Abstract
The endoplasmic reticulum (ER) is a target for endogenously generated reactive oxygen species (ROS) during aging. We have previously shown that the ER chaperones, protein disulfide isomerase (PDI) and immunoglobulin heavy chain binding protein (BiP) are oxidatively modified within the livers of aged mice. In this study we assess the functional consequences of the age-dependent oxidation of these two proteins. Specific activity measurements, performed on purified protein samples obtained from young and aged mouse livers, show definitive decreases in BiP ATPase activity and dramatic reductions in PDI enzymatic activity with age. Overall, these results suggest that protein folding and other activities mediated through PDI and BiP are diminished during aging. Furthermore, the relative loss of these chaperone-like activities could directly contribute to the age-dependent accumulation of misfolded proteins, a characteristic of the aging phenotype.
Keywords: Aging, BiP, Carbonylation, Chaperone Modification, PDI, Protein Folding, Oxidative Stress
Introduction
The endoplasmic reticulum (ER) is a focal point for intracellular protein assembly. Within the lumen of the ER, proteins are folded, post-translationally modified, and assembled into protein complexes before they are exported to the cytoplasm, other organelles, the cell surface, or secreted from the cell altogether [1-3]. Within the ER, protein disulfide isomerase (PDI) and immunoglobulin heavy chain binding protein (BiP) belong to a cadre of chaperones and other enzymes that catalyze the proper folding and assembly of proteins.
The immunoglobulin heavy chain binding protein (BiP), is a member of the Hsp70 chaperone family [4]. BiP interacts with newly synthesized polypeptides through a C-terminal binding domain that preferentially interacts with linear stretches of amino acids that contain alternating hydrophobic and aromatic residues [5]. Peptide binding at the C-terminus is coupled with ATP binding at the protein’s N-terminal domain where the ATP substrate induces a conformational change that triggers peptide release [6,7]. BiP also plays a central role in the unfolded protein stress response (UPR) [7]. Under normal low stress conditions, BiP interacts with the luminal domains of IRE1, PERK, and ATF6 [4,8,9]. Increase in the burden of unfolded protein disrupts these interactions, and triggers the cascade of UPR signaling [7,10]. BiP also interacts with a variety of other chaperones and folding enzymes, participating in one of the major chaperone complexes within the ER [11].
PDI is a disulfide isomerase that mediates the oxidative folding and unfolding of polypeptides within the ER [12,13]. It also catalyzes the disulfide bond isomerization of a variety of substrates in vitro [14,15]. In the cell, however, PDI is directly oxidized through the Ero-1 pathway and functions as a sulfhydryl oxidase, catalyzing disulfide bond formation for polypeptide substrates in the process of folding into native structures [16]. PDI also functions in the large network of ER chaperones that include BiP, Grp94, ERp72, and other folding enzymes [1,11]. The concerted activities of these proteins are required for ER protein quality control [1].
The accumulation of misfolded proteins is a hallmark of the phenotype of aged tissues and is thought to be a causative factor in a variety of age-associated diseases such as Parkinson’s and Alzheimer’s disease. However, the molecular events that lead to protein accumulation during aging are not fully understood. Recent reports have emphasized that the ER is a major target for oxidative stress [16-18] and we have previously shown that PDI and BiP are oxidatively modified by carbonylation in aged mouse liver [19]. We hypothesize, therefore, that one of the factors that contributes to protein misfolding during aging is the loss of the chaperone-like activities mediated through PDI and BiP. Here we test this hypothesis by investigating the functional consequences of the age-related oxidation of these proteins.
Materials and Methods
Mice
Young (3-5 months) and aged (20-24 months) male C57BL/6 mice from the National Institute on Aging colonies (Bethesda, MD) were obtained through Charles River Laboratories (Wilmington, MA). Mice were maintained with a 12 hour light/dark cycle and fed ad libitum on a standard chow diet for at least one week before use. Mice were sacrificed by cervical dislocation. The livers were harvested and snap frozen in liquid nitrogen until analysis.
