Abstract
Staphylococcus aureus becomes resistant to methicillin by acquiring a genomic island, known as staphylococcal chromosome cassette mec (SCCmec), which contains the methicillin resistance determinant, mecA. SCCmec is site-specifically integrated into the staphylococcal chromosome at a locus known as the SCCmec attachment site (attB). In an effort to gain a better understanding of the potential that methicillin-sensitive S. aureus (MSSA) isolates have for acquiring SCCmec, the nucleotide sequences of attB and surrounding DNA regions were examined in a diverse collection of 42 MSSA isolates. The chromosomal region surrounding attB varied among the isolates studied and appears to be a common insertion point for acquired foreign DNA. Insertions of up to 15.1 kb were found containing open reading frames with homology to enterotoxin genes, restriction-modification systems, transposases, and several sequences that have not been previously described in staphylococci. Two groups, containing eight and four isolates, had sequences found in known SCCmec elements, suggesting SCCmec elements may have evolved through repeated DNA insertions at this locus. In addition, the attB sequences of the majority of MSSA isolates in this collection differ from the attB sequences of strains for which integrase-mediated SCCmec insertion or excision has been demonstrated, suggesting that some S. aureus isolates may lack the ability to site-specifically integrate SCCmec into their chromosomes.
Methicillin-resistant Staphylococcus aureus (MRSA) arose from methicillin-susceptible S. aureus (MSSA) upon acquisition of a genomic island known as staphylococcal chromosome cassette mec (SCCmec) (17). The first MRSA was isolated shortly after the introduction of methicillin in 1959. Since then, the rates of MRSA infections in the hospital, as well as disease in the community, have continued to rise (28). Currently, MRSA isolates are responsible for 59% of S. aureus infections encountered in U.S. emergency departments (24) and 59.5% of S. aureus infections in intensive-care units (1), and in one study, the rates of MRSA disease in the community more than doubled between 2002 and 2004 (6). The increasing burden of MRSA disease is likely due to two factors: clonal expansion of existing MRSA lineages and the conversion of successful MSSA lineages to MRSA by the transfer of SCCmec into these backgrounds.
SCCmec is a variable genetic element that contains the methicillin resistance determinant, mecA, and a site-specific recombinase gene, ccrAB or ccrC, and may contain additional resistance determinants (15-17, 22). The targets of β-lactam antibiotics are transpeptidases or penicillin-binding proteins (PBPs) that cross-link bacterial cell walls. mecA encodes an alternative PBP (PBP2a) that has a reduced affinity for β-lactam antibiotics and is able to maintain cell wall synthesis when the bacterium's other PBPs are inhibited (4, 20). ccrAB and ccrC are site-specific recombinase genes that catalyze the chromosomal excision and integration of SCCmec (15-17, 25). SCCmec elements exist in six major isotypes (SCCmec I to VI), and variations of each type have been recognized (15-17, 22, 26, 27). In addition, non-mecA-containing SCC elements have been discovered. All of these elements are precisely integrated into the same SCCmec chromosomal attachment site, attB, which is found at the end of an open reading frame of unknown function, orfX. They contain homologues of the SCCmec recombinase genes, ccrA and ccrB, and are flanked by the 15-bp directly repeated sequences characteristic of SCCmec elements (13, 18, 21, 23).
Using multilocus sequence typing in conjunction with SCCmec typing, Enright et al. found that MRSA strains are present in only 38 of the 162 S. aureus genetic backgrounds (10). The presence of MRSA in only a subset of possible S. aureus genetic lineages suggests that these select lineages are uniquely capable of acquiring SCCmec. Lineages that do not contain SCCmec may lack the DNA sequences necessary for the element's chromosomal integration. More specifically, these lineages may not contain an intact attB or accessory regions necessary for CcrAB-mediated integration of SCCmec. It is also possible that the attB sequences of some lineages are otherwise occupied with non-mecA-containing SCC elements. In the present study, we examined the nucleotide sequence of attB and surrounding regions from a collection of 42 MSSA isolates, representing diverse genetic backgrounds, to gain a better understanding of these isolates' potentials for acquiring SCCmec.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the MSSA isolates used in this study, as well as their relevant characteristics. Isolates were chosen from available collections, and genetic diversity was sought by ensuring that all isolates were of different spa type and represented a wide range of multilocus sequence types (STs). Isolates NRS104, NRS198, NRS199, and NRS204 were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus. All four strains were originally isolated from patients in Great Britain in the 1940s. Strains 6520 and 6881 were kindly provided by Fred Tenover at the Centers for Disease Control and Prevention. All other MSSA isolates were from the collection of isolates at Duke University recovered between 1997 and 2004 from patients with either bacteremia or skin infections or colonizing the nares of healthy carriers. The MRSA strains N315, NRS384 (USA300-like PFGE type strain), MRSA252, and COL were all obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus. J39 was a gift from Henry Chambers at the University of California at San Francisco.
TABLE 1.
