details on construction and a list of the oligonucleotides used can be found in Table S1 in the supplemental material.
FIG. 1.
Genetic analysis of biofilm-forming revertants derived from S. epidermidis CSF41498 icaC::IS256 insertion mutants. Southern hybridizations of EcoRI-restricted chromosomal DNA were performed using icaC-specific (A) and IS256-specific (B) DNA fragments as probes (19). Lanes 1, S. epidermidis RP62A; lanes 2, S. epidermidis CSF41498 wild type; lanes 3, S. epidermidis CSF41498-1; lanes 4, S. epidermidis CSF41498-2; lanes 5 to 8, S. epidermidis CSF41498-1-derived revertants; lanes 9 to 12, S. epidermidis CSF41498-2-derived revertants.
Alteration of TSD homology abolishes precise excision.
In the next step, we tested whether precise excision from the chromosome might occur as a result of a recombination process, with the 8-bp TSDs serving as homologous regions. To answer this question, we introduced a 3-bp deletion (GGT) into the right TSDs of CSF41498-1 and CSF41498-2, thereby reducing the homology between the TSDs to 3 bp (GGTGGTTA/GGTTACAT). After serial passages of the corresponding mutant strains CSF41498-3 and CSF41498-4 (Table 1), biofilm-positive revertants were isolated as black, rough colonies on CRA. However, despite extensive PCR screening in seven independent experiments, we could not detect precise excision of IS256 in any of the biofilm-forming revertants. Analyses of these strains indicated that they had switched from PIA-mediated biofilm formation to the expression of a protein-mediated biofilm, which is described elsewhere (12). Our data demonstrate that a reduction of TSD homology abolishes precise excision or at least diminishes the process to a point below the detection limit of this experimental setup.
Quantification of IS256 excision events.
To quantify the rate of excision events, we constructed shuttle plasmid pTH1, carrying a spectinomycin resistance cassette (spc) inactivated by an artificial IS256 insertion, including 8-bp TSDs (Table 1). The plasmid was transformed into the spectinomycin-sensitive S. epidermidis CSF41498 background, and the resulting strain was screened for the occurrence of spectinomycin-resistant colonies that had lost the IS256 insertion. This positive selection approach allowed us to analyze a considerably larger number of colonies than in the previous approach and to calculate the precise excision rates per cell and generation. The data underline that precise excision is a rare event, on the order of magnitude of 10−11 per cell and generation when using vector pTH1 (Table 2). This low excision rate suggests that a host-mediated recombination process is behind precise IS256 excision. Also, Southern blot analysis revealed no new IS256 insertions in the chromosome in these strains (data not shown).
TABLE 2.
Excision rates of IS256 from the spc gene, using a plasmid-based system
| Plasmid | Right/left TSD sequencea | IS256 sequence | Excision frequency (per cell generation)b |
|---|---|---|---|
| pTH1c | AGATCTAT/AGATCTAT | Full length | 1.5 × 10−11 |
| pTH2d | AGATCTAT/AGATCTAT | 166-bp fragment | 9.8 × 10−11 |
| pTH3c | AGATCTAT/AGATATTT | Full length | <2.0 × 10−13 |
| pTH4c | AGATCTAT/AGATTTGG | Full length | <4.7 × 10−13 |
| pTH5c | AGATCTAT/AGTTGGAA | Full length | <1.2 × 10−13 |
Mismatches between the TSDs are shown in bold.
Data are medians for all experiments.
Experiments were carried out in triplicate.
Experiments were repeated six times.
Next, we wanted to analyze the minimal homology requirement of the IS256 TSDs for precise excision. Therefore, we introduced deletions into the right-hand TSD of spc::IS256, leading to mismatches of two, three, and four base pairs and resulting in plasmids pTH3, pTH4, and pTH5, respectively (Table 1). No precise excision event was detected in any of the TSD mutants, and the probability of precise IS256 excision was calculated to be <10−13 per cell and generation (Table 2).
These data support the hypothesis that the microhomologies at either end of the element are a necessary prerequisite for excision. At the present stage of experimental work, we cannot decide unambiguously how TSD-dependent IS256 excision proceeds on the molecular level. However, the data collected so far strongly imply a host-mediated recombination process as the underlying mechanism. Since RecA-dependent recombination requires sequence homologies of 300 bp or more and site-specific recombination relies on site-specific integrases along with defined DNA recognition sites (2, 10), it is rather unlikely that one of these mechanisms mediates precise IS256 excision. In contrast, illegitimate recombination by so-called replicational slippage would provide a model for this phenomenon. Precise excision was described for other transposons as well (5, 9), and for Tn10, replicational slippage was suggested to be the underlying mechanism (7). Replicational slippage requires short homologous DNA stretches located in close vicinity to each other on the same DNA molecule. In Escherichia coli, eight nucleotides appears to be the minimal length required for slippage (1, 8), and slight sequence differences were already shown to reduce the deletion frequency dramatically (15). The same seems to hold true for IS256 in S. epidermidis in this report, as eight homologous base pairs was sufficient for precise excision, while reduction to six base pairs abolished the process to a point below the detection limit.
