Abstract
The coenzyme A (CoA)-activated C5-dicarboxylic acids mesaconyl-CoA and β-methylmalyl-CoA play roles in two as yet not completely resolved central carbon metabolic pathways in bacteria. First, these compounds are intermediates in the 3-hydroxypropionate cycle for autotrophic CO2 fixation in Chloroflexus aurantiacus, a phototrophic green nonsulfur bacterium. Second, mesaconyl-CoA and β-methylmalyl-CoA are intermediates in the ethylmalonyl-CoA pathway for acetate assimilation in various bacteria, e.g., in Rhodobacter sphaeroides, Methylobacterium extorquens, and Streptomyces species. In both cases, mesaconyl-CoA hydratase was postulated to catalyze the interconversion of mesaconyl-CoA and β-methylmalyl-CoA. The putative genes coding for this enzyme in C. aurantiacus and R. sphaeroides were cloned and heterologously expressed in Escherichia coli, and the proteins were purified and studied. The recombinant homodimeric 80-kDa proteins catalyzed the reversible dehydration of erythro-β-methylmalyl-CoA to mesaconyl-CoA with rates of 1,300 μmol min−1 mg protein−1. Genes coding for similar enzymes with two (R)-enoyl-CoA hydratase domains are present in the genomes of Roseiflexus, Methylobacterium, Hyphomonas, Rhodospirillum, Xanthobacter, Caulobacter, Magnetospirillum, Jannaschia, Sagittula, Parvibaculum, Stappia, Oceanicola, Loktanella, Silicibacter, Roseobacter, Roseovarius, Dinoroseobacter, Sulfitobacter, Paracoccus, and Ralstonia species. A similar yet distinct class of enzymes containing only one hydratase domain was found in various other bacteria, such as Streptomyces species. The role of this widely distributed new enzyme is discussed.
The phototrophic Chloroflexus aurantiacus, a thermophilic green nonsulfur bacterium, lives in hot sulfur-containing springs of neutral to slightly alkaline pH (6, 8). There, it forms visible orange microbial mats and is the dominant organism that grows photoautotrophically (43). However, in the dark or in the presence of organic substrates in the light, the bacterium switches to heterotrophic growth. This niche seems to favor a special type of central carbon metabolism that allows a flexible response to the available carbon and electron sources. There are different lines of evidence that autotrophic CO2 fixation proceeds via the 3-hydroxypropionate cycle (12, 22, 25, 39, 40, 43). This new metabolic pathway uses bicarbonate rather than CO2 and allows the simultaneous assimilation of small organic fermentation products, such as acetate, propionate, or succinate, which may be produced by fermenting bacteria underneath the microbial mat.
In brief, in a first cycle, one acetyl-coenzyme A (CoA) and two bicarbonate molecules are converted to l-malyl-CoA by a series of steps driven by ATP as the energy source and NADPH as the reductant (1, 15, 21, 27, 39) (Fig. 1A). Bicarbonate fixation proceeds via acetyl-CoA and propionyl-CoA carboxylation. l-Malyl-CoA is then cleaved by l-malyl-CoA lyase to glyoxylate and acetyl-CoA, thus closing the first cycle (21, 39). In a second cycle, glyoxylate and propionyl-CoA are disproportionated to acetyl-CoA and pyruvate, which can be used as a precursor for biosynthesis (12, 22, 23). In a first reaction, propionyl-CoA and glyoxylate are condensed to β-methylmalyl-CoA by the same lyase that catalyzes the last reaction of the first cycle (21, 22). β-Methylmalyl-CoA is dehydrated to mesaconyl-CoA by a postulated β-methylmalyl-CoA dehydratase/mesaconyl-CoA hydratase. The subsequent enzymes in the pathway, converting β-methylmalyl-CoA via mesaconyl-CoA to citramalate, have not been characterized. Citramalate appears to be activated by CoA transfer to the corresponding thioester, followed by the cleavage of citramalyl-CoA into acetyl-CoA and pyruvate (16, 17). Thus, three molecules of bicarbonate are converted to one molecule of pyruvate. Most of the involved enzymes catalyze freely reversible reactions, and only very few pace-making enzymes catalyze virtually irreversible reactions.
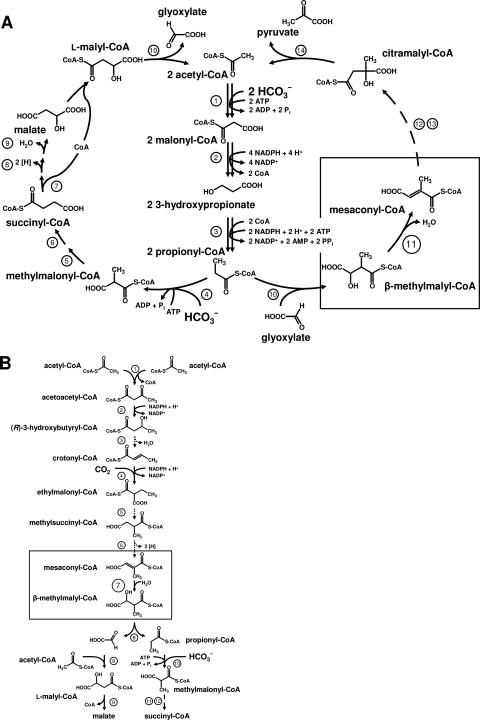
FIG. 1.
Proposed roles of mesaconyl-CoA hydratase/β-methylmalyl-CoA dehydratase in two bacterial pathways of central carbon metabolism. The boxes mark the postulated reactions catalyzed by the enzyme. (A) Proposed 3-hydroxypropionate cycle of autotrophic CO2 fixation in C. aurantiacus. 1, acetyl-CoA carboxylase; 2, malonyl-CoA reductase; 3, propionyl-CoA synthase; 4, propionyl-CoA carboxylase; 5, methylmalonyl-CoA epimerase; 6, methylmalonyl-CoA mutase; 7, succinyl-CoA:l-malate CoA transferase; 8, succinate dehydrogenase; 9, fumarate hydratase; 10, l-malyl-CoA lyase/β-methylmalyl-CoA lyase; 11, mesaconyl-CoA hydratase; 12, unknown reactions; 13, succinyl-CoA:citramalate CoA-transferase; 14, citramalyl-CoA-lyase. (B) Proposed alternate glyoxylate cycle for acetate assimilation in isocitrate-lyase-negative bacteria, such as R. sphaeroides. This pathway, termed the ethylmalonyl-CoA pathway, also serves to assimilate acetyl-CoA produced from methanol and CO2 in methylotrophic bacteria, such as M. extorquens, which uses the serine cycle for formaldehyde fixation during growth on methanol. 1, β-ketothiolase; 2, acetoacetyl-CoA reductase; 3, crotonase; 4, crotonyl-CoA carboxylase/reductase; 5, ethylmalonyl-CoA mutase; 6, methylsuccinyl-CoA dehydrogenase; 7, mesaconyl-CoA hydratase; 8, l-malyl-CoA lyase/β-methylmalyl-CoA lyase; 9, malyl-CoA hydrolase; 10, propionyl-CoA carboxylase; 11, methylmalonyl-CoA epimerase; 12, methylmalonyl-CoA mutase.