Characteristics of strains
| Strain | spa typea | spa repeat profile | STb | Genetic groupc | attB structured | Notes |
|---|---|---|---|---|---|---|
| 15658 | 529 | W1-A1-K1-A1-O1-M1-Q1-Q1-Q1 | 30 | 1a | 15580 | |
| 15671 | 18 | W1-F1-K1-A1-O1-M1-Q1 | 30 | 1a | 15580 | |
| 15677 | 33 | W1-G1-K1-A1-K1-A1-O1-M1-Q1-Q1 | 30 | 1a | 15580 | |
| 15580 | 468 | X1-K1-A1-K1-A1-O1-M1-Q1-Q1 | 30 | 1a | 15580 | 15.1 kb, depicted in Fig. 4 |
| 15637 | 208 | X1-K1-B1-Q1-B1-B1-M1-M1 | 30 | 1a | 12.5-kb long-range PCR product, sequence unknown | |
| NRS104 | 99 | X1-K1-K1-A1-K1-A1-O1-M1-Q1-Q1 | 30 | 1a | 15580 | |
| NRS204 | 251 | W1-G1-K1-K1-A1-K1-A1-O1-M1-Q1 | 30 | 1a | 15580 | |
| 6520 | 43 | W1-G1-K1-A1-K1-A1-O1-M1-Q1 | 30 | 1a | 15580 | |
| 6881 | 43 | W1-G1-K1-A1-K1-A1-O1-M1-Q1 | 30 | 1a | 15580 | |
| 15578 | 42 | A2-A1-K1-B1-E1-M1-B1-K1-B1 | 45 | 1a | ND | Does not contain reverse primer binding site |
| 15651 | 220 | X1-K1-A1-K1-E1-M1-B1-K1-B1 | 45 | 1a | ND | Does not contain reverse primer binding site |
| 15630 | 93 | X1-K1-B1 | 47 | 1a | ND | Does not contain reverse primer binding site |
| 3304 | 558 | I2-Z2-E1-G1-M1-M1-M1-J1-H2-M1 | 121 | 1b | 3298 | 6.9 kb, depicted in Fig. 3 |
| 15649 | 466 | I2-Z2-E1-M1-M1-J1-H2-M1 | 121 | 1b | 3298 | |
| 3298 | 411 | I2-Z2-E1-G1-M1-J1-H2-M1 | 121 | 1b | 3298 | |
| 15585 | 17 | Z1-D1-M1-D1-M1-N1-K1-B1 | 59 | 1b | 15585 | 6.9 kb, depicted in Fig. 3 |
| 15601 | 526 | U1-M1-B1 | 188 | 2 | 8 kb LR-PCR product, sequence unknown | |
| 15594 | 509 | T1-J1-A1-I3-J1-A1-B1-B1-B1 | 9 | 2 | 8325 | Prototypical unoccupied attB as found in S. aureus 8325 |
| 15668 | 2 | T1-J1-M1-B1-M1-D1-M1-G1-M1-K1 | 5 | 2 | 8325 | |
| 15634 | 514 | T1-K1-B1-M1-D1-M1-G1-M1-K1 | 5 | 2 | 8325 | |
| 15602 | 37 | U1-K1-G1-J1-B1 | 109 | 2 | 8325 | |
| 15611 | 139 | Y1-G1-F1-M1-B1-L1-O1 | 8 | 2 | 8325 | |
| 15681 | 7 | Y1-H1-G1-C1-M1-B1-Q1-B1-L1-O1 | 8 | 2 | 8325 | |
| 15589 | 1 | Y1-H1-G1-F1-M1-B1-Q1-B1-L1-O1 | 8 | 2 | 8325 | |
| 15579 | 46 | Y1-M1-B1-Q1-B1-L1-O1 | 195 | 2 | 8325 | |
| NRS198 | 1 | Y1-H1-G1-F1-M1-B1-Q1-B1-L1-O1 | 8 | 2 | 8325 | |
| 15607 | 505 | I2-F1-K1-B1-P1-E1 | 1 | 2 | 15575 | 6.3 kb, depicted in Fig. 4 |
| 15575 | 515 | U1 | 1 | 2 | 15575 | |
| 15576 | 35 | U1-J1-F1-K1-B1-P1-E1 | 1 | 2 | 15575 | |
| 15591 | 175 | U1-J1-F1-K1-P1-E1 | 1 | 2 | 15575 | |
| 15647 | 154 | Y1-C2-F1-M1-B1-Q1-B1-L1-O1-O1 | 6 | 2 | 15580 | 10.5-kb long-range PCR product, sequence unknown |
| 15679 | 105 | U1-J1-G1-F1-M1-B1-B1-B1-P1-B1 | 97 | 2 | 15584 | 9.8 kb, depicted in Fig. 3 |
| 15584 | 527 | U1-M1-F1-M1-B1-B1-P1-B1 | ND | 2 | 15584 | |
| 15604 | 141 | U1-J1-F1-Q1-P1-L1-M1 | 12 | 2 | 15604 | 10 kb, depicted in Fig. 3 |
| 15653 | 21 | U1-J1-G1-B1-B1-G1-G1-J1-A1-G1-J1 | 15 | 2 | 15653 | 5.6 kb, depicted in Fig. 3 |
| 15673 | 151 | I2-G1-B1-B1-G1-G1-J1-A1-G1-J1 | 15 | 2 | 15666 | 3.8 kb, depicted in Fig. 3 |
| 15666 | 152 | U1-J1-F1-G1-M1-D1-M1-G1-G1-M1 | 72 | 2 | 15666 | |
| 3294 | 118 | U1-J1-G1-B1-B1-G1-E1-G1-J1-A1-G1-J1 | 15 | 2 | 15666 | |