Preparations of ER proteins
Liver homogenates were prepared separately from three young and three aged mice [19]. ER proteins were prepared from the livers of individual mice with minor modifications [19]. Briefly, the ER pellet was resuspended in purification buffer (25 mM KCl, 5 mM MgCl2, 50 mM Tris-HCl, pH 7.5) with protease and phosphatase inhibitors and 0.5 % Triton X-100 to permeabilize microsomes. The suspension was allowed to stand on ice for 20 minutes and then centrifuged for 90 minutes at 105,000 × g. The supernatant contained the enriched ER proteins used in this study. Protein was quantified with the Bradford protein assay (Bio-Rad), using BSA as a standard. Pools of young and aged ER protein were created by combining equal amounts of samples from three young and three aged mouse ER fractions. Protein pools from both age groups were used for protein purification.
Purification of PDI and BiP
The purification of PDI and BiP from mouse livers was based on a previously published method with slight modifications [20]. All column chromatography steps were performed on a dual pump HPLC system (ESA Biosciences) equipped with a UV-Vis detector (UV-Vis Model 528, ESA Biosciences) and a Gilson FC 204 fraction collector. Solubilized ER protein extracts from mouse livers were applied to an anion exchange column (5 ml UNO-Q-Sepharose column, Bio-Rad). After application, the column was washed with 30 mL of 25 mM Tris (pH 8.0), developed with a shallow 20 mL linear NaCl gradient (0-250 mM NaCl in 25 mM Tris pH 8.0), followed by a steep 20 ml NaCl gradient (250 mM – 1 M NaCl in 25 mM Tris pH 8.0). The flow rate equaled 1 mL/min throughout purification and fractions were collected at a rate of one fraction per minute. Fractions containing PDI and BiP were identified by resolving a small aliquot of these fractions on large format 26 well acrylamide gels (Bio-Rad), transferring the gels to Immobilon-P (Millipore) PVDF membranes and probing membranes with antibodies specific for either PDI or BiP. Protein samples containing PDI and BiP were pooled and applied to a 2 ml Bio-Scale ceramic hydroxyapatite (HAP) column (Bio-Rad). After the column was washed with 5 ml of low phosphate buffer (5 mM sodium phosphate, pH 7.4), protein was eluted with a shallow 20 minute linear sodium phosphate (pH 7.4) gradient (5 mM – 150 mM) followed by a steep 20 minute linear sodium phosphate (pH 7.4) gradient (150 mM – 500 mM). Throughout HAP chromatography, the flow rate equaled 0.5 mL/min and fractions were collected at a rate of one fraction every minute. SDS-PAGE/immunoblotting procedures were again used to locate fractions containing PDI and BiP.
Fractions containing PDI and BiP from young and aged mice were pooled and concentrated by centrifugal concentration (Icon® 20 kDa MWCO, Pierce). Protein concentrations were then determined with the bicinchonic acid (BCA) protein assay (Pierce) using bovine serum albumin as a standard.
PDI activity measurement
PDI specific activity was quantified by measuring the GSH-dependent, PDI-catalyzed reduction of insulin with the coupled glutathione reductase assay [21]. Prior to analysis, DTT was added to both samples to a final concentration of 10 mM and the samples were incubated for 4 hrs at 4° C to reduce active site thiols. Each assay was performed as follows: Buffer components were added to a cuvette to give final concentrations of 0.2 M sodium phosphate (pH 7.5), 5 mM EDTA, 3.7 mM GSH, 0.12 mM NADPH, and 16 units/mL of glutathione reductase. The reaction buffer was allowed to stand undisturbed for 30 seconds to allow conversion of contaminating GSSG to GSH. Bovine insulin (30 μM) was added to the cuvette and the uncatalyzed reaction was recorded for two minutes at 340 nm. PDI was then added to a concentration of 10 μg/mL and the reaction was monitored for an additional two minutes. The uncatalyzed background rate was subtracted from the catalyzed rate to produce the final values. The PDI specific activities reported here are average values calculated from five independent measurements of young and aged protein samples. All PDI activity measurements were made at 25° C in a Beckman Coulter DU530 spectrophotometer.