The assimilation of acetate requires a separate pathway to replenish intermediates of the citric acid cycle used for cell carbon biosynthesis. Over the last 5 decades, several organism have been shown not to use the glyoxylate cycle for acetate assimilation (3, 4, 10, 19, 20, 29, 34). This was revealed by the lack of isocitrate lyase activity in cell extracts of these organisms or by the absence of the corresponding gene in recently determined genome sequences. Examples are Rhodobacter sphaeroides, Methylobacterium extorquens, and Rhodospirillum rubrum. Other organisms apparently contain an isocitrate lyase gene but do not express it under certain growth conditions, even when grown on acetate. Examples are Rhodobacter capsulatus (34), Streptomyces collinus (20), and Paracoccus versutus (formerly Thiobacillus versutus) (9).
We postulated a new pathway for acetate assimilation termed the ethylmalonyl-CoA pathway, which consists of two parts (2, 13, 14) (Fig. 1B). In the first part, two molecules of acetyl-CoA are converted to a C4 compound, as suggested for M. extorquens (30). The C4 intermediate, crotonyl-CoA, is reductively carboxylated to ethylmalonyl-CoA. Ethylmalonyl-CoA appears to be converted via methylsuccinyl-CoA to mesaconyl-CoA. A gene was identified by transposon mutagenesis in R. sphaeroides and proposed to encode an enzyme that hydrates mesaconyl-CoA to β-methylmalyl-CoA (2). The mutant no longer grew on acetate as a sole carbon source; however, growth was rescued by the addition of glyoxylate. The effect of glyoxylate was expected, since β-methylmalyl-CoA is cleaved to glyoxylate and propionyl-CoA. The second part of the proposed ethylmalonyl-CoA pathway consists of propionyl-CoA carboxylation and conversion to succinate by conventional reactions; glyoxylate is assimilated by condensation with another molecule of acetyl-CoA to form l-malyl-CoA or l-malate. In methylotrophs growing with methanol, glyoxylate is converted to glycine, which serves as an acceptor molecule for formaldehyde assimilation.
Hence, the interconversions of mesaconyl-CoA and β-methylmalyl-CoA play important roles in the two pathways of central carbon metabolism. In the autotrophic pathway, β-methylmalyl-CoA dehydration to mesaconyl-CoA is required, whereas in the acetate assimilation pathway, the reverse reaction, mesaconyl-CoA hydration to β-methylmalyl-CoA, is essential.
In this investigation, we cloned and heterologously expressed the putative genes for mesaconyl-CoA hydratase from C. aurantiacus and R. sphaeroides and purified the recombinant proteins. Indeed, they both catalyze the postulated hydration/dehydration reaction (enoyl-CoA hydratase reaction). The identification and characterization of this enzyme represents an important argument in favor of the proposed new pathways.
MATERIALS AND METHODS
Materials.
Chemicals were obtained from Fluka (Neu-Ulm, Germany), Sigma-Aldrich (Deisenhofen, Germany), Merck (Darmstadt, Germany), Serva (Heidelberg, Germany), or Roth (Karlsruhe, Germany). Biochemicals were from Roche Diagnostics (Mannheim, Germany), Applichem (Darmstadt, Germany), or Gerbu (Craiberg, Germany). Materials for cloning and expression were purchased from MBI Fermentas (St. Leon-Rot, Germany), New England Biolabs (Frankfurt, Germany), Novagen (Schwalbach, Germany), Genaxxon Bioscience GmbH (Biberach, Germany), MWG Biotech AG (Ebersberg, Germany), or Qiagen (Hilden, Germany). Materials and equipment for protein purification were obtained from GE Healthcare (Freiburg, Germany) or Millipore (Eschborn, Germany). [1-14C]propionate and [U-13C]propionate were obtained from Hartmann Analytic (Braunschweig, Germany) and Cambridge Isotope Laboratories, Inc. (Andover, MA), respectively.
Bacteria and growth conditions.
C. aurantiacus strain OK-70-fl (DSMZ 636) was grown phototrophically in a 12-liter glass fermentor as described before (27). Cells were also grown anaerobically under photoheterotrophic conditions on modified minimal medium D supplemented with 0.25% (wt/vol) Casamino Acids, 0.1% (wt/vol) yeast extract, and trace elements (8). The cells were stored under liquid nitrogen until they were used. E. coli strain BL-21(DE3) (41), strain Sure, and strain DH5α were grown at 37°C in Luria-Bertani (LB) medium (36). Ampicillin was added to E. coli cultures to a final concentration of 0.1 mg/ml. R. sphaeroides strain 2.4.1 (DSMZ 158) was grown anaerobically in the light (5,000 lx) in 2-liter bottles at pH 6.7 and 30°C on minimal medium (2) containing 10 mM acetate or succinate. For large-scale production of recombinant enzymes, E. coli BL-21 was grown at 37°C in a 200-liter fermentor in LB medium containing 0.1 mg/ml ampicillin, and the cells were induced for 4 h at an optical density of 0.6 to 0.8 by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Syntheses.
Propionyl-CoA was synthesized by different methods, depending on the purpose.
(i) Unlabeled propionyl-CoA.
Propionyl-CoA was synthesized from its anhydride (37, 38) by a modified method previously described (21), and the dry powder was stored at −20°C.
(ii) [1-14C]propionyl-CoA.
[1-14C]propionyl-CoA was synthesized enzymatically using 3-hydroxypropionyl-CoA synthetase from Metallosphaera sedula (B. Alber and G. Fuchs, unpublished results). The reaction mixture (1 ml) contained 100 mM 2-(N-morpholino)propanesulfonic acid/KOH (MOPS-K+) buffer (pH 8.0), 5 mM MgCl2, 5 mM ATP, 0.3 U (μmol min−1) of 3-hydroxypropionyl-CoA synthetase, 2 mM [1-14C]propionate (0.5 MBq), and 0.3 mM CoA. The reaction was carried out at 70°C. CoA was replenished five times after consumption because of the inhibiting effect of higher concentrations on the enzyme. The consumption of CoA was monitored spectrophotometrically at 412 nm using Ellman's reagent: a test mixture (25 μl) contained 12.5 μl 200 mM MOPS-K+ (pH 7.5), 6.25 μl H2O, 1.25 μl Ellman's reagent [10 mM 5-5′-dithiobis(2-nitrobenzoic acid)], 0.5 M potassium phosphate (pH 7.2), 1 mM EDTA, and 5 μl of reaction mixture. The synthesis of propionyl-CoA was stopped by adding 50 μl of 99% formic acid. The precipitated protein was removed by centrifugation. The reaction mixture was purified of ATP and free acids by application to a Strata-X 33-μm polymeric reversed-phase column (500 mg; 6 ml; Phenomenex Ltd., Aschaffenburg, Germany). The column was conditioned with 4 ml of 100% methanol and equilibrated with 6 ml 2% methanol in 40 mM K2HPO4/50 mM formic acid (pH 4.2). After the reaction mixture was added, the column was washed with 6 ml of 2% methanol in 40 mM K2HPO4/formic acid (pH 4.2). [1-14C]propionyl-CoA was eluted with 4 ml of 80% methanol, dried in a Speedvac concentrator, and stored at −20°C.
(iii) [1,2,3-13C]propionyl-CoA.
[1,2,3-13C]propionyl-CoA was synthesized enzymatically using 3-hydroxypropionyl-CoA synthetase from M. sedula. The reaction mixture (5 ml) contained 100 mM MOPS-K+ buffer (pH 8.0), 5 mM MgCl2, 5 mM ATP, 1.5 U of 3-hydroxypropionyl-CoA synthetase, 2 mM [1,2,3-13C]propionate, traces of [1-14C]propionate (0.1 MBq), and 0.3 mM CoA. The reaction was carried out at 70°C. CoA was replenished five times after consumption, as described above. The reaction was stopped by the addition of 250 μl of 99% formic acid. The precipitated protein was removed by centrifugation. The reaction mixture was purified of ATP and free acids by application to a Strata-X 33-μm polymeric reversed-phase column (1 g; 12 ml; Phenomenex Ltd., Aschaffenburg, Germany). The column was equilibrated with 12 ml of 2% methanol in 20 mM ammonium formate (pH 4.0). After the reaction mixture was added, the column was washed with 20 ml of 2% methanol in 20 mM ammonium formate, pH 4. [1,2,3-13C]propionyl-CoA was eluted with 8 ml of 80% methanol, dried in a Speedvac concentrator, and stored at −20°C.