| 15682 | 549 | Z1-G1-F1-G1-U2-D1-M1-G1-M1 | 25 | 2 | 15682 | 7.4 kb, depicted in Fig. 4 |
| 15680 | 507 | T1-G2-M1-F1-B1-B1-B1 | 20 | 2 | ND | No long-range PCR amplification product |
| 15639 | 519 | U1-J1-G1-B1-B1-G1-G1-B1-B1-G1-G1-J1-A1-G1-J1 | 15 | 2 | ND | Does not contain reverse primer binding site |
| NRS199 | Z1-O3-M1-O1-M1-O1-M1 | NRS199 | 5 kb, depicted in Fig. 4 |
spa typing uses sequence repeats in the spa gene to determine the evolutionary relatedness of S. aureus isolates (29).
Multilocus sequence typing (9).
Isolates broadly grouped according to their evolutionary relatedness (5).
Designation corresponds to the attB region in sequenced MSSA isolates. ND, not determined.
PCR and Southern analysis.
The primers used for PCR and sequencing are listed in Table 2. PCRs were performed using the PCR Master Mix kit (Qiagen, Valencia, CA), according to the manufacturer's suggested parameters, on total cellular DNA isolated using the QIAamp DNA Mini kit (Qiagen, Valencia, CA). Southern analysis was performed on DNA separated by pulsed-field gel electrophoresis (PFGE). Preparation of genomic DNA and separation by PFGE were adapted from the method of Bannerman et al. (3) as previously described (11). Southern analysis was performed as described by Hovis et al. using a Hybond-N membrane and an [α-32P]dATP-labeled probe (14).
TABLE 2.
Primers
| Primer | Sequence | Description |
|---|---|---|
| orfXfor | GAGAAATATTGGAAGCAAGCC | Detects orfX |
| orfXrev | CGCATAATCTTAAATGCTCTG | |
| mecAfor | CTCATATAGCTCATCATACACTTTACC | Detects mecA |
| mecArev | CACTTATTTTAATAGTTGTAGTTGTCGG | |
| cc1rev | CGTAATGTCATTGAGTTGC | Detects ccrA |
| cc4for | GCTATGTCACTAAAAAGGGTAAACC | |
| cc4rev | GCATTCATGTTTTTAGGACAGACG | Detects ccrB |
| cc1for | CGTACCATGTTCATATCTTAAGC | |
| ccrCF | CAGTAATGTCAAGATGTCGATGAATGC | Detects ccrC |
| ccrCR | CCGTCGACATACCATATTATTGCCG | |
| I1-F | GTTCCAGACGAAAAAGCACCAG | Amplifies attB |
| I1-R | CATTTTATGAGTCTCGCAAATTGTCAG | |
| colA | GTTCCCAGTAGCAACCTTCC | Amplifies the circular, excised SCCmec element in S. aureus 450 M |
| colB | CAATGAAAGCTTGGAAGAAGGGC | |
| unirev | GCACAGTGGGAATTAATCGAAGC | Reverse primer for long-range PCR |
| 252R | CCACTATTTAACTGACTTGATATACC | Alternate reverse primer for long-range PCR |
| uniF | GCTTCAAATTCATCTAGTAGTGC | Amplifes unirev binding site |
| uniR | GATAAAAGATTTAATGCCACTGATG | |
| a252F | CCTTCAAAAATAAATGTATGGTC | Amplifies 252R binding site |
| a252R | CATCATAAATACAATTAATACGTTGAC | |
| LE1F | CTAATGCTCAATGCATTTTCTTCAG | PCR primer walking of the region outside of SCCmec type II in strain MRSA252 |
| LE1R | GCATAGCGAAGCCATTTAATAGCG | |
| LE2F | CGTAGTCATCAAAGTTTGATTCAGC | |
| LE2R | GGGAGGCGTCAAAATTTGAGG | |
| LE3F | CATTTCGAAAGCGCCAGCTAATCTC | |
| LE3R | CATATGTAGGTAGTAAAATTTTTAAAAGC | |
| LE4F | GCAATATGCCATAATGCTATCTCC | |
| LE4R | GATAGATTATAATGATACAACATTGG | |
| LE5F | GTAATACAAACTGAAAGCAAGGG | |
| LE5R | GATGATGTTACAACAAGCTCTGG |
Long-range PCR and primer walk sequencing.