BiP activity measurement
The ATPase activity of BiP was quantified using an end-point assay that measured the release of organic phosphate during BiP catalyzed hydrolysis [22]. Assay mixtures contained 20 mM HEPES (pH 7.0), 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT and 1 mM [γ-32P] ATP. Activity assays were initiated by adding BiP (1 μM, final concentration) to the reaction mixture and incubating at 37° C for 30 minutes; the volume of each reaction equaled 20 μL. The reaction was quenched by adding an equal volume of 4% SDS to each sample and briefly vortexing each tube. Inorganic phosphate released during catalysis was extracted by adding 50 μL fresh phosphate reagent (4 N sulfuric acid, 4% ammonium molybdate, and 0.02 M silicotungstic acid) and 200 μL of 35:65 isobutanol:xylene (v/v) to each reaction and vortexing each tube for 10 seconds (2x) to form aqueous and organic phases. The upper phase (50 μL) was removed and analyzed by liquid scintillation counting. Each activity measurement was repeated three times and average values were calculated for young and aged BiP samples.
Detection of oxidized PDI and BiP
The relative abundance of oxidative modifications (carbonyls) in young and aged protein samples were determined using the Oxyblot kit [23,24; Intergen Company]. Carbonylated proteins were detected following the manufacturer’s recommendations with slight modification. Briefly, 0.5 μg of the purified protein from each age group was derivatized with 2,4-dinitrophenylhydrazine for exactly 10 minutes after which the proteins were resolved by SDS-PAGE, and transferred to a PVDF membrane. Blots were developed using a primary antibody specific for the 2,4-dinitrophenylhydrazone (DNP) moiety and blots visualized with an appropriate HRP-conjugated secondary antibody. Blots were quantified by densitometry and the Coomassie Blue stained membrane was used to normalize for sample loading variation.
Results
Purification of PDI and BiP
To purify PDI and BiP from the livers of young and aged mice, a two column chromatography approach was developed (Fig. 1, Materials and Methods). Solubilized ER proteins were first fractionated by anion exchange chromatography (Fig. 1A). PDI eluted between 35 and 40 minutes within the initial shallow salt gradient and BiP eluted between 45 and 50 minutes at the beginning of the steeper salt gradient.
Fig. 1.
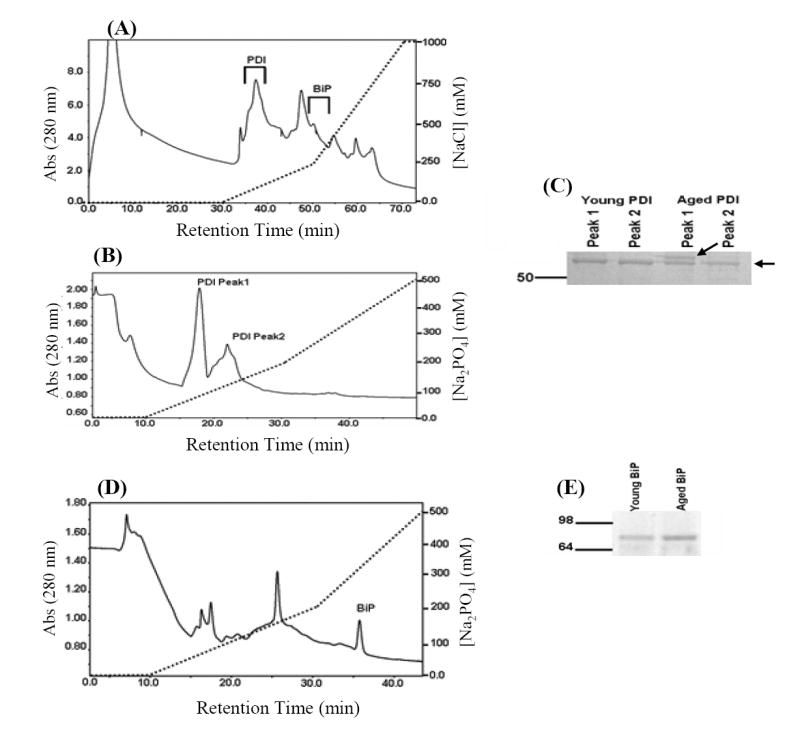
Purification of PDI and BiP from young and aged mouse liver. (A) Solubilized ER extracts from young and aged mouse liver were fractionated by anion exchange chromatography (Q-sepharose) as decribed in Materials and methods. Fractions containing PDI and BiP were located by Western blotting and the elution positions of the proteins are indicated on the figure. (B) Q-sepharose chromatography fractions containing PDI were pooled and applied to a hydroxyapatite column for further purification as described in Materials and methods. Fractions containing PDI, as assessed by Western blotting, corresponded to the two abundant peaks indicated on the figure. (C) Aliquots (250 ng) from young and old PDI samples were resolved by SDS-PAGE and proteins visualized by Coomassie Blue staining. The purification scheme yielded two protein species, identified by arrows on the figure. The higher molecular weight species is more prominent in the aged sample relative to the young sample. (D) Q-Sepharose fractions containing BiP were applied to a hydroxyapatite column and developed with the same elution protocol described above. BiP was identified by Western blotting procedures and corresponded to the peak labeled in the figure. (E) An aliquot of the young and aged BiP protein pools (100 ng) was resolved by SDS-PAGE and proteins visualized by Coomassie Blue staining. The above chromatographs (A, B, and C) were obtained from fractionation of the young protein sample, fractionation of the aged sample yielded a similar elution profiles.