Preparation of cell extract.
Cells were suspended in a onefold volume of 50 mM MOPS-K+ buffer (pH 7.5) containing 4 mM MgCl2 and 0.2 mg DNase I per ml of cell suspension and passed twice through a chilled French pressure cell at 137 kPa. The lysate was ultracentrifuged for 1 h at 100,000 × g at 4°C. Extracts (100,000 × g supernatant) were stored at −20°C in the presence of 20% glycerol.
Enzyme assays.
Enzyme assays were performed at 30°C (R. sphaeroides) or 55°C (C. aurantiacus). One unit corresponds to 1 μmol substrate converted per minute.
(i) HPLC analysis.
The routine assay mixture (0.4 ml) contained 100 mM MOPS-K+ buffer (pH 7.5), 4 mM MgCl2, 2.5 mM propionyl-CoA, traces of [1-14C]propionyl-CoA (50 kBq), 15 mM glyoxylate, and recombinant l-malyl-CoA/β-methylmalyl-CoA lyase. After 10 min of preincubation at 55°C, a 100-μl sample was withdrawn, and the reaction was started by addition of the enzyme. In the case of the Rhodobacter enzyme, the reaction mixture was cooled to 30°C before the addition of the enzyme. After 10 min, an additional 100-μl sample was withdrawn. The reaction was stopped by the addition of 10 μl of ≥99% formic acid. The precipitated protein was removed by centrifugation, and the supernatants were analyzed for CoA thioesters by high-pressure liquid chromatography (HPLC) using a reversed-phase column (LiChroCART 125-4 RP 18e; end capped; 5 μm; 125 by 4 mm; Merck, Darmstadt, Germany). A 30-ml gradient from 1 to 10% acetonitrile in 40 mM K2HPO4/50 mM formic acid buffer (pH 4.2) with a flow rate of 1 ml min−1 was used. CoA thioesters were detected at 260 nm. Retention times were as follows: free organic acids, 2 to 5 min; CoA, 14 min; β-methylmalyl-CoA, 16 min; mesaconyl-CoA, 19 min; and propionyl-CoA, 28 min.
(ii) Coupled spectrophotometric assay.
Transformations were followed spectrophotometrically at 290 nm. Preincubation for 15 min at 55°C with recombinant l-malyl-CoA/β-methylmalyl-CoA lyase, excess of glyoxylate, and saturating concentrations of propionyl-CoA were used to generate the substrate β-methylmalyl-CoA from propionyl-CoA and glyoxylate. We used an estimated differential absorption coefficient at 290 nm (Δɛ290) (mesaconyl-CoA minus methylmalyl-CoA) of 2,150 M−1 cm−1 for the spectrophotometric determination of mesaconyl-CoA formation. The routine assay mixture (0.5 ml) contained 200 mM MOPS/K+ buffer (pH 7.5), 4 mM MgCl2, recombinant l-malyl-CoA/β-methylmalyl-CoA lyase (non-rate limiting), 10 mM glyoxylate, 2.5 mM propionyl-CoA, and protein fraction. Either substrate could be used to start the reaction in a controlling assay. The buffers used to determine the pH optimum were 140 mM 2-(N-morpholino)ethanesulfonic acid (MES)/K+ buffer (pH 5.5 to 6.5), 140 mM MOPS/K+ buffer (pH 6.5 to 7.5), and 140 mM N-[Tris-(hydroxymethyl)-methyl]-3-aminopropane sulfonic acid/K+ buffer (pH 7.5 to 9.0).
Conversion of propionyl-CoA and glyoxylate, sample preparation, and product analysis.
To purify and identify the product of the reaction, the routine assay was modified.
(i) Samples for HPLC-ESI-MS analysis.
The reaction mixture (0.4 ml) contained 15 mM NH4HCO3/HCOOH, pH 7.5, 3 mM MgCl2, 2.5 mM propionyl-CoA, 7.5 mM glyoxylate, 0.07 U l-malyl-CoA/β-methylmalyl-CoA lyase, and 2.7 U mesaconyl-CoA hydratase. The reaction was started by adding glyoxylate. After 10 min of incubation, a 0.2-ml sample was withdrawn, 10 μl of ≥99% formic acid was added on ice, protein was removed by centrifugation, and the supernatant was used directly for electron spray ionization-mass spectrometry (ESI-MS) analysis (HPLC solvent system 3).
(ii) Sample preparation for TLC and TLC analysis.
To the reaction mixture (0.2 ml) as described above, 25 kBq of [1-14C]propionyl-CoA was added. Samples (10 μl) were retrieved at intervals, the reaction was stopped by adding 1 μl of 1% formic acid on ice, and the protein precipitate was removed by centrifugation. The supernatant was dried, and the residue was treated with 10 μl of 0.1 M KOH for 40 min at 75°C. Then, 1 μl of 5% formic acid was added, and 5 μl was analyzed by thin-layer chromatography (TLC). As a control, mesaconate, citraconate, and itaconate were treated in the same way as the enzyme assay samples. TLC was performed using silica gel 60 F254 plates (20 by 20 cm; Merck, Darmstadt, Germany) and diisopropyl ether/formic acid/water (90/7/3 [vol/vol/vol]) as a solvent (2-h development at room temperature). Authentic compounds (0.2 μmol) were cochromatographed. The Rf values were as follows: mesaconate, 0.84; citraconate, 0.25; itaconate, 0.52. The plates were dried for 30 min at 90°C; then, radioactivity was detected by phosphorimaging using FUJI-BAS100X phosphorimager plates. Acid spots were detected by spraying the plates with 0.05% (wt/vol) bromocresol green in ethanol/water (1/4 [vol/vol]).
(iii) Samples for NMR analysis.
A reaction mixture of 10 ml contained 15 mM NH4HCO3/HCOOH, pH 7.5, 3 mM MgCl2, 2.5 mM [1,2,3-13C]propionyl-CoA, 400 kBq [1-14C]propionyl-CoA, 7.5 mM glyoxylate, 1.2 U l-malyl-CoA/β-methylmalyl-CoA lyase, and 45 U mesaconyl-CoA hydratase. The reaction was started by adding glyoxylate. After 20 min of incubation, 250 μl of 4% formic acid was added to obtain a pH of 4. The sample was put on ice, and then it was centrifuged at 15,800 × g, and the supernatant was applied to a 5-g Isolute SPE C18 column (70 ml; end capped; Separtis GmbH, Grenzach-Wyhlen, Germany), which was run at a flow rate of 0.8 to 1.2 ml min−1. The column was equilibrated with 120 ml of 1% methanol in 20 mM ammonium formate/formic acid buffer, pH 4.0, the sample was applied, the column was washed with 100 ml of this buffer, and CoA esters were eluted with 100 ml of 80% aqueous methanol. The eluate was reduced to 10 ml in a vacuum rotary evaporator and then lyophilized. The dry powder was used for nuclear magnetic resonance (NMR) analysis. The reaction course and the purification were followed in parallel by HPLC analysis and scintillation counting of radioactivity in the fractions. Purification of CoA thioesters resulted in a 70 to 80% yield.