Long-range PCR was performed using the Platinum PCR Supermix High Fidelity (Invitrogen, Carlsbad, CA) according to the manufacturer's specifications. Amplification products were gel extracted and purified using the Qiagen Gel Extraction kit. Both strands of the amplification product were sequenced using a primer-walking approach by the DNA Facility of the Iowa State University Office of Biotechnology (available at http://www.dna.iastate.edu/). All DNA sequences were confirmed by PCR primer walking these chromosomal regions from genomic DNA isolated from the appropriate MSSA isolates.
Sequence analysis.
Sequence data were assembled using the Contig Express portion of the Invitrogen VectorNTI software. Nucleotide homology searches were performed using Blastn, available on the NCBI website (http://www.ncbi.nlm.nih.gov/). Open reading frames were predicted and translated using the VectorNTI software default parameters. Protein homology searches were performed using Blastp, available on the NCBI website.
Nucleotide sequence accession numbers.
The DNA sequences surrounding the SCCmec insertion sites from the 10 MSSA isolates studied have been deposited in GenBank. The accession numbers are as follows: EU272077 for strain NRS199, EU272078 for strain 3298, EU272079 for strain 15575, EU272080 for strain 15580, EU272081 for strain 15584, EU272082 for strain 15585, EU272083 for strain 15604, EU272084 for strain 15653, EU272085 for strain 15666, and EU272086 for strain 15682.
RESULTS
Diversity of the isolate collection.
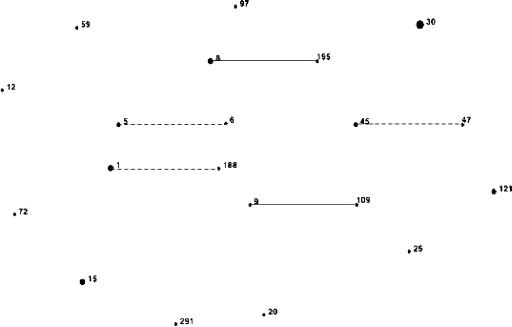
As shown in Table 1, each isolate examined in this study was unique, as determined by the spa type (29). In addition, the isolates represented all three suggested phylogenetic groups (1a, 1b, and 2) described by Cooper and Feil and 21 different STs (5, 9). Figure 1 shows the eBURST diagram, based on multilocus sequence typing, for this collection of isolates. Five related groups of S. aureus isolates were identified. The rest of the isolates were completely unrelated.
FIG. 1.
eBURST diagram of all S. aureus isolates examined in this investigation. The interrelatedness, based on multilocus sequence typing, of all isolates in this investigation is shown as calculated using eBURST with the default settings. The dot size is representative of the number of isolates with a given ST. Any relationship between STs is indicated by a solid line for double-locus variants and a dotted line for single-locus variants. Only five related isolate groups were present in this study; ST8 and ST195 (group 1), ST9 and ST109 (group 2), ST 45 and ST47 (group 3), ST1 and ST188 (group 4), and ST5 and ST6. All other isolates are unrelated.
Detection of genes associated with SCCmec.
The collection consisted of 42 S. aureus isolates that were methicillin susceptible (as determined by their failure to grow on 6 μg/ml oxacillin). To better characterize the SCCmec chromosomal region, PCR and Southern blot analysis were used to detect mecA, ccrAB, ccrC, and orfX. None of the strains contained mecA, ccrAB, or ccrC. The absence of ccrAB, ccrC, or their remnants suggested that non-mecA-containing SCC elements were not present in this collection, as all known non-mecA-containing SCC elements contain recombinase genes. Every isolate in this collection was found to contain the orfX gene. The nucleotides encoding the C-terminal 5 amino acids of orfX comprise the 15-bp core sequence of SCCmec attB. This sequence is directly repeated upon SCCmec insertion, flanking each end of the integrated element. The presence of orfX suggests that each of these strains has the potential to acquire SCCmec.
attB.
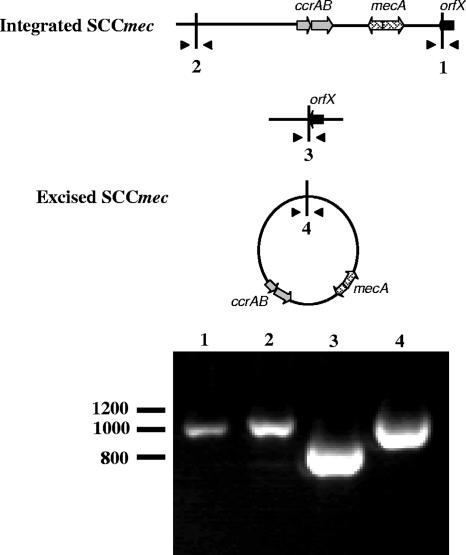
For this study, S. aureus 8325 was considered the MSSA representative of a strain with an unoccupied and functional attB site. This is because type I SCCmec has been introduced into 8325 derivatives by both transduction and transformation and integrated into the orfX attachment site (Fig. 2). In addition, as seen in Fig. 2, SCCmec can be excised from this strain (450 M) by plasmid-encoded ccrAB, resulting in regeneration of the 8325 attB sequence. Using PCR primers (I1-F and I1-R), an 850-bp fragment containing attB was amplified from strain 8325. The same fragment with identical or nearly identical DNA sequence was also amplified from 9 of the 42 MSSA isolates examined in this study (Table 1) (accession no. NC_007795).