Fractions containing PDI were pooled and applied to a hydroxyapatite column (Fig. 1B). In both young and aged samples PDI eluted as two peaks (Fig. 1B). This chromatographic behavior has been attributed to inter-subunit disulfide bonding between PDI molecules [25]. Treatment of the samples with DTT reduces the disulfide bonds without affecting the specific activity of PDI within the different peaks. Thus, we combined protein fractions eluted at these peaks from hydroxyapatite columns and used it as purified PDI in all further studies. Equal quantities of purified PDI (250 ng) from each sample were resolved by SDS-PAGE and proteins were visualized by Coomasie Blue staining (Fig. 1C). The stained gel shows that highly purified PDI was obtained from both young and aged mouse liver extracts. Interestingly a higher molecular weight protein band is more prominent in the aged sample relative to the young sample (Fig. 1C). Both protein bands were immunoreactive with an anti-PDI antibody (data not shown).
Q-Sepharose chromatography fractions that contained BiP were fractionated by hydroxyapatite chromatography (Fig. 1D). BiP eluted as a single peak at ~35 minutes. Aliquots of the young and aged BiP samples (100 ng) were resolved by SDS PAGE and proteins visualized by Sypro Ruby Staining (Fig. 1E). The stained gel shows that highly purified BiP was obtained from young and aged mouse liver.
Increased protein carbonylation correlates with age-associated decrease in PDI activity
We have previously shown an age-dependent carbonylation of PDI in the mouse liver [19]. After derivitization of the purified PDI samples with 2,4-dinitrophenylhydrazine, Western blot analysis was used to assess the extent of carbonylation of these samples (Fig. 2A). Densitometric analysis of PDI protein in young and aged samples (annotated with an arrow on the figure) indicates that PDI from old livers contains approximately 4.5 times more carbonyl modifications relative to that from young livers. The higher levels of carbonylation in the aged PDI correlates with decreased PDI activity, i.e., ~45% relative to the young enzyme, as measured by GSH-dependent insulin reduction (Fig. 2B).
Fig. 2.