(iv) HPLC systems.
A C18 column (Grom-Sil 120 ODS-4 HE; 5 μm; 125 by 4 mm; Merck) was used, combined with photodiode detection and flowthrough radioactivity detection using solid scintillation detection (Ramona 2000; Raytest, Straubenhardt, Germany). The output signal of solid scintillation detection yields arbitrary radioactivity units that can be normalized when the amount of radioactivity in a compound applied to the HPLC column is known from liquid scintillation counting. The reaction rate was calculated by using the 260-nm absorption integrals of the individual CoA thioester peaks. Calculations were based on the assumption that the molar 260-nm absorption coefficient of the thioesters was 16,400 M−1 cm−1, as for acetyl-CoA (11). With [14C]propionyl-CoA as a precursor, calculations were based on the radioactivity integrals of the substrate and the products.
(a) Solvent 1.
Specifications for solvent 1 were as follows: 40 ml acetonitrile gradient, 2 to 10% (vol/vol) in 40 mM K2HPO4/HCOOH, pH 4.2; flow rate, 1 ml min−1; retention times, CoA, 16.7 min, β-methylmalyl-CoA, 20.4 min, mesaconyl-CoA, 25.0 min, and propionyl-CoA, 33.7 min. This system was used for routine product analysis.
(b) Solvent 2.
Specifications for solvent 2 were as follows: 40 ml acetonitrile gradient, 2 to 10% (vol/vol) in 40 mM ammonium acetate buffer, pH 6.7; flow rate, 1 ml min−1; retention times, β-methylmalyl-CoA, 10.9 min, mesaconyl-CoA, 11.6 min, propionyl-CoA, 26.2 min. This system was used to determine the pH dependence of the HPLC separation system.
(c) Solvent 3.
Specifications for solvent 3 were as follows: 32 ml acetonitrile gradient, 2 to 10% (vol/vol) in 40 mM ammonium acetate buffer, pH 4.2; flow rate, 0.8 ml min−1. This system was used for ESI-MS analysis of the product.
NMR spectroscopy and liquid chromatography-MS.
NMR spectra were recorded with a Bruker Avance DRX-400 spectrometer at 27°C in methanol-D4 as the solvent. Chemical shifts were recorded and reported in ppm relative to methanol-d4 (1H: δ = 3.31, 13C: δ = 49.15) as the internal standard. Inadequate measurements were performed with the following values: SF = 100.624 MHz, D1 = 6 s, Aq = 0.1 s, PW90 = 8.8 μs −3 db, NS = 160, TD − F2 = 4,096, and TD − F1 = 128.
HPLC-MS was performed on an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany) interfaced with an Applied Biosystems API 2000 triple quadrupole (Applied Biosystems, Foster City, CA) using the separation conditions described above. The temperature of the Turbo-Ionspray auxiliary gas was 430°C, and the ionization voltage was −5,500 V. The samples were analyzed with a mass range of 100 to 1,200 Da.
Heterologous expression and purification of recombinant enzymes. (i) Recombinant l-malyl-CoA/β-methylmalyl-CoA lyase (Mcl) from C. aurantiacus.
The mcl gene was cloned in E. coli DH5α as described below. The recombinant enzyme was purified as described elsewhere (21).
(ii) Recombinant mesaconyl-CoA hydratases.
The mesaconyl-CoA hydratase (mch) genes were heterologously expressed in E. coli BL-21, and the recombinant N-terminally His10-tagged enzymes were purified.
Chloroflexus enzyme.
Extract of 3 g (fresh weight) E. coli cells (100,000 × g supernatant) was heat precipitated for 10 min at 70°C, followed by centrifugation at 15,800 × g.The supernatant was applied at a flow rate of 1 ml min−1 to a 1-ml Ni-Sepharose Fast Flow Column (HisTrap FF; GE Healthcare), which was equilibrated with buffer A (20 mM MOPS-K+ buffer, pH 7.4, containing 250 mM KCl). The column was washed with buffer A at a flow rate of 1 ml min−1. A step of 100 mM imidazole in buffer A was used to elute unwanted proteins. The His10-tagged enzyme was eluted with 500 mM imidazole in buffer A.
Rhodobacter enzyme.
Extract of 3 g (fresh weight) E. coli cells (100,000 × g supernatant) was directly applied to the affinity column and treated as described above. Active fractions were combined and stored in the presence of 20% glycerol at −20°C.
Gel filtration chromatography was used to estimate the native molecular mass. A 120-ml HiLoadSuperdex 200 16/60 column (GE Healthcare) was run with a flow rate of 1.5 ml min−1; the running buffer was 50 mM MOPS-K+ buffer (pH 7.2), 150 mM KCl. The column was calibrated with blue dextran (2,000 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (66 kDa), and vitamin B12 (1.5 kDa).
Cloning of genes in E. coli BL-21(DE3).
Standard protocols were used for purification, preparation, cloning, transformation, and amplification of DNA (5, 36). Plasmid DNA was isolated with the QIAprep Spin Miniprep Kit (Qiagen). Oligonucleotides (restriction sites are in boldface) were designed upstream (5′-AGTCCGTCATATGAGCGCTAAAACC-3′; NdeI) and downstream (5′-TAGAGGATCCCGACCAGTCATCC-3′; BamHI) of the C. aurantiacus mch gene. Oligonucleotides were designed upstream (5′-CAAGCTGGGAGACCACCATATGAAGAC-3′; NdeI) and downstream (5′-GTGATCACAAGCTTCGGGCCTGTG-3′; HindIII) of the R. sphaeroides mch gene. Pfu polymerase (Genaxxon; 1 μl per 40-μl assay mixture) was used for PCR. PCR conditions for the mch gene from C. aurantiacus were as follows: 25 cycles of 45 s of denaturation at 95°C, 1 min of primer annealing at 55°C, and 3 min of elongation at 72°C. The PCR-amplified DNA was treated with NdeI and BamHI, and the mch gene was ligated into pET16b. PCR conditions for the mch gene from R. sphaeroides were as follows: 25 cycles of 45 s of denaturation at 95°C, 60 s of primer annealing at 55°C, and 2 min of elongation at 72°C. The PCR-amplified DNA was treated with NdeI and HindIII, and the mch gene was ligated into pET16b. The plasmids were transformed into E. coli DH5α and then into E. coli BL-21. For the l-malyl-CoA/β-methylmalyl-CoA lyase (mcl) gene from C. aurantiacus, two oligonucleotides (restriction sites are in boldface) were designed upstream (5′-CGTATGCACTCCCGGGAATGATGAG-3′; SmaI) and downstream (5′-CCTTGCTGCAGCGTCACAGAC-3′; PstI) of the gene. PCR conditions were as follows: 25 cycles of 4 min of denaturation at 95°C, 1 min of denaturation at 95°C, 60 s of primer addition at 56°C, 150 s of synthesis at 72°C, 60 s of primer addition at 56°C, 10 min of synthesis at 72°C, and a pause at 4°C. The PCR-amplified DNA was treated with SmaI and PstI, and the mcl gene was ligated into pTrc99A (42). The construct, as well as the original plasmid, was transformed into E. coli DH5α.
DNA sequencing and computer analysis.
Inserts of both constructs used for expression were fully sequenced by G. L. Igloi (Institut Biologie II, Universität Freiburg, Germany). DNA and amino acid sequences were analyzed by the BLAST network service at the National Center for Biotechnology Information (Bethesda, MD), on the local C. aurantiacus server (http://genome.jgi-psf.org/draft_microbes/chlau/chlau.home.html) at the Department of Energy Joint Genome Institute (Walnut Creek, CA), and with the program Clone Manager 7 (SciEd Software, Cary, NC).