Narrowing the distance between the TSDs results in an increase of precise excision rates.
Last, we tested whether the spatial distance between the target site homologies has an impact on excision frequencies. To this end, we constructed pTH2 (Table 1). In this case, we truncated the IS256 copy on pTH1 considerably, leaving only the noncoding regions (NCRs) of the element on either end (containing the left and right inverted repeats [IRL and IRR, respectively]) as well as the 8-bp TSDs intact, separated by 166 bp of DNA consisting of the 101-bp left NCR, a 15-bp transposase gene remnant, and the 50-bp right NCR. The excision frequency of this truncated IS256 version in pTH2 (Table 2) showed a significant increase compared to that for pTH1 carrying a full-length IS256 copy (one-tailed Mann-Whitney U test; P < 0.025). From other studies, it is known that a decrease of the distance between repeats increases replicational slippage rates significantly (2, 4). Apparently, this also applies to IS256. Thus, the model of replicational slippage provides an interesting hypothesis for future experimental work to investigate precise IS256 excision in molecular detail.
Concluding remarks.
Our data substantiate that IS256-mediated phase variation involves two distinct processes, namely, transposition of IS256 and transposase-independent precise excision. The frequencies of these processes differ dramatically, by at least 5 orders of magnitude, with precise excision being an extremely rare event. Interestingly, we found that with respect to biofilm formation, S. epidermidis CSF41498 is obviously able to compensate for the low reversion rate due to precise excision by frequent switching to an alternative, protein-mediated biofilm, highlighting again the functional importance of the biofilm lifestyle for S. epidermidis (12). Thus, the latter mechanism appears to be of more biological significance than precise excision of IS256. Taken together, the data obtained in this study add to our understanding of IS activity and function in bacterial genomes and their impact on the generation of genetic and phenotypic variety.
Supplementary Material
Acknowledgments
We are grateful to Katja Dietrich, Elena Katzowitsch, and Katharina Schlereth for excellent technical assistance, and we thank James P. O'Gara (University College Dublin, Ireland) for providing strain Staphylococcus epidermidis CSF41498.
This study was funded by Deutsche Forschungsgemeinschaft grants IGK 587/2, SFB479, and SFB/TR34 as well as by the Federal Ministry of Education and Research (Network Pathogenomics-Plus; grant PTJ-BIO//03U213B).
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albertini, A. M., M. Hofer, M. P. Calos, and J. H. Miller. 1982. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 29319-328. [DOI] [PubMed] [Google Scholar]
- 2.Bi, X., and L. F. Liu. 1994. recA-independent and recA-dependent intramolecular plasmid recombination: differential homology requirement and distance effect. J. Mol. Biol. 235414-423. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81361-367. [DOI] [PubMed] [Google Scholar]
- 4.Chedin, F., E. Dervyn, R. Dervyn, S. D. Ehrlich, and P. Noirot. 1994. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol. Microbiol. 12561-569. [DOI] [PubMed] [Google Scholar]
- 5.Collins, J., G. Volckaert, and P. Nevers. 1982. Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene 19139-146. [DOI] [PubMed] [Google Scholar]
- 6.Dyke, K. G., S. Aubert, and N. el Solh. 1992. Multiple copies of IS256 in staphylococci. Plasmid 28235-246. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich, S. D., H. Bierne, E. d'Alencon, D. Vilette, M. Petranovic, P. Noirot, and B. Michel. 1993. Mechanisms of illegitimate recombination. Gene 135161-166. [DOI] [PubMed] [Google Scholar]
- 8.Farabaugh, P. J., U. Schmeissner, M. Hofer, and J. H. Miller. 1978. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 126847-863. [DOI] [PubMed] [Google Scholar]
- 9.Foster, T. J., V. Lundblad, S. Hanley-Way, S. M. Halling, and N. Kleckner. 1981. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell 23215-227. [DOI] [PubMed] [Google Scholar]
- 10.Grindley, N. D., K. L. Whiteson, and P. A. Rice. 2006. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75567-605. [DOI] [PubMed] [Google Scholar]
- 11.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 201083-1091. [DOI] [PubMed] [Google Scholar]
- 12.Hennig, S., S. Nyunt Wai, and W. Ziebuhr. 2007. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297117-122. [DOI] [PubMed] [Google Scholar]
- 13.Kozitskaya, S., S.-H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 721210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 1844709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovett, S. T., and V. V. Feschenko. 1996. Stabilization of diverged tandem repeats by mismatch repair: evidence for deletion formation via a misaligned replication intermediate. Proc. Natl. Acad. Sci. USA 937120-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice, L. B., and S. H. Marshall. 1994. Insertions of IS256-like element flanking the chromosomal beta-lactamase gene of Enterococcus faecalis CX19. Antimicrob. Agents Chemother. 38693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziebuhr, W., C. Heilmann, F. Götz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziebuhr, W., V. Krimmer, S. Rachid, I. Loessner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32345-356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.