Abstract
Activation of the transcription factor nuclear factor kappa B (NF-κB) is controlled by proteolysis of its inhibitory subunit (IκB) via the ubiquitin-proteasome pathway. Signal-induced phosphorylation of IκBα by a large multisubunit complex containing IκB kinases is a prerequisite for ubiquitination. Here, we show that FWD1 (a mouse homologue of Slimb/βTrCP), a member of the F-box/WD40-repeat proteins, is associated specifically with IκBα only when IκBα is phosphorylated. The introduction of FWD1 into cells significantly promotes ubiquitination and degradation of IκBα in concert with IκB kinases, resulting in nuclear translocation of NF-κB. In addition, FWD1 strikingly evoked the ubiquitination of IκBα in the in vitro system. In contrast, a dominant-negative form of FWD1 inhibits the ubiquitination, leading to stabilization of IκBα. These results suggest that the substrate-specific degradation of IκBα is mediated by a Skp1/Cull 1/F-box protein (SCF) FWD1 ubiquitin-ligase complex and that FWD1 serves as an intracellular receptor for phosphorylated IκBα. Skp1/Cullin/F-box protein FWD1 might play a critical role in transcriptional regulation of NF-κB through control of IκB protein stability.
The ubiquitin-proteasome pathway is a key mechanism for substrate-specific degradation to control the abundance of a number of proteins (1, 2). The formation of ubiquitin-protein conjugates involves three components that participate in a cascade of ubiquitin-transfer reactions: a ubiquitin-activation enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The specificity in protein ubiquitination often derives from the E3 ubiquitin ligases. Proteins polyubiquitinated by these enzymes are subjected to degradation by the 26S proteasome. Recent genetic and biochemical studies in yeast have led to the identification of a class of E3 ligases [termed the Skp1/Cull 1/F-box (SCF) complex] required for degradation of cyclins and their inhibitors (3, 4). Not only cell-cycle related proteins but also an increasing number of molecules in other biological systems of yeast have been identified as the substrates of the SCF E3 complexes. The SCF complex consists of invariable components, such as Skp1 and Cdc53, and variable components called F-box proteins that bind to Skp1 through the F-box motif (5–7). F-box proteins serve as receptors for the target protein, which usually is phosphorylated (6, 7). The physiological roles of the SCF complex have not yet been elucidated in multicellular organisms.
Also, in mammals, a number of short-lived regulatory proteins have been shown to undergo ubiquitination. Among them, one of the most extensively studied molecules is IκB, an inhibitor protein that associates with the dimeric nuclear factor kappa B (NF-κB) transcription factor to retain NF-κB in the cytoplasm. Transcription factors of the NF-κB Rel family are critical regulators of genes that function in inflammation, cell proliferation, and apoptosis (8–10). NF-κB exists in the cytoplasm of resting cells but enters the nucleus in response to various stimuli, including viral infection, ultraviolet irradiation, and inflammatory cytokines, such as tumor necrosis factor α and interleukin-1. These external signals induce phosphorylation of IκB by a large molecular-mass kinase complex [termed the IκB kinase (IKK) complex], and the signal-induced phosphorylation is a prerequisite for subsequent ubiquitination of IκB (11, 12). Degradation of multiubiquitinated IκB by the 26S proteasome allows liberated NF-κB to translocate into the nucleus, leading to transcriptional activation of a number of NF-κB-responsible genes (8–10). Recent studies have focused on the molecular mechanism underlying phosphorylation of IκB in response to the external stimuli (13–23). The molecular mechanism by which IκB is marked specifically for degradation by ubiquitination in response to phosphorylation, however, is still unclear.