Other methods.
To quantify radioactivity in TLC, the labeled spots (detected by phosphorimaging) were scratched off the plates and the powder (or any liquid sample) was carefully suspended by shaking it in 3 ml Rotiscint 2200 cocktail (Roth, Karlsruhe, Germany). 14C was measured by liquid scintillation counting using external standardization. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5%) was performed as described by Laemmli (32). Molecular mass standards were as follows: rabbit phosphorylase b, 97 kDa; bovine serum albumin, 67 kDa; egg ovalbumin, 45 kDa; lactate dehydrogenase, 34 kDa; carbonic anhydrase, 29 kDa; and lysozyme, 14 kDa. Proteins were visualized by Coomassie blue staining (44). Protein was measured by the method of Bradford (7) using bovine serum albumin as a standard.
RESULTS
Identification of the putative mesaconyl-CoA hydratase gene.
Previous work on the novel pathway of acetate assimilation in R. sphaeroides identified an essential gene that codes for an enzyme of the (R)-enoyl-CoA hydratase family (2). Similar genes were found in other bacteria that have in common with R. sphaeroides the absence of a functional glyoxylate cycle for acetate assimilation. However, the overall sequence identity of these proteins to the characterized member of this so-called MaoC or FkbR2 family, an R-specific 3-hydroxyacyl-CoA dehydratase from Aeromonas caviae (18), is only in the range of 20%. Furthermore, the expected molecular mass of the encoded enzyme is about 38 kDa, which is larger than the masses of normal enoyl-CoA hydratases (14 to 28 kDa). A similar gene was found in C. aurantiacus next to the gene coding for l-malyl-CoA/β-methylmalyl-CoA lyase. The gene products from C. aurantiacus and R. sphaeroides had 58% identical amino acid sequences. They showed two hydratase domains, which exhibited only 25% identity on the amino acid level. Normally, enoyl-CoA hydratases contain only one such domain. This suggests gene duplication and also explains the larger size of the subunits compared to other enoyl-CoA hydratases.
Cloning and expression.
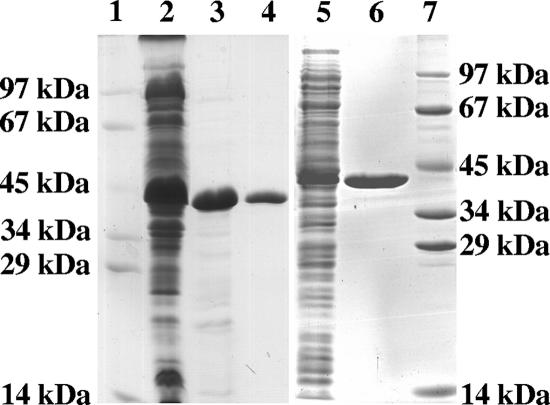
The putative 1.06-kb Chloroflexus mesaconyl-CoA hydratase gene coded for a 38-kDa protein (352 amino acids), and the 1.02-kb Rhodobacter gene coded for a 37-kDa protein (343 amino acids). The genes from both bacteria were amplified, and the expected PCR products were cloned into the expression vector pET16b, resulting in coding for N-terminal His10-tagged proteins with altered molecular masses of about 40 kDa. Both plasmids were transformed into E. coli DH5α and then transformed into E. coli BL-21(DE3) for expression. As a control, the expression vector without an insert was also transformed. Both genes were heterologously expressed, and the enzymes were soluble, as deduced from an induced protein band around 40 kDa in SDS-PAGE of the soluble cell fraction (Fig. 2).
FIG. 2.
Denaturing PAGE (12.5%) of various purification steps of heterologously expressed mesaconyl-CoA hydratase of C. aurantiacus and R. sphaeroides from extracts of 3 g E. coli cells. Lanes 1 and 7, molecular mass standards; lanes 2 to 4, Chloroflexus enzyme; lane 2, extract (100,000 × g supernatant) of induced E. coli cells (45 μg protein); lane 3, heat precipitation fraction (20 μg protein); lane 4, affinity chromatography fraction (8 μg protein); lanes 5 and 6, Rhodobacter enzyme; lane 5, extract (100,000 × g supernatant) of induced E. coli cells (30 μg protein); lane 6, affinity chromatography fraction (8 μg protein). The gel was stained with Coomassie brilliant blue R-250.
Spectrophotometric enzyme assay.
The recombinant l- malyl-CoA/β-methylmalyl-CoA lyase from C. aurantiacus was used in a coupled spectrophotometric assay to enzymatically convert propionyl-CoA and glyoxylate to β-methylmalyl-CoA, the substrate of the postulated mesaconyl-CoA hydratase/β-methylmalyl-CoA dehydratase. The further transformation of β-methylmalyl-CoA to other coenzyme A thioesters was tested with E. coli extracts containing overproduced putative mesaconyl-CoA hydratase. Both extracts, which contained either the Chloroflexus or the Rhodobacter gene product, catalyzed a reaction that was associated with an increase in absorbance above 260 nm, with a maximum at around 284 nm. Because protein absorption interfered at this wavelength, we used 290 nm to follow the reaction spectrophotometrically (Fig. 3). The reaction was dependent on propionyl-CoA, glyoxylate, and l-malyl-CoA/β-methylmalyl-CoA lyase. In contrast, extracts of wild-type E. coli or of E. coli carrying the plasmid without an insert were inactive.
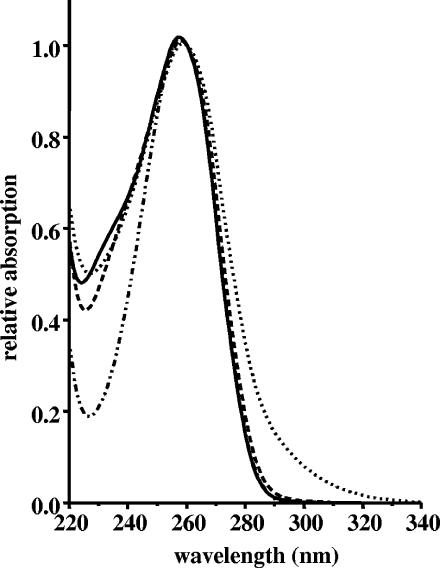
FIG. 3.
UV spectra of coenzyme A (.._..), propionyl-CoA (—), β-methylmalyl-CoA (—), and the product of the hydratase reaction, mesaconyl-CoA (…….). The spectra were recorded in 40 mM K2HPO4/formic acid buffer, pH 4.2, and were normalized to the same absorption at 260 nm.
Confirmation of a β-methylmalyl-CoA-transforming enzyme activity.
HPLC analysis revealed that β-methylmalyl-CoA was consumed almost completely and a new product was formed (Fig. 4). At equilibrium, the ratio of the concentrations of β-methylmalyl-CoA to the product was near 1:10. The product still had the 260-nm adenine nucleotide absorbance maximum characteristic of CoA, propionyl-CoA, or β-methylmalyl-CoA. However, it had an additional absorbance shoulder at longer wavelengths (Fig. 3). The estimated molar 290-nm absorption coefficient of this product (most likely mesaconyl-CoA) was 2,350 M−1 cm−1, that of the substrate β-methylmalyl-CoA was 200 M−1 cm−1, and that of propionyl-CoA was approximately 200 M−1 cm−1. This estimation is based on the assumption that the molar absorption coefficients at 260 nm (ɛ260) of the substrate and the product are very similar to that of normal CoA esters, such as acetyl-CoA (16,400 M−1 cm−1; for comparison, the ɛ260 of CoA is 16,800 M−1 cm−1) (11). The Δɛ290 (mesaconyl-CoA minus β-methylmalyl-CoA) of 2,150 M−1 cm−1 was used to quantify the β-methylmalyl-CoA consumption and product formation reaction in the spectrophotometric assay.