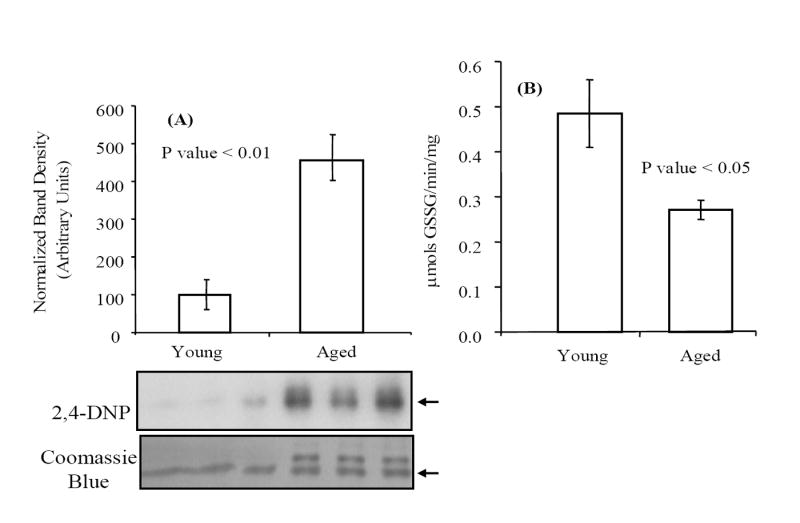
The age-associated decrease in PDI specific activity correlates with increased carbonylation. (A) PDI purified from young and aged mouse livers was analyzed for carbonyl content by Western blot analysis. Protein samples (~200 ng) from each age group were derivatized with 2,4-dinitrophenylhydrazine, as described in Materials and methods, resolved on SDS-PAGE and transferred to a PVDF membrane. Relative levels of carbonylation were compared by probing blots with an anti-DNP antibody; the Coomassie Blue stained membrane was used to normalize protein loading. Densitometric analysis was conducted on the band present in both young and aged samples (indicated with arrow on the figure). (B) Specific activity for young and aged PDI was measured as GSH-dependent reduction of insulin as described in Materials and methods. The average values reported for each age group were obtained from five independent experiments and error bars represent calculated standard errors of mean.
Increased protein carbonylation correlates with age-associated decrease in the BiP ATPase activity
We have previously shown that within mouse liver, BiP is carbonylated in an age-dependent fashion [19]. Densitometric analyses of Western blots of DNP-derivatized BiP demonstrated that BiP from old livers contains approximately 2 times more carbonyl modifications relative to that from young livers (Fig. 3). The higher levels of carbonylation in the aged BiP correlates with ~20% decrease of BiP ATPase activity, as measured by [γ-32P] phosphate release (Fig. 3B).
Fig. 3.
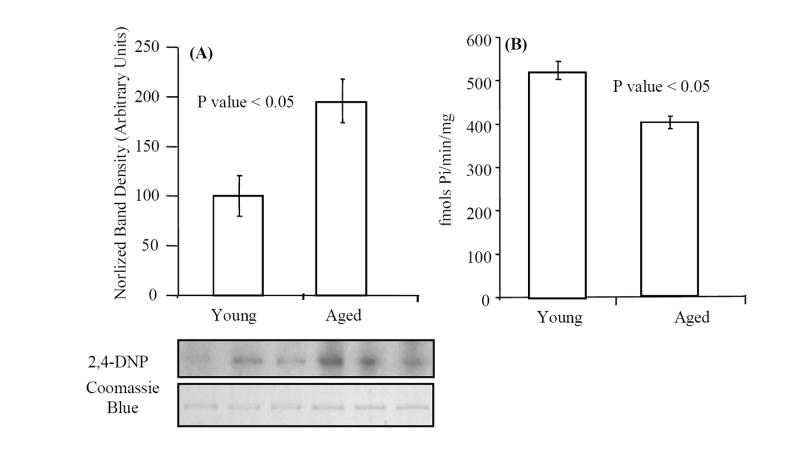
The age-associated decrease in BiP specific activity correlates with increased carbonylation. (A) BiP purified from young and aged mouse liver was analyzed for carbonyl content by Western blot analysis. Protein samples (~100 ng) from each age group were derivatized with 2,4-dinitrophenylhydrazine, as described in Materials and methods, resolved on SDS-PAGE and transferred to a PVDF membrane. Relative levels of carbonylation were compared by probing blots with an anti-DNP antibody; the Coomassie Blue stained membrane was used to normalize protein loading. Densitometric analysis was conducted for determination of fold change in carbonylation of BiP with age. (B) The ATPase activity of BiP was quantified using hydrolysis of [γ-32P] ATP as described in Materials and methods. Average activity values were calculated from three independent experiments and error bars represent calculated standard errors of mean.
Discussion
The oxidizing environment of the ER makes resident proteins potential targets for reactive oxygen species [16-18]. We have previously shown that levels of carbonylated PDI and BiP are much higher in livers of aged mice compared to young mice. Additionally, several recent studies have shown that BiP and PDI protein levels decrease with age, further suggesting that chaperone dysfunction could be a principle cause of increased protein misfolding and accumulation of oxidatively modified proteins, a physiological basis for the progression of aging characteristics [26,27]. In this study we have shown that increase in age-related oxidative modification of PDI and BiP correlates to a decrease in their chaperone-like activities, thus, suggesting a decline in tissue function due to ER-dysfunction.