Another representative protein that is ubiquitinated is β-catenin, which has an essential role in the Wingless/Wnt signaling cascade, as well as being a central component of the cadherin cell-adhesion complex (24, 25). Deregulated accumulation of β-catenin as a result of mutations either in the adenomatous polyposis coli tumor-suppression protein or in β-catenin itself is believed to initiate colorectal neoplasia (26). The abundance of β-catenin is regulated by the ubiquitin-dependent proteolysis system (27, 28). Surprisingly, there is partial similarity between IκB and β-catenin around the phosphorylation site that is prerequisite for ubiquitination. Recently, it was reported that the mutation of the F-box/WD40-repeat protein Slimb in the fly leads to accumulation of the Armadillo protein, a homologue of human β-catenin, suggesting that Slimb might be involved in Armadillo degradation (29). These clues prompted us to examine whether the mammalian homologue of Slimb is involved in IκB and β-catenin degradation. Here, we report the identification of an F-box protein, designated FWD1, as a mammalian homologue of Slimb, and show that FWD1 mediates ubiquitination of IκB and β-catenin (M.K., S.H., M.S., M.M., N. Ishida, K.H., I. Nakamichi, A. Kikuchi, K.-i.N., and K.N., unpublished work) as an intracellular receptor associated with the core complex of SCF E3 ubiquitin ligase.
METHODS
Transfection, Immunoprecipitation, and Immunoblot Analysis.
HeLa or 293T cell lines were transfected by the calcium phosphate method (30) or lipofection (Lipofectamine, GIBCO/BRL). After 48 h, cells were lysed with buffer containing 50 mM Tris⋅HCl (pH 7.6), 300 mM NaCl, 0.5% Triton X-100, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 mM iodoacetamide, 1 mM PMSF, 0.4 mM Na3VO4, 0.4 mM EDTA, 10 mM NaF, and 10 mM sodium pyrophosphate. Immunoprecipitates with antibodies were separated on SDS/PAGE and visualized by using immunoblot analysis with the following antibodies at a concentration of 1 μg/ml: anti-Myc Ab (9E10, Boehringer Mannheim), anti-Flag Ab (M5, IBI), anti-HA Ab (16B12, Babco, Richmond, CA), anti-AU1 Ab (AU1, Babco), or anti-Ub Ab (1B3, MBL, Nagoya, Japan).
Metabolic Labeling.
Transfected 293T cells were metabolically labeled with Tran-35S-label (ICN) at a concentration of 100 μCi/ml for 1 h and chased. Cell lysates were immunoprecipitated with anti-Myc Ab followed by protein G Sepharose (Amersham Pharmacia), separated on SDS/PAGE, exposed, and quantified by BAS-2000 (Fuji).
Immunofluorescence Staining.
HeLa cells were grown on glass coverslips in growth medium. Cells were transfected by using the calcium phosphate method (30) and prepared as described (31). Briefly, cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature and incubated with anti-Flag Ab (M5) or anti-p65/Rel A Ab (SC-109, Santa Cruz Biotechnology) at a concentration of 1 μg/ml in PBS containing 0.1% BSA and 0.1% saponin for 1 h at room temperature, followed by incubation with dichlorotriazinyl aminofluorescein-labeled anti-mouse Ig (Chemicon) or Cy3-labeled anti-rabbit Ig (Amersham) at a dilution of 1:500 for 1 h at room temperature, respectively. Cells were covered with a drop of GEL/MOUNT (Biomeda, Foster City, CA), viewed, and photographed with a Nikon Eclipse E800M microscope with a color chilled 3CCD camera C5810 (Hamamatsu Photonics, Hamamatsu City, Japan).
RESULTS
Identification of FWD1.
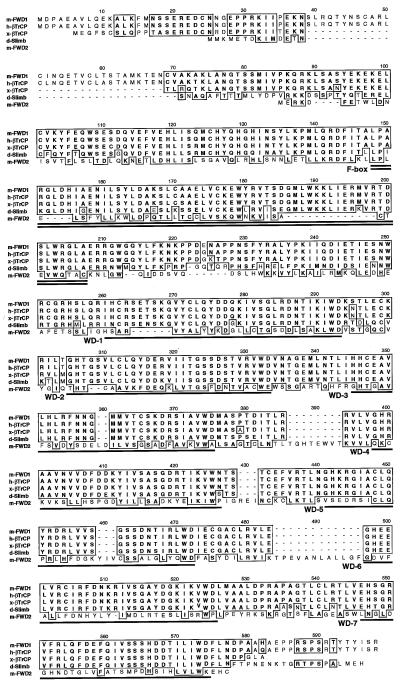
As a result of our search of a mouse expression sequence tag database, we identified a clone with significant homology with Slimb. Full-length DNA sequencing indicated that this clone encodes a protein with features of an F-box domain followed by seven WD40 repeats (Fig. 1). This clone has an extensive similarity with Slimb (D. melanogaster; ref. 29), βTrCP (X. laevis; ref. 32), and human βTrCP (H. sapiens; ref. 33). To avoid inconsistent nomenclature, we designated this clone FWD1 (F-box/WD40-repeat protein 1), based on its structural characteristics. Such prominent conservation of FWD1 throughout evolution suggests its biological importance. We also found another F-box/seven WD40-repeat protein in the mouse expression sequence tag database that had been identified as an F-box protein of unknown function, called MD6 (5). Because of its structural similarity to FWD1, we refer to it as FWD2.