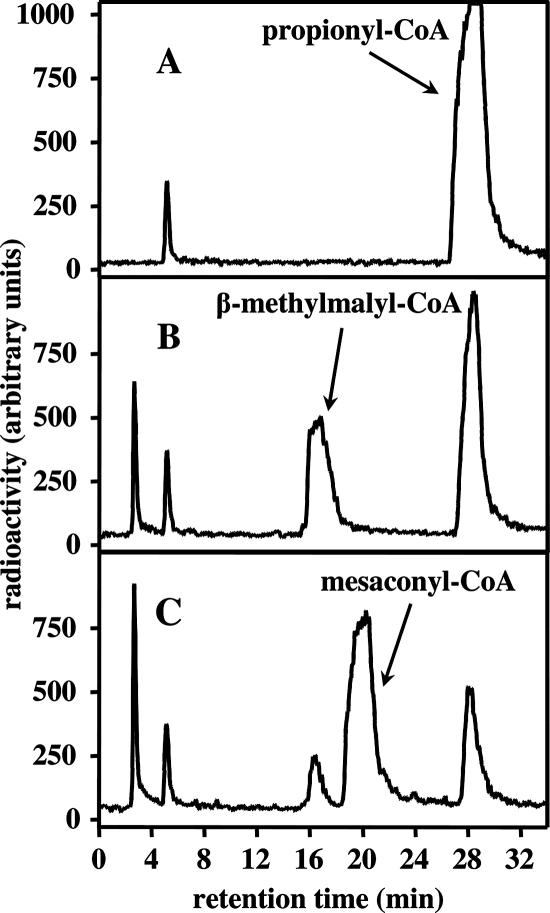
FIG. 4.
HPLC separation of [14C]mesaconyl-CoA as the product of the mesaconyl-CoA hydratase from [14C]β-methylmalyl-CoA and [14C]propionyl-CoA. [14C]β-methylmalyl-CoA is enzymatically formed from [14C]propionyl-CoA and glyoxylate with recombinant l-malyl-CoA/β-methylmalyl-CoA lyase. CoA and its derivatives were detected at 260 nm. Free acids of the corresponding CoA thioesters were eluted between 2 and 5 min. (A) Before addition of glyoxylate to the assay mixture. (B) Ten minutes after addition of glyoxylate. (C) Formation of mesaconyl-CoA after another 10 minutes of incubation with recombinant mesaconyl-CoA hydratase. The sensitivities of 14C detection by solid-phase scintillation counting in the different runs were the same, and therefore, the signals can be directly compared. For the conditions, see Materials and Methods.
Enzyme activity in cell extract of C. aurantiacus.
Using excess amounts of recombinant l-malyl-CoA/β-methylmalyl-CoA lyase from C. aurantiacus, saturating concentrations of [14C]propionyl-CoA, and an excess of glyoxylate, the specific activity of the enzyme system that produced labeled mesaconyl-CoA from labeled propionyl-CoA in cell extract of C. aurantiacus could be estimated (55°C). The specific enzyme activity in extracts of autotrophically grown cells amounted to 20 to 30 nmol min−1 mg protein−1, depending on the batch of cells. The specific enzyme activity was 10- to 15-fold lower in heterotrophically grown cells.
Purification of recombinant enzymes.
Most of the protein overproduced in E. coli was in the soluble cell fraction, and purification started from the 100,000 × g supernatant. For the recombinant Chloroflexus enzyme, E. coli extract was heated for 10 min at 70°C; the enzyme from this moderate thermophile remained active and in the supernatant (Fig. 2). The supernatant (or cell extract in the case of the recombinant Rhodobacter enzyme) was then chromatographed on an Ni-Sepharose high-performance affinity column. From 3 g E. coli cells (wet weight), 6.4 mg of Chloroflexus enzyme with a yield of 87% or 4 mg of Rhodobacter enzyme with a yield of 52% was obtained (Table 1).
TABLE 1.
Purification of recombinant His-tagged mesaconyl-CoA hydratase of C. aurantiacus and R. sphaeroides from 3 g of E. coli cellsa
| Purification step | Total enzyme activity (μmol min−1) | Total protein (mg) | Sp act (μmol min−1 mg protein−1) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | −/10,500 | −106 | −/99 | −/100 | −/1 |
| Heat precipitation | 9,300/− | 16.5/− | 560/− | 100/− | 1− |
| Ni-Sepharose high performance | 8,100/5,500 | 6.4/4.0 | 1,300/1,400 | 87/52 | 2.3/14.0 |
The first number in each column refers to the C. aurantiacus enzyme, and the second number (boldface) in each column refers to the R. sphaeroides enzyme. For the enzyme assay, see Materials and Methods. −, not determined for the Chloroflexus enzyme or not applicable for the Rhodobacter enzyme.
Properties of mesaconyl-CoA hydratases.
The specific activity of the Chloroflexus enzyme at 55°C was 1,300 μmol min−1 mg−1, and that of the Rhodobacter enzyme at 30°C was 1,400 μmol min−1 mg−1. The enzyme turnover per dimeric enzyme based on specific activities for the Chloroflexus enzyme was 1,700 s−1, and that for the Rhodobacter enzyme was 1,900 s−1. Both enzymes exhibited optimal activity at pH 7.5 and half-maximal activity at pH 6.5 and 8.5 (data not shown). The UV spectra of the enzymes showed absorptions only at 280 nm. SDS-PAGE revealed that the enzymes were composed of approximately 40-kDa subunits. Gel filtration indicated a molecular mass of the native enzymes of about 80 kDa, suggesting a homodimeric structure.
Isolation and structure elucidation of the product of the transformation of 13C-labeled β-methylmalyl-CoA as mesaconyl-CoA.
The 13C-enriched product of the conversion of [1,2, 3-13C]propionyl-CoA and glyoxylate by recombinant l-malyl-CoA/β-methylmalyl-CoA lyase and recombinant mesaconyl-CoA hydratase of C. aurantiacus or R. sphaeroides was analyzed by HPLC-MS. The product had a virtual molecular mass of 877.9 Da, as determined by ESI-MS (negative ion mode). This corresponds to a molecular mass of 878.9 Da, which is close to the expected value of 878.6 Da for mesaconyl-CoA.