FIG. 2.
Site-specific chromosomal integration and excision of SCCmec in S. aureus 450 M. S. aureus 450 M is an 8325 derivative containing SCCmec. Schematics of the 450 M chromosome with SCCmec integrated or excised are shown at the top, with triangles indicating primer binding sites, numbers indicating the region amplified in the corresponding lane of the gel, and vertical lines indicating attachment sites. Select open reading frames are indicated as reference points. Shown in lanes 1 and 2 are PCR amplification of the right and left SCCmec attachment sites using primers I1F/colA and I1R/colB, respectively, indicating that SCCmec is site-specifically integrated into the chromosome of 450 M. Shown in lane 3 is PCR amplification of the chromosomal junction from which SCCmec was site-specifically excised by CcrAB, using primers I1F and I1R. Lane 4 contains the PCR amplification product of the excised, circular SCCmec element using primers colA and colB.
Structure surrounding attB in other MSSA isolates.
Long-range PCR amplification of the region surrounding attB was performed in the 33 MSSA isolates that did not contain the unoccupied 8325 attB. One of the long-range PCR primers (orfXfor) was designed to bind in orfX. The reverse primer (unirev) was anchored in an open reading frame with homology to a tRNA dihydrouridine synthase (annotated as MW0054 on the MW2 genome sequence, accession no. NC_003923). This was the first predicted open reading frame on the non-orfX side of SCCmec that was common to all genome-sequenced MRSA and MSSA strains. The results are shown in Table 1. Long-range PCR amplification of the attB genetic region yielded products, ranging in size from 3.7 to 15.1 kb, in 28 of the 33 uncharacterized MSSA isolates. Four of the five isolates (MSSA 15630, 15578, 15651, and 15639) that long-range PCR failed to amplify did not contain sequences to which the reverse primer (unirev) could bind, while the fifth (MSSA 15601) may have had too much intervening sequence for long-range PCR amplification. Further characterization of the attB sites in these strains was not performed. Amplification products of the same size were digested with HindIII, BglII, and EcoRI, and the sizes of the restriction fragments were compared. Restriction fragments of identical size were found in groups of eight, four, three, three, and two isolates. While the presence of identical-size fragments does not conclusively establish identical sequence, we assumed that those groups with fragments of identical size after digestion with three restriction endonucleases were highly related. Each of the eight remaining isolates had a unique restriction pattern. The nucleotide sequences of one representative of each of the five groups and five of the eight unique amplification products were determined.
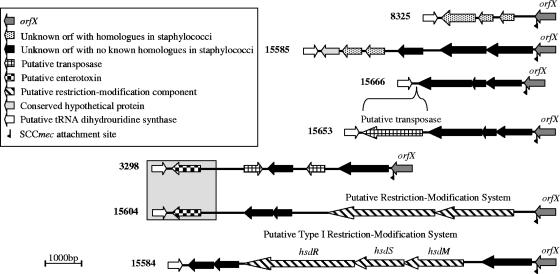
A schematic of the attB region of S. aureus 8325 is shown in Fig. 3. This region is approximately 3 kb and contains three predicted open reading frames. The same size PCR fragment and three open reading frames were found in all nine 8325-like attB sites. However, these three open reading frames were not found in any of the other sequenced MSSA isolates. A schematic of the attB genetic regions from seven isolates is shown in Fig. 3 to illustrate the plasticity of this region. It is notable that genes were found that have no homologues in any other staphylococcal genome and that homologues of restriction modification systems found in Lactococcus lactis (15604) and Staphylococcus haemolyticus (15584 group) were also detected. Although identical genes and sequences were shared among some isolates, the majority of the sequence in this region is entirely unrelated among these strains.
FIG. 3.
Schematic of the genetic regions surrounding attB in seven MSSA isolates. The nucleotide sequences surrounding attB in strains 8325, 15585, 15666, 15653, 3298, 15604, and 15584 are depicted. Block arrows represent predicted open reading frames, and their fill patterns indicate their putative functions, as shown in the key. Where possible, the predicted function was assigned based on homology. Open reading frames with similar predicted functions are not identical in the different isolates. Each region is flanked by orfX (gray arrow) and a conserved predicted open reading frame (white arrow). 15666 and 15653 differ only by the insertion of a putative transposase into 15653 that is not present in 15666. 3298 and 15604 share a region at the left end that encodes a putative enterotoxin (boxed in gray). Open reading frames labeled hsdS, hsdR, and hsdM are predicted components of a type 1 restriction-modification system.