Figure 1.
FWD1 is an F-box/WD40-repeat protein related to βTrCP and Slimb. Alignment of the amino acid sequences of FWD1 (Mus musculus), βTrCP (Homo sapiens and Xenopus laevis), Slimb (Drosophila melanogaster), and FWD2 (M. musculus) is shown. Similar amino acids among more than three members are boxed. The seven copies of the WD40 repeats in all four proteins are underlined; the single F-box in each protein is double-underlined. The overall identity to FWD1 is 99% (for human βTrCP), 86% (for Xenopus βTrCP), 70% (for Drosophila Slimb), and 13% (for Mus FWD2).
FWD1 Is a Component of a Mammalian SCF Complex.
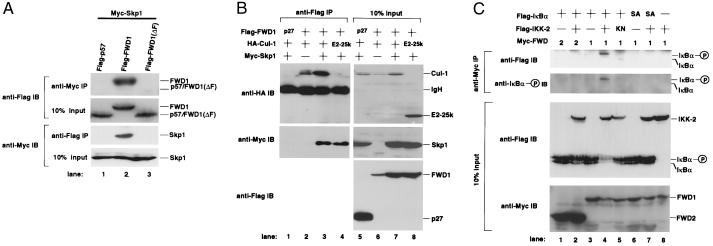
A recent report indicated that human Skp1 specifically associated with Cul1, a homologue of yeast Cdc53, but not with other cullin family members (34). Therefore, we molecularly cloned mouse Skp1 and Cul1 and examined whether mouse Skp1, Cul1, and FWD1, constituted an SCF trimolecular complex in vivo. A coimmunoprecipitation experiment showed that FWD1, as well as other F-box proteins, formed a complex with mouse Skp1 and Cul1 in mammalian cells (Fig. 2 A and B). The interaction between FWD1 and Skp1 was detected without introduction of Cul1, and this association was also confirmed by the yeast two-hybrid system and by recombinant baculoviral proteins (data not shown). These results indicate that FWD1 is associated directly with Skp1 through the F-box domain. Indeed, FWD1 (ΔF), which lacks an F-box domain (amino acids 148–192), did not interact with Skp1 (Fig. 2A). FWD1 weakly associated with Cul1 without introduction of Skp1 (Fig. 2B), although it remains possible that FWD1 and Cul1 indirectly interact through endogenous Skp1. Together, these observations suggest that FWD1 is a component of a mammalian SCF complex (referred to as SCFFWD1).
Figure 2.
FWD1 associates with Skp1, Cul1, and IκBα. (A) FWD1 binds to Skp1. Cells (293T) were transfected with expression plasmids encoding Myc-Skp1 and Flag-p57 (lane 1), Flag-FWD1 (lane 2), or Flag-FWD1 (ΔF) [lane 3; FWD1 (ΔF) is mutant FWD1 that lacks an F-box domain]. Cell lysates were immunoprecipitated via a Myc tag on Skp1 or via a Flag tag on p57, FWD1, or FWD1 (ΔF), then immunoblotted, and probed with anti-Flag (Upper) or anti-Myc (Lower) antibodies; 10% of the input lysates was also immunoblotted and probed with antibodies to show the expression level of those proteins. The position of each protein is indicated. (B) Coimmunoprecipitation of Skp1 and Cul1 with FWD1. Cells (293T) were transfected as indicated at the top of each lane. As controls, Flag-p27 (lane 1) and HA-E2-25k (lane 4) were used as indicated. Cell lysates were immunoprecipitated via a Flag tag on FWD1, then immunoblotted, and probed with anti-HA (Top), anti-Myc (Middle), and anti-Flag (Bottom); 10% of the input lysates was also shown (Right, lanes 5–8) in identical order. (C) FWD1 associates with phosphorylated IκBα. Cells (293T) were transfected with expression plasmids as indicated at the top of each lane. SA indicates the mutant IκBα in which both Ser-32 and Ser-36 are replaced with Ala, and KN indicates the kinase-negative mutant IKK-2 (K44M). Myc-FWD2 (lanes 1 and 2) or Myc-FWD1 (lanes 3–8) also were introduced. Cell lysates were immunoprecipitated via Myc tag on FWD2 or FWD1, then immunoblotted, and probed with anti-Flag to detect total IκBα or anti-phosphorylated IκBα (Upper). IKK-2 seems to interact weakly with FWD1, probably through IκBα, but it is invisible in this figure (data not shown); 10% of the input lysates also was immunoblotted and probed with anti-Flag to show the expression levels of IκBα and IKK-2 or anti-Myc for FWD1/2 (Lower). The positions of native and phosphorylated IκBα are indicated.