The product was isolated by HPLC and analyzed by one- and two-dimensional NMR spectroscopy. Mesaconyl-CoA was characterized through proton NMR signals at 0.87 ppm (s, 3H, 10″ CH3), 1.10 (s, 3H, 11″ CH3), 2.26 (ddd, J = 130.0 Hz, 5.8 Hz, 1.5 Hz, 3H, *CH3 [mesaconyl]), 2.46 (m, 2H, H6″), 3.13 (q, J = 6.5 Hz, 2H, H9″), 3.40 (t, J = 6.4 Hz, 2H, H8″), 3.51 (t, J = 6.8 Hz, 2H, H5″), 3.64 (m, 1H, H1″A), 4.06 (m, 1H, H1″B), 4.11 (s, 1H, H3″), 4.32 (s, br, 2H, H5′), 4.51 (s, br, 1H, H4′), 4.91 (m, 1H, H2′), 5.01 (s, br, 1H, H3′), 6.16 (d, J = 5.8 Hz, 1H, H1′), 6.70 (“t,” br, J = 8.0 Hz, 1H [mesaconyl]), 8.24 (s, 1H, H2), 8.61 (s, 1H, H8). The 13C-enriched carbons (C1, C2, and α-methyl) of mesaconyl-CoA gave signals at 14.3 ppm (dd, J = 42.6 Hz, 2.3 Hz), 149.4 (dd, J = 55.5 Hz, 42.6 Hz), and 195.3 (dd, J = 55.5Hz, 2.3 Hz), indicating the incorporation of the intact propionyl 13C-enriched carbon chain. The corresponding signal in the proton NMR at 2.26 ppm of the 13C-enriched α-methyl group (13C NMR, 14.3 ppm) was identified through a gradient heteronuclear single quantum coherence experiment. Moreover, the signal at 6.70 ppm (proton NMR, β-CH) showed a cross-peak to the three signals at 14.3, 149.4, and 195.3 ppm (13C NMR) in the heteronuclear multiple-bond correlation experiment, verifying the mesaconyl-CoA structure. The CH2 group at 3.13 ppm (proton NMR) gave a cross-peak with the 13C-enriched carbonyl-C at 195.3 ppm (13C NMR) in the heteronuclear multiple-bond correlation experiment, verifying that the CoA residue was attached at C-1 of mesaconyl-CoA.
DISCUSSION
Properties of mesaconyl-CoA hydratase.
We have identified and characterized a new enzyme of the enoyl-CoA hydratase family, mesaconyl-CoA hydratase. The reversible reaction catalyzed is as follows: erythro-β-methylmalyl-CoA → mesaconyl-CoA + H2O. The EC number is 4.2.1.x. The enzyme belongs to the carbon-oxygen lyases (EC 4.2.), subclass hydrolyases (4.2.1.). Its systematic name is erythro-β-methylmalyl-CoA hydrolyase. Enzymes of this type, like the classical enoyl-CoA hydratase (EC 4.2.1.17), normally catalyze reversible reactions of the type (3S)-3-hydroxyacyl-CoA → trans-2 (or 3)-enoyl-CoA + H2O; the (3R)-3-hydroxyacyl-CoA compounds are converted to the cis compounds. However, there are enoyl-CoA hydratases of the MaoC family that act on the (3R)-3-hydroxyacyl-CoA compound and still produce the trans compound. Mesaconyl-CoA hydratase belongs to this MaoC enzyme family. The stereochemistry of erythro-β-methylmalyl-CoA corresponds to the 3R stereoconfiguration.
Role of mesaconyl-CoA hydratase in the autotrophic 3-hydroxypropionate cycle.
Mesaconyl-CoA hydratase functions in autotrophic CO2 fixation of C. aurantiacus and converts β-methylmalyl-CoA to mesaconyl-CoA. Mesaconyl-CoA is then transformed to citramalate or citramalyl-CoA (16, 21-23). Citramalyl-CoA cleavage by citramalyl-CoA lyase regenerates the starting molecule, acetyl-CoA, and releases pyruvate as the CO2 fixation product and precursor molecule for all biosynthetic routes (17). The autotrophic 3-hydroxypropionate cycle is thus closed (Fig. 1A). The following features corroborate the importance of the enzyme for autotrophic growth. The specific enzyme activity in extracts of autotrophically grown cells (20 to 30 nmol min−1 mg protein−1) was on the order of the estimated minimal enzyme rate (12 nmol min−1 mg protein−1) that could explain the slow autotrophic growth (21). The 10- to 15-fold up-regulation under autotrophic conditions is in line with this function. In C. aurantiacus, the mch gene is located directly downstream of the gene coding for l-malyl-CoA/β-methylmalyl-CoA lyase (mcl). Unfortunately, mutant studies with this filamentous bacterium are difficult and are not available.
Role of mesaconyl-CoA hydratase in the ethylmalonyl-CoA pathway.
Mesaconyl-CoA hydratase is also crucial in converting mesaconyl-CoA to β-methylmalyl-CoA in the novel acetate assimilation pathway termed the ethylmalonyl-CoA pathway (14) (Fig. 1B). Here, mesaconyl-CoA is formed from two molecules of acetyl-CoA and one molecule of CO2, and cleavage of β-methylmalyl-CoA yields glyoxylate and propionyl-CoA (34). In R. sphaeroides, transposon mutagenesis showed that inactivation of the mch gene resulted in an inability to grow on acetate. Growth, however, was rescued by the addition of glyoxylate (2). Furthermore, the mch gene product was identified among seven proteins that were up-regulated during growth with acetate compared to growth with glucose supplied as the sole carbon source, supporting its role in acetate assimilation (2). The corresponding mch gene in M. extorquens was annotated as meaC (31); it is upregulated when cells are shifted from growth on succinate to methylotrophic growth (33). In Streptomyces coelicolor, the mch gene is clustered together with the genes for crotonyl-CoA carboxylase/reductase (ccr) and other genes postulated to be involved in acetyl-CoA assimilation (14, 20, 34). Interestingly, erythro-β-methylmalate and mesaconate were identified in R. rubrum when extracts were incubated with glyoxylate and propionyl-CoA (35).
Occurrence of mesaconyl-CoA hydratase, a member of the MaoC family of (R)-enoyl-CoA hydratases.
Enzymes of the MaoC family of (R)-enoyl-CoA hydratases catalyze the hydration/dehydration of trans-2-enoyl-CoA/(R)-3-hydroxyacyl-CoA, e.g., as part of the polyhydroxyalkanoate biosynthetic pathway. Other enzymes are the putative MaoC dehydratase, the peroxisomal hydratase-dehydrogenase-epimerase protein, and the fatty acid synthase β-subunit. The structure of the monomer includes a five-strand antiparallel β-sheet wrapped around a central α-helix, referred to as a hot dog fold. Proteins with a hot dog fold belong to a conserved family of proteins found mostly in Eubacteria and Archaebacteria. The active site lies within a substrate-binding tunnel formed by the homodimer (24). Interestingly, mesaconyl-CoA hydratase is derived from this class of enzymes by gene duplication, and the native enzyme is a homodimer. The reason for the gene duplication event is not obvious. In the database, enzymes with two (R)-enoyl-CoA hydratase domains are found, but they are only distantly related to mesaconyl-CoA hydratase.
Genes coding for enzymes very similar to the mesaconyl-CoA hydratases (ca. 38-kDa subunits) with two (R)-enoyl-CoA hydratase domains described here were found in Roseiflexus, Methylobacterium, Hyphomonas, Rhodospirillum, Xanthobacter, Caulobacter, Magnetospirillum, Jannaschia, Sagittula, Parvibaculum, Stappia, Oceanicola, Loktanella, Silicibacter, Roseobacter, Roseovarius, Dinoroseobacter, Sulfitobacter, Paracoccus, and Ralstonia species (Fig. 5). Many members of this group have additional genes, which are postulated to play a role in the new acetate assimilation pathway (14) (Fig. 1B), and the gene for isocitrate lyase is missing. Interestingly, several of these bacteria appear to live in the ocean surface waters and demethylate dimethylsulfoniopropionate to methylmercaptopropionate (26). These bacteria may act as methylotrophs, similar to M. extorquens. This conclusion is corroborated by the presence of hydroxypyruvate reductase and l-malyl-CoA lyase genes in many of the available genomes. These genes are characteristic of the serine cycle of formaldehyde assimilation, e.g., in M. extorquens. In this pathway, acetyl-CoA is formed from one molecule of formaldehyde and one molecule of HCO3−. Acetyl-CoA is then assimilated via the ethylmalonyl-CoA pathway. The mch gene in Roseiflexus sp. is probably required for autotrophic CO2 fixation via the 3-hydroxypropionate cycle, as in C. aurantiacus and Chloroflexus aggregans (Fig. 1A) (28). A similar yet distinct class of enzymes containing only one hydratase domain (ca. 20-kDa subunit) was found in various other bacteria, such as Streptomyces sp. Nevertheless, this shorter version of the enzyme in Streptomyces sp. is postulated to be involved in acetyl-CoA assimilation via the ethylmalonyl-CoA pathway (see above) (Fig. 1B) (14).