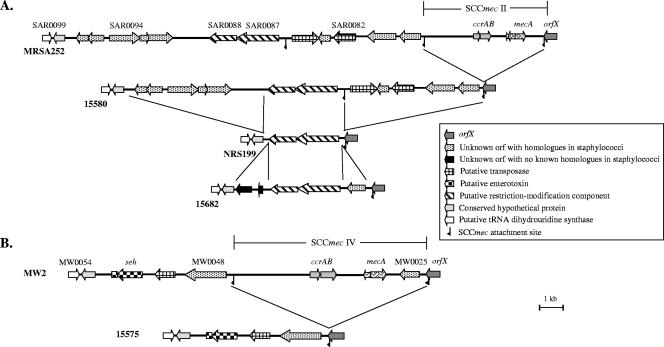
An evolutionary relationship between sequences inserted at attB in MSSA and those in MRSA containing SCCmec can be seen in Fig. 4. Both the type II SCCmec region from the genome-sequenced strain MRSA252 (accession no. NC_002952) (13) and the type IV SCCmec region from MW2 (accession no. NC_003923) (2) have sequences at their non-orfX termini that are identical to those found in MSSA. All 15.1 kb from the eight isolates in the 15580 group were found adjacent to the MRSA252 SCCmec element, while the 6.3 kb in the four isolates of the 15575 group were adjacent to the MW2 SCCmec element. In addition, as shown in Fig. 4A, three unique sequences show an evolutionary relationship to those in the 15580 group. There is also a 15-bp sequence, identical to the direct repeat that is the core of the SCCmec attachment site, that comes after the first 6 kb of sequence flanking the non-orfX end of MRSA252, as well as 15580. This core sequence is repeated as a result of ccrAB-mediated SCCmec insertion.
FIG. 4.
Comparison of the chromosomal regions of MRSA252, 15580, NRS199, and 15682, as well as MW2 and 15575. (A) The chromosomal region containing SCCmec and the surrounding sequence of MRSA252 is shown, along with similar regions from MSSA strains 15580, NRS199, and 15682. MRSA252 contains SCCmec type II and a 6-kb region encoding two regions homologous to transposases inserted into attB. The left end of this chromosomal region contains two open reading frames with homology to restriction and modification genes. Strain 15580 differs from MRSA252 by the absence of SCCmec. Strains NRS199 and 15682 contain the two open reading frames with homology to restriction and modification genes, but the other regions are absent in these strains. (B) The chromosomal region containing SCCmec and surrounding sequence in MW2 is shown, along with a similar region from MSSA 15575. MW2 carries SCCmec type IV. The region outside of SCCmec contains an open reading frame also present within the right end of SCCmec (MW0048 and MW0025, respectively). This region also contains a putative transposase, staphylococcal enterotoxin H (seh), and a truncated region with homology to staphylococcal enterotoxin O (seo). 15575 differs from MW2 by the absence of SCCmec type IV. Each region is flanked by orfX (gray arrow) and a conserved predicted open reading (white arrow). Brackets are used to show the sequence difference between strains. SCCmec elements are not drawn to scale.
Comparison of attB sequences.
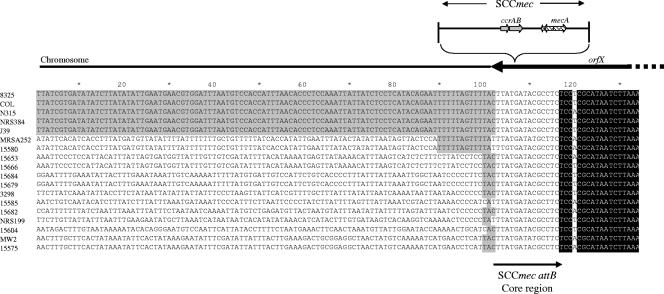
The CcrAB- or CcrC-mediated recombination events that result in the chromosomal integration or excision of SCCmec are dependent on the nucleotide composition of the 15-bp core sequence of the SCCmec attachment site, as it is the site of DNA strand exchange (15). However, it is likely that the recombinases also bind to DNA outside of this 15-bp core sequence. Therefore, these regions may also be important for integration or excision of SCCmec. Figure 5 shows the nucleotide sequence alignment of attB and surrounding regions from 18 S. aureus strains. attB from S. aureus 8325 represents the nine MSSA isolates with an 8325-like attB. SCCmec could be excised from strains N315, COL, NRS384 (USA300-like PFGE type), and J39 by introducing plasmid-borne ccrAB. Following SCCmec excision, the attB region was PCR amplified from these strains, and the nucleotide sequences are shown in Fig. 5. The sequence of attB for MW2 has been inferred from the published genome sequence (2). The sequences surrounding attB from the 10 sequenced MSSA isolates are also included. The sequences of orfX and the 15 core nucleotides were highly conserved among all S. aureus isolates. However, the region on the non-orfX side of the core sequence varied. This region was nearly identical in 8325, N315, COL, NRS384 (USA300-like PFGE type), and J39—all strains in which integration or excision of SCCmec has been demonstrated. Strain MW2 does not share this sequence and is not capable of CcrAB-mediated SCCmec excision (25). The 10 MSSA sequences also do not share this sequence and, similarly, may not be able to acquire SCCmec.