Association of FWD1 with Phosphorylated IκBα.
We examined whether FWD1 binds IκBα. A coimmunoprecipitation assay showed that FWD1 was associated with IκBα only when IKK-2 was present (Fig. 2C). FWD1-IκBα association was not observed with a kinase-negative mutant of IKK-2(K44M), suggesting the requirement of IKK-2 activity for the association. FWD1 interacted preferentially with the phosphorylated IκBα, which was confirmed by using an antibody specific for IκBα phosphorylated at Ser-32. Furthermore, FWD1 could not bind to the mutant IκBα in which two serines (Ser-32 and Ser-36) were replaced with alanines, even in the presence of IKK-2. This interaction between FWD1 and IκBα was confirmed by a reciprocal coimmunoprecipitation experiment (data not shown). We also detected the complex formation of Skp1-FWD1-IκBα-NF-κB (p65/RelA) in vivo (data not shown). Hence, FWD1 binds to IκBα when it is complexed with NF-κB. Other mammalian F-box proteins such as mouse Skp2 and FWD2 did not bind to IκBα, confirming that the interaction between FWD1 and IκBα is specific (Fig. 2C and data not shown). It should be noted that the abundance of IκBα protein was reduced significantly with FWD1 and IKK-2 (see Fig. 2C, lane 4; Fig. 3A, lane 3), which might reflect an accelerated turnover rate of IκBα (see Fig. 4). Collectively, these data show that FWD1 associates preferentially with IκBα phosphorylated on Ser-32 and Ser-36.
Figure 3.
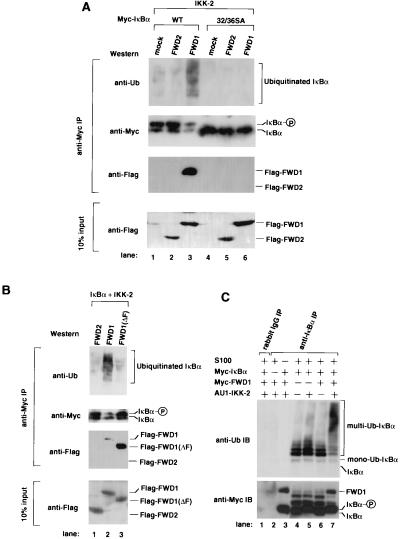
FWD1 facilitates IκBα ubiquitination. (A) FWD1 promotes IκBα ubiquitination in vivo. Cells (293T) were transfected with expression plasmids alone (mock; lanes 1 and 4) or plasmids encoding Flag-FWD2 (lanes 2 and 5) or Flag-FWD1 (lanes 3 and 6) in combination with wild-type IκBα (lanes 1 to 3) or 32/36 SA mutant IκBα (lanes 4 to 6). An expression plasmid encoding IKK-2 also was transfected in all lanes. Cell lysates were immunoprecipitated via a Myc tag on wild-type or mutant IκBα, then immunoblotted, and probed with anti-ubiquitin, anti-Myc to detect total IκBα, and anti-Flag to detect FWD1 or FWD2, which is associated with IκBα; 10% of the input lysates was immunoblotted and probed with anti-Flag to show the expression level of FWD1 and FWD2. (B) The F-box of FWD1 is required for ubiquitination but not for association with IκBα. Cells (293T) were transfected with expression plasmids encoding Flag-FWD2 (lane 1), Flag-FWD1 (lane 2), or Flag-FWD1 (ΔF) (lane 3). Expression plasmids encoding Myc-IκBα and IKK-2 were transfected in all lanes. Cell lysates were immunoprecipitated via a Myc tag on IκBα, then immunoblotted, and probed with anti-ubiquitin, anti-Myc to detect whole amount of IκBα, and anti-Flag to detect FWD2, FWD1, or FWD1 (ΔF). (C) In vitro ubiquitination of IκBα is facilitated dramatically by recombinant FWD1. Purified recombinant proteins were generated as described and mixed in combination as indicated with S100 lysate from NIH 3T3 cells. Reaction mixtures were immunoprecipitated with anti-rabbit IgG (lane 1) or anti-IκBα (lanes 2 to 7), then subjected to immunoblotting, and probed with anti-ubiquitin (Upper) or anti-Myc to indicate IκBα and FWD1 in the precipitates (Lower).