PDI and BiP belong to different classes of ER-protein folding enzymes, disulfide isomerases and ATP utilizing chaperones, respectively. Because other proteins with similar activities are present in the ER, activity measurements must be conducted on purified protein samples. The two column purification procedures we developed for this study facilitated preparation of highly enriched PDI and BiP (Fig. 1), and measurement of their activities. Our results suggest that the activities of both chaperone proteins are altered by carbonylation (Fig. 2B and 3B), thus suggesting that the loss of function of these chaperones may be due to altered structure caused by oxidative modification. It is interesting that though these enzymes were purified from the same tissue pools, we measured rather large differences in relative levels of oxidative modification between PDI and BiP from young and aged samples. Thus, the age-associated decrease in PDI and BiP activity correlates with increased levels of their carbonylation suggesting that within in vivo settings, different proteins are variably susceptible to ROS-induced damage. Furthermore, the data also suggest that protein structural changes due to oxidative damage may cause this decrease in activity (Fig. 2 and 3).
Roles for BiP and PDI in normal aging and age-related diseases
Over the span of several decades, experiments, mainly conducted in vitro, have demonstrated reduced enzyme activity, decreased structural stability and an increased propensity to aggregate that generally accompany protein oxidation [28-34]. These experiments provided much of the evidence that has popularized the free radical theory of aging as a direct relationship between protein oxidation and reduced protein function. Based on our studies we propose that the loss of activity from oxidatively damaged ER chaperone proteins is consistent with the decline of tissue function that occurs in normal aging and age-related diseases. Specifically, several lines of evidence suggest that PDI may play a central role in the progression of such catastrophic neurodegenerative diseases as Parkinson’s and Alzheimer’s diseases [35].
Our studies add credence to the current theories that integrate the loss of chaperone function into theories regarding aging and several age-related diseases [36,37]. We suggest that chronic oxidative stress, a hallmark of normal aging, result in the accumulation of misfolded proteins by two mechanisms. One source of misfolded proteins is the direct oxidation of proteins by ROS as some proteins, such as PDI and BiP, are intrinsically susceptible to oxidative modification and the oxidized forms of these proteins may accumulate with age. Secondly, the folding efficiency of nascent, de novo synthesized proteins may also decrease as chaperone activity decreases with age. In addition to PDI and BiP, the loss of chaperone activity of many other folding enzymes such as ERp55, ERp57, ERp72, and calnexin has been reported to decrease with age [26]. Thus, the burden of misfolded, aggregation-prone proteins may increase with age but the capacity to mitigate this folding stress will decrease with age. We suggest that decreased ER chaperone activity and increased folding stress is one of the underlying mechanisms of aging. Interestingly, decreased chaperone activity could impact the function of multiple systems and complexes throughout the cell.
In addition to the oxidative damage of ER chaperones, PDI AND bIp, our lab has observed similar damage and accumulation of proteins of the aged mouse kidney mitochondrial electron transport chain complexes that correspond with loss of activity [38] and suggests increased mitochondrial dysfunction with aging. Thus, we propose that the oxidative modifications of PDI and BiP will be additive to the mitochondrial dysfunction for the development of age-associated decline in tissue function. In conclusion, our study provides important insight into physiological effects of oxidative modification on PDI and BiP function and their possible role in ROS-mediated ER stress in aging.