FIG. 5.
Nucleotide alignment of attB genes and surrounding regions from 18 S. aureus strains. The thick black arrow indicates the portion of the orfX coding region shown. The thin black arrow marks the 15-bp core sequence of the SCCmec attachment site. This sequence is present at the carboxyl terminus orfX and, when SCCmec inserts, is directly repeated at the other end of the element. SCCmec and its point of chromosomal insertion are depicted above the alignment (not drawn to scale). The region shaded in light gray represents a conserved sequence present outside of attB in all S. aureus strains known to undergo CcrAB-mediated SCCmec integration or excision. The conserved sequence of orfX is shaded in black, while strains are indicated on the left. The attB region of strain MW2 was inferred based on the genome sequences of attL and attR, since SCCmec did not excise in MW2.
DISCUSSION
This analysis of sequences integrated at the attB site of the S. aureus chromosome in MSSA isolates was prompted in part by the finding that the genome of MSSA476 (NC002953) contained an SCCmec-like genomic island including the genes for the ccrAB integrase/excisase. However, the current study found that strain to be the exception. None of the 42 strains examined contained any ccrAB sequences or any other sequences found within the SCCmec element. Instead, 79% of the isolates contained DNA of both staphylococcal and nonstaphylococcal origin that may provide clues to both the evolution and mechanism of acquisition of SCCmec, as well as the clustering of the MRSA genotype within a few strain types and clonal lineages.
Previous studies have shown that SCCmec can be excised and circularized by ccrAB, acting at the core 15-bp sequence comprising the terminus of orfX and duplicated at the other end of the element upon insertion (15-17, 25). Following excision, the sequences remaining are identical to those in 8325 for 102 bp following orfX. However, SCCmec from MRSA strain MW2 and other strains of ST1 lineage could not be excised and contained sequences at the non-orfX end of the element that differed from the 102 bp found in 8325 and in MRSA, with an excisable SCCmec (25). It was postulated that sequences outside the 15-bp core were necessary for CcrAB-mediated excision and integration. The current study identified, by DNA sequence, 10 different variants at the attB site that do not contain sequences conserved in strains known to undergo integration or excision of SCCmec. It is possible that this sequence is required for CcrAB- or CcrC-mediated SCCmec insertion. The absence of this sequence in the majority of MSSA isolates studied may explain the absence of SCCmec in many MRSA lineages. In support of this hypothesis, of the nine MSSA isolates with an 8325-like attB sequence, four belong to ST8, two to ST5, and one to ST9. ST5, -8, and -9 are found within the prominent MRSA clonal complexes, indicating that isolates with these genetic backgrounds have acquired SCCmec in the past (8, 10). In contrast, among the 10 isolates representing the non-8325 attB sequences depicted in Fig. 5, only two (ST59 and -72) have ever been associated with MRSA lineages. Those two ST lineages may have acquired SCCmec by non CcrAB-mediated mechanisms, or sequences required for CcrAB-mediated insertion may be gained or lost in particular lineages.
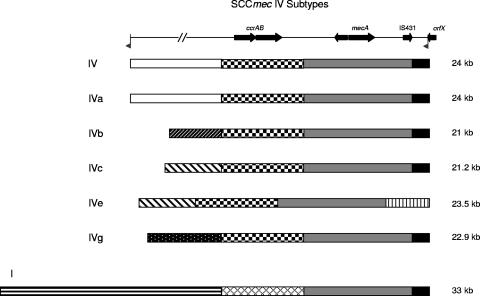
In contrast, there are two MSSA groups, shown in Fig. 4, that have sequences and genetic backgrounds associated with SCCmec acquisition. Both the eight isolates in MSSA group 15580 and the four in group 15575 have sequences at the attB site that are identical to those found following the duplicated 15-bp core sequence at the non-orfX end of SCCmec in the genome-sequenced strains MRSA252 and MW2. In the case of MRSA252 and group 15580, the first 13 bp are present after the 15-bp core attachment sequence. Thus, SCCmec in MRSA252 may have been inserted into a 15580 MSSA background by a Ccr-mediated mechanism. This hypothesis is further supported by all isolates in the 15580 group belonging to ST30 or an ST30-related type (ST291), a single locus variant of the MRSA252 type (ST36), and a member of the same clonal complex (CC30) (11, 13). However, another possible mechanism for SCCmec acquisition is illustrated by MW2 and MSSA group 15575. We have previously shown not only that SCCmec could not be excised from MW2, but also that the sequences next to orfX were exactly duplicated at the non-orfX end of the inserted SCCmec element (25). This suggests acquisition by homologous recombination with insertion duplication of the homologue. There is abundant evidence for gain or loss of DNA in this region of the staphylococcal chromosome by recombination. Figures 3 and 4 show the common occurrence of similar genes and sequences at the attB site, with additional blocks of sequence that could have been gained or lost by recombination. In addition, there is considerable homology among the variants of type IV SCCmec, as well as between types IV and I SCCmec (12, 30). Figure 6 depicts multiple subtypes of SCCmec type IV, as well as SCCmec type I, all of which share a common core region but differ in blocks of sequence at the left and right ends that could have been gained or lost by recombination. All of these data may suggest that the attB region is not only a common site for acquisition of foreign DNA, but also fosters frequent recombinational rearrangement of acquired sequences.