FIG. 5.
Phylogenetic tree of homologues of mesaconyl-CoA hydratase based on amino acid sequences. Note that members of this group of enzymes have approximately double the sizes of normal enoyl-CoA hydratases and contain two enoyl-CoA hydratase domains. They are formed by duplication of genes containing only one enoyl-CoA hydratase domain (tree not shown). The tree topography and evolutionary distances are given by the neighbor-joining method with Poisson correction. The scale bar represents a difference of 0.1 substitution per site. The numbers at the nodes indicate the percentage bootstrap values for the clade of this group in 1,000 replications. Only numbers above 50% were considered to be significant.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, and Evonik Degussa GmbH (supported by the state Nordrhein-Westfalen and cofinanced by the European Union).
Thanks are due to Nasser Gad'on, Christa Ebenau-Jehle, and Volker Brecht (all Freiburg) for invaluable expert technical assistance.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 27712137-12143. [DOI] [PubMed] [Google Scholar]
- 2.Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61297-309. [DOI] [PubMed] [Google Scholar]
- 3.Albers, H., and G. Gottschalk. 1976. Acetate metabolism in Rhodopseudomonas gelatinosa and several other Rhodospirillaceae. Arch. Microbiol. 11145-49. [DOI] [PubMed] [Google Scholar]
- 4.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 6.Blankenship, R. E., M. T. Madigan, and C. E. Bauer. 1995. Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 8.Castenholz, R. W. 1969. Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33476-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claasen, P. A. M., and A. J. B. Zehnder. 1986. Isocitrate lyase activity in Thiobacillus versutus grown anaerobically on acetate and nitrate. J. Gen. Microbiol. 1323179-3185. [Google Scholar]
- 10.Cutinelli, C., G. Ehrensvärd, L. Reio, E. Saluste, and R. Stjernholm. 1951. Acetic acid metabolism in Rhodospirillum rubrum under anaerobic conditions. Arkiv Kemi. 3315-322. [Google Scholar]
- 11.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 12.Eisenreich, W., G. Strauss, U. Werz, G. Fuchs, and A. Bacher. 1993. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 215619-632. [DOI] [PubMed] [Google Scholar]
- 13.Ensign, S. A. 2006. Revisiting the glyoxylate cycle: alternate pathway for microbial acetate assimilation. Mol. Microbiol. 61274-276. [DOI] [PubMed] [Google Scholar]
- 14.Erb, T. J., I. A. Berg, V. Brecht, M. Müller, G. Fuchs, and B. E. Alber. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: The ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. USA 10410631-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedmann, S., A. Steindorf, B. E. Alber, and G. Fuchs. 2006. Properties of succinyl-coenzyme A:L-malate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J. Bacteriol. 1882646-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedmann, S., B. E. Alber, and G. Fuchs. 2006. Properties of succinyl-coenzyme A:D-citramalate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J. Bacteriol. 1886460-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedmann, S., B. E. Alber, and G. Fuchs. 2007. Properties of R-citramalyl-coenzyme A lyase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J. Bacteriol. 1892906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukui, T., and Y. Doi. 1997. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J. Bacteriol. 1794821-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottschal, J. C., and J. G. Kuenen. 1980. Mixotrophic growth of Thiobacillus A2 on acetate and thiosulfate as growth limiting substrates in the chemostat. Arch. Microbiol. 12633-42. [Google Scholar]
- 20.Han, L., and K. A. Reynolds. 1997. A novel alternate anaplerotic pathway to the glyoxylate cycle in streptomycetes. J. Bacteriol. 1795157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herter, S., A. Busch, and G. Fuchs. 2002. l-Malyl-coenzyme A lyase/β-methylmalyl-coenzyme A lyase from Chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO2 fixation. J. Bacteriol. 1845999-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herter, S., G. Fuchs, A. Bacher, and W. Eisenreich. 2002. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J. Biol. Chem. 27720277-20283. [DOI] [PubMed] [Google Scholar]
- 23.Herter, S., J. Farfsing, N. Gad'on, C. Rieder, W. Eisenreich, A. Bacher, and G. Fuchs. 2001. Autotrophic CO2 fixation in Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 1834305-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holden, H. M., M. M. Benning, T. Haller, and J. A. Gerlt. 2001. The crotonase superfamily: divergently related enzymes that catalyze different reactions involving acyl coenzyme A thioesters. Acc. Chem. Res. 34145-157. [DOI] [PubMed] [Google Scholar]
- 25.Holo, H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151252-256. [Google Scholar]
- 26.Howard, E. C., J. R. Henriksen, A. Buchan, C. R. Reisch, H. Bürgmann, R. Welsh, W. Ye, J. M. Gonzalez, K. Mace, S. B. Joye, R. P. Kiene, W. B. Whitman, and M. A. Moran. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314649-652. [DOI] [PubMed] [Google Scholar]
- 27.Hügler, M., C. Ménendez, H. Schägger, and G. Fuchs. 2002. Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Bacteriol. 1842404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klatt, C. G., D. A. Bryant, and D. M. Ward. 2007. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environ. Microbiol. 92067-2078. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg, H. L., and J. Lascelles. 1960. The formation of isocitratase by the Athiorhodaceae. J. Gen. Microbiol. 23511-517. [DOI] [PubMed] [Google Scholar]
- 30.Korotkova, N., and M. E. Lidstrom. 2001. Connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 1831038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korotkova, N., M. E. Lidstrom, and L. Chistoserdova. 2005. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 1871523-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 33.Laukel, M., M. Rossignol, G. Borderies, U. Volker, and J. A. Vorholt. 2004. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and nonmethylotrophic conditions. Proteomics 41247-1264. [DOI] [PubMed] [Google Scholar]
- 34.Meister, M., S. Saum, B. E. Alber, and G. Fuchs. 2005. l-Malyl-coenzyme A/β-methylmalyl-coenzyme A lyase is involved in acetate assimilation of the isocitrate lyase-negative bacterium Rhodobacter capsulatus. J. Bacteriol. 1871415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osumi, T., and H. Katsuki. 1977. A novel pathway for l-citramalate synthesis in Rhodospirillum rubrum. J. Biochem. 81771-778. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Simon, E. 1957. S-Succinyl coenzyme A. Biochem. Prep. 530-32. [Google Scholar]
- 38.Stadtman, E. R. 1957. Preparation and assay of acyl coenzyme A and other thiol esters; use of hydroxylamine. Methods Enzymol. 3931-941. [Google Scholar]
- 39.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215633-643. [DOI] [PubMed] [Google Scholar]
- 40.Strauss, G., W. Eisenreich, A. Bacher, and G. Fuchs. 1992. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing archaebacterium Thermoproteus neutrophilus and in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 205853-866. [DOI] [PubMed] [Google Scholar]
- 41.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 42.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 821074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meer, M. T. J., S. Schouten, J. W. de Leeuw, and D. M. Ward. 2000. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ. Microbiol. 2428-435. [DOI] [PubMed] [Google Scholar]
- 44.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182157-159. [DOI] [PubMed] [Google Scholar]