Figure 4.
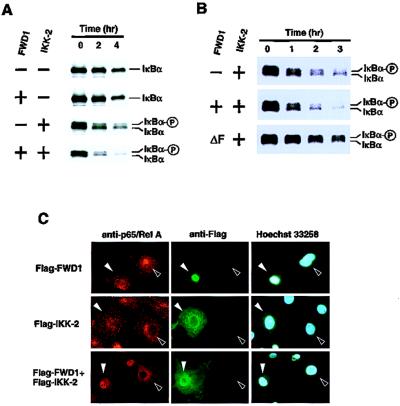
FWD1 induces rapid degradation of IκBα. (A) Pulse–chase analysis of the turnover rate of IκBα radiolabeled with [35S]methionine/cysteine in 293T cells that were transfected with expression plasmids alone or plasmids encoding FWD1, IKK-2, or FWD1/IKK-2, in combination with the Myc-tagged IκBα. Cell lysates were immunoprecipitated via a Myc tag on IκBα, then subjected to SDS/PAGE, and autoradiographed. (B) A dominant-negative form of FWD1 inhibits the degradation induced by IKK-2. Wild-type FWD1 displayed promoted degradation induced by IKK-2. In contrast, FWD1 (ΔF) had a significant inhibitory effect on IKK-2-induced degradation. (C) FWD1 facilitates nuclear translocation of NF-κB. Cos7 cells were transfected with expression plasmids encoding Flag-tagged FWD1 (Top), IKK-2 (Middle), or both (Bottom). After 48 h, the cells were fixed and stained with anti-p65/RelA to examine the subcellular distribution of p65/RelA (Left), with anti-Flag to identify the transfected cells (Center), and with Hoechst 33258 dye to show nuclei (Right). Filled arrowheads indicate the transfected cells, and open arrowheads indicate nontransfected cells. Introduction of either FWD1 or IKK-2 alone is not sufficient for nuclear translocation of p65/RelA, whereas coexpression of FWD1 and IKK-2 leads to translocation of p65/RelA to the nucleus (Bottom Left).
Facilitated Ubiquitination by FWD1.
If FWD1 is involved in IκBα ubiquitination, the introduction of FWD1 into cells might alter the ubiquitination, as well as the stability of IκBα. In the presence of IKK-2, cells transfected with FWD1 had a higher degree of ubiquitination on IκBα compared with mock or FWD2 transfectants (Fig. 3A). This increase in multiubiquitinated IκBα and the decrease in native IκBα were not observed without the introduction of IKK-2 (data not shown). Moreover, the mutant IκBα (32/36 SA) was neither phosphorylated nor ubiquitinated, suggesting that FWD1-induced ubiquitination requires prior phosphorylation on Ser-32 and Ser-36. FWD1 (ΔF) did not promote ubiquitination, whereas it remained complexed with IκBα (Fig. 3B). Hence, the association between FWD1 and IκBα is necessary but not sufficient for the ubiquitination. Because FWD1 (ΔF) did not interact with Skp1, it is highly likely that wild-type FWD1 attracts phosphorylated IκBα in the proximity of the Skp1/Cul1 E3 components leading to multiubiquitination of IκBα.