FIG. 6.
DNA acquisition and deletion within SCCmec. A schematic of a generic SCCmec element is shown at the top, with block arrows representing selected open reading frames and flags indicating the left and right SCCmec attachment sites. The sequences of SCCmec types IV, IVa, IVb, IVc, IVe, IVg, and I are depicted in cartoon fashion (GenBank accession numbers NC_003923, AB063172, AB063173, AB096217, AJ810121, DQ106887, and NC_002951, respectively). Regions of the SCCmec elements with >90% nucleotide identity are filled with the same color/pattern, while different fill patterns indicate entirely different blocks of sequence. Each of these elements contains a core sequence including IS431, mecA, and a truncated mecRI (shown in gray). All SCCmec type IV elements share a region encoding ccrA2B2 (checkered), while SCCmec type I contains a homologous, yet distinct, region encoding ccrA1B1 (cross-hatched). The left and right ends of SCCmec vary among types and subtypes in a manner consistent with the insertion or deletion of large blocks of nonhomologous sequence.
This study suggests that the acquisition of SCCmec by integrase-mediated insertion at attB may be impacted by the genetic makeup of the region. However, it also suggests that acquisition of mecA may occur by mechanisms other than integrase-mediated insertion. Finally, acquisition and loss of mecA or SCCmec may be dynamic events, and the fingerprints found in this study may represent either isolates that have not yet acquired the genes or isolates that have acquired and lost them by excision or recombination. Other studies have shown SCCmec and/or mecA to confer fitness costs on the organism (7, 19). Therefore, the acquisition of SCCmec and mecA may be advantageous only to an organism in the presence of β-lactam antibiotics, and these genetic elements may be lost once the selective pressure is removed.
Acknowledgments
This work was supported by grant 5R01AI35705-13 from the National Institutes of Allergy and Infectious Diseases.
We thank Paige Fox, Qixun Zhao, and Alastair Monk for their technical assistance and input.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Anonymous. 2004. National Nosocomial Infections Surveillance (NNIS) System Report; data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32470-485. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers, H. F. 2003. Solving staphylococcal resistance to beta-lactams. Trends Microbiol. 11145-148. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, J. E., and E. J. Feil. 2006. The phylogeny of Staphylococcus aureus—which genes make the best intra-species markers? Microbiology 1521297-1305. [DOI] [PubMed] [Google Scholar]
- 6.Crum, N. F., R. U. Lee, S. A. Thornton, O. C. Stine, M. R. Wallace, C. Barrozo, A. Keefer-Norris, S. Judd, and K. L. Russell. 2006. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. Am. J. Med. 119943-951. [DOI] [PubMed] [Google Scholar]
- 7.Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bachi. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 482295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C. 2003. The evolution of a resistant pathogen—the case of MRSA. Curr. Opin. Pharmacol. 3474-479. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, P. M., M. W. Climo, and G. L. Archer. 2007. Lack of relationship between purine biosynthesis and vancomycin resistance in Staphylococcus aureus: a cautionary tale for microarray interpretation. Antimicrob. Agents Chemother. 511274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanssen, A. M., and J. U. Ericson Sollid. 2006. SCCmec in staphylococci: genes on the move. FEMS Immunol. Med. Microbiol. 468-20. [DOI] [PubMed] [Google Scholar]
- 13.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovis, K. M., J. V. McDowell, L. Griffin, and R. T. Marconi. 2004. Identification and characterization of a linear-plasmid-encoded factor H-binding protein (FhbA) of the relapsing fever spirochete Borrelia hermsii. J. Bacteriol. 1862612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 482637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 441549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1852711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama, Y., H. Z. Zhang, D. Hong, and H. F. Chambers. 2003. Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1855465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol 9870-876. [DOI] [PubMed] [Google Scholar]
- 21.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 1843623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 461147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 481823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355666-674. [DOI] [PubMed] [Google Scholar]
- 25.Noto, M. J., and G. L. Archer. 2006. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob. Agents Chemother. 502782-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 404289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira, D. C., C. Milheirico, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 503457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, L. B. 2006. Antimicrobial resistance in gram-positive bacteria. Am. J. Infect Control 34S11-S199, S64-S73. [DOI] [PubMed] [Google Scholar]
- 29.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39309-317. [DOI] [PubMed] [Google Scholar]