Acknowledgments
This publication was supported by U.S.P.H.S. grant 1P01 AG021830 awarded by the National Institute on Aging, and the National Institue on Aging 1 P30 AG024832-03 Claude D. Pepper Older Americans Independence Center grant, and by the UTMB Sealy Center on Aging. J.E.N. would like to thank the Kempner Foundation and the National Institutes of Environmental Health Sciences, Training Grant (T32-07254) for their fellowship support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Levine T, Rabouille C. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr Opin Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 3.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 4.Dorner AJ, Wasley LC, Kaufman RJ. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 6.Wei J, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- 7.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 8.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 9.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 10.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 11.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- 13.Laboissiere MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J Biol Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- 14.Mayer M, Kies U, Kammermeier R, Buchner J. BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J Biol Chem. 2000;275:29421–29425. doi: 10.1074/jbc.M002655200. [DOI] [PubMed] [Google Scholar]
- 15.Vinci F, Ruoppolo M, Pucci P, Freedman RB, Marino G. Early intermediates in the PDI-assisted folding of ribonuclease A. Protein Sci. 2000;9:525–535. doi: 10.1110/ps.9.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 18.Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 19.Rabek JP, Boylston WH, Papaconstantinou J., III Carbonylation of ER chaperone proteins in aged mouse liver. Biochem Biophys Res Commun. 2003;305:566–572. doi: 10.1016/s0006-291x(03)00826-x. [DOI] [PubMed] [Google Scholar]
- 20.Rowling PJ, McLaughlin SH, Pollock GS, Freedman RB. A single purification procedure for the major resident proteins of the ER lumen: endoplasmin, BiP, calreticulin and protein disulfide isomerase. Protein Expr Purif. 1994;5:331–336. doi: 10.1006/prep.1994.1049. [DOI] [PubMed] [Google Scholar]
- 21.Morjana NA, Gilbert HF. Effect of protein and peptide inhibitors on the activity of protein disulfide isomerase. Biochemistry. 1991;30:4985–4990. doi: 10.1021/bi00234a021. [DOI] [PubMed] [Google Scholar]
- 22.Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 23.Levine RL, Wehr N, Williams JA, Stadtman ER, Shacter E. Determination of carbonyl groups in oxidized proteins. Methods Mol Biol. 2000;99:15–24. doi: 10.1385/1-59259-054-3:15. [DOI] [PubMed] [Google Scholar]
- 24.Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert HF. Protein disulfide isomerase. Methods Enzymol. 1998;290:26–50. doi: 10.1016/s0076-6879(98)90005-2. [DOI] [PubMed] [Google Scholar]
- 26.Erickson RR, Dunning LM, Holtzman JL. The effect of aging on the chaperone concentrations in the hepatic, endoplasmic reticulum of male rats: the possible role of protein misfolding due to the loss of chaperones in the decline in physiological function seen with age. J Gerontol A Biol Sci Med Sci. 2006;61:435–443. doi: 10.1093/gerona/61.5.435. [DOI] [PubMed] [Google Scholar]
- 27.Paz GM, Vela J, Castano A, Ramos B, del Rio JC, Vitorica J, Ruano D. Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol Aging. 2006;27:973–982. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Chao CC, Ma YS, Stadtman ER. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci U S A. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies KJ. Protein damage and degradation by oxygen radicals. I. general aspects. J Biol Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 30.Friguet B, Szweda LI, Stadtman ER. Susceptibility of glucose-6-phosphate dehydrogenase modified by 4-hydroxy-2-nonenal and metal-catalyzed oxidation to proteolysis by the multicatalytic protease. Arch Biochem Biophys. 1994;311:168–173. doi: 10.1006/abbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 31.Oliver CN, Ahn BW, Moerman EJ, Goldstein S, Stadtman ER. Age-related changes in oxidized proteins. J Biol Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 32.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 33.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci U S A. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarian CS, Rebrin I, Sohal RS. Aconitase and ATP synthase are targets of malondialdehyde modification and undergo an age-related decrease in activity in mouse heart mitochondria. Biochem Biophys Res Commun. 2005;330:151–156. doi: 10.1016/j.bbrc.2005.02.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura T, Gu Z, Lipton SA. Contribution of glutamatergic signaling to nitrosative stress-induced protein misfolding in normal brain aging and neurodegenerative diseases. Aging Cell. 2007;6:351–359. doi: 10.1111/j.1474-9726.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 38.Choksi KB, Nuss JE, Boylston WH, Rabek JP, Papaconstantinou J. Age-related increases in oxidatively damaged proteins of mouse kidney mitochondrial electron transport chain complexes. Free Radic Biol Med. 2007;43:1423–1438. doi: 10.1016/j.freeradbiomed.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]