The results of the in vivo assays were reproduced in vitro. IκBα, IKK-2, and FWD1 were produced in insect cells infected with their recombinant baculoviruses and purified. The purified recombinant proteins were mixed with NIH 3T3 cell S100 lysate as a source of E1, E2, and other possible constituents, incubated, and immunoprecipitated with anti-IκBα Ab, followed by immunoblotting with an anti-ubiquitin antibody (Fig. 3C). Although mono-, di-, and tri-ubiquitination were observed when IκBα was mixed with the S100 lysate alone, multiple ubiquitination was evoked exclusively with the mixture of S100 lysate, IKK-2, and FWD1. Purified recombinant FWD1 and phosphorylated IκBα associated each other in vitro, suggesting that this binding is direct (Fig. 3C).
Accelerated Degradation of IκBα by FWD1.
A pulse–chase experiment was performed to determine whether FWD1 affects the turnover rate of IκBα as well as the static level of IκBα. The introduction of FWD1 alone had a marginal effect on the turnover rate, whereas IKK-2 slightly accelerated it (Fig. 4A). Combination of wild-type FWD1 with IKK-2 significantly accelerated the turnover rate of IκBα. The introduction of a dominant-negative mutant FWD1 (ΔF) into the cells significantly inhibited the degradation of IκBα evoked by IKK-2 (Fig. 4B). Next, we hypothesized that elimination of IκBα by FWD1 facilitates the nuclear translocation of NF-κB. To test the hypothesis, an immunofluorescence assay was performed (Fig. 4C). Introduction of either FWD1 or IKK-2 did not lead to efficient translocation of p65/RelA, a subunit of NF-κB, whereas combined expression of FWD1 and IKK-2 induced apparent nuclear translocation. Taken together, these data show that FWD1 may control the protein stability of IκBα through ubiquitination.
DISCUSSION
Protein degradation via the ubiquitin-proteasome system seems highly selective and precisely timed, and protein degradation allows an instant switch from one functional program to another (1–4). Although E3 ubiquitin ligases are thought to have a critical role in the determination of substrate-specificity, there have been few E3s whose correlations to specific targets have been identified in higher eukaryotes. As far as we know, there has been no biochemical indication of SCF E3 complex involvement in substrate-specific ubiquitination in multicellular organisms. Our data suggest that SCFFWD1 is a bona fide IκB ubiquitin ligase. A possibility remains, however, that other E3s also are involved in IκB ubiquitination. One approach to address this issue would be to create FWD1 gene-ablated mice.
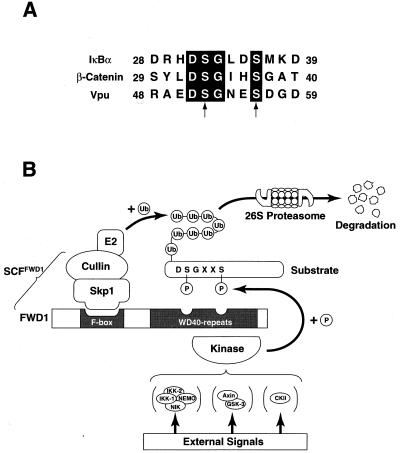
We also found that FWD1 is involved in ubiquitination of phosphorylated β-catenin as expected from genetic evidence in the Drosophila Slimb mutant (M.K., S.H., M.S., M.M., N. Ishida, K.H., I. Nakamichi, A. Kikuchi, K.-i.N., and K.N., unpublished work). In addition, human βTrCP, a homologue of FWD1, interacts with HIV-1 protein Vpu (33), although it remains unclear whether human βTrCP induces ubiquitination of Vpu. Because previous studies in yeast suggest that F-box proteins are likely to recognize multiple substrates (6, 7, 35–38), it does not seem unusual that SCFFWD1 also serves as a ubiquitin ligase for β-catenin and Vpu. There is a common structural feature among IκBα, β-catenin, and Vpu (Fig. 5A): signal-induced phosphorylation occurs on two closely located serines at positions 32 and 36 (IκBα), 33 and 37 (β-catenin), and 52 and 56 (Vpu), suggesting that FWD1 recognizes a DSGXXS motif whose serines are phosphorylated (Fig. 5B). The association between FWD1 and the substrates is specific, because other F-box proteins neither interact nor promote ubiquitination. What, then, distinguishes these substrates? Possible candidates are the kinases that phosphorylate each substrate. IκBα is phosphorylated by the IKK complex (13–23); β-catenin is phosphorylated by the GSK-3β/Axin complex (39); and Vpu is phosphorylated by the casein kinase II (40, 41). Phosphorylated DSGXXS motifs on these substrates might be the signal for the common ubiquitination pathway through FWD1. Another possibility is the differential use of ubiquitin-conjugating enzymes (E2s). These possibilities remain to be tested in the future.
Figure 5.
A model for the function of SCFFWD1 in ubiquitination. (A) Alignment of amino acid sequences necessary for association with FWD1 in IκBα (human), β-catenin (mouse), and Vpu (HIV). Serines subject to phosphorylation are indicated by arrows. (B) External signals activate the corresponding kinase, which phosphorylates the substrates at the DSGXXS motif. The phosphorylated substrate is attracted by FWD1. FWD1 links the target protein to the Skp1/Cullin/E2 ubiquitination apparatus, leading to the formation of a multiple ubiquitin chain. The multiubiquitinated protein is subject to the proteolysis by the 26S proteasome.
The NF-κB family of transcription factors plays versatile roles in immune and stress responses, inflammation, and apoptosis (8–10). A number of previous reports suggested that phosphorylation- and ubiquitination-dependent degradation of the inhibitory molecule IκB is a key event for activation of the NF-κB pathway. It is recognized generally that substances modifying NF-κB functions are valuable lead compounds for the development of drugs for inflammatory and neurodegenerative diseases, as well as for cancer. Because ubiquitination of IκBα by SCFFWD1 seems highly specific, inhibitors of the interaction between FWD1 and IκBα are possible candidates for therapeutic drugs.
Acknowledgments
We thank T. Takemori, Y. Kanegae, T. Kawabe, and S. Tanaka for the plasmids and cell lines used in this study; A. Yamanaka, M. Miura, N. Ishida, I. Nakamichi, N. Nishimura, S. Matsushita, K. Shimoharada, R. Yasukochi, and other laboratory members for technical assistance; M. Kimura and A. Takimoto for secretarial assistance; and Tazim Verjee for careful preparation of this manuscript. This work was supported in part by grants from the Ministry of Education, Science, Sports, and Culture of Japan, the Toray Science Foundation, the Uehara Memorial Foundation, and the Mochida Memorial Foundation, and by Public Health Service/National Institutes of Health, Institute on Aging Grant AG05628-14 (to R.A.G.).
ABBREVIATIONS
- NF-κB
nuclear factor kappa B
- IκB
inhibitory subunit of NF-κB
- IKK
IκB kinase
- SCF
Skp1/Cull 1/F-box
- FWD1
F-box/WD40-repeat protein 1
- SCFFWD1
SCF protein FWD1
- FWD1 (ΔF)
mutant FWD1 lacking an F-box domain
Footnotes
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 2.Weissman A M. Immunol Today. 1997;18:189–198. doi: 10.1016/s0167-5699(97)84666-x. [DOI] [PubMed] [Google Scholar]
- 3.Elledge S J, Harper J W. Biochim Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 4.Krek W. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 5.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 6.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 7.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 8.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 10.May M J, Ghosh S. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 11.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 12.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 14.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 15.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 16.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 17.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, et al. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 18.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 19.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zandi E, Chen Y, Karin M. Science. 1998;281:1360–1363. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]
- 21.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 22.Cohen L, Henzel W J, Baeuerle P A. Nature (London) 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 23.Rothwarf D M, Zandi E, Natoli G, Karin M. Nature (London) 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 24.Kirkpatrick C, Peifer M. Curr Opin Genet Dev. 1995;5:56–65. doi: 10.1016/s0959-437x(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 25.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 26.Peifer M. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 27.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orford K, Crockett C, Jensen J P, Weissman A M, Byers S W. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 29.Jiang J, Struhl G. Nature (London) 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 30.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheng Y, Axel R. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 31.Hatakeyama S, Jensen J P, Weissman A M. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 32.Spevak W, Keiper B D, Stratowa C, Castanon M J. Mol Cell Biol. 1993;13:4953–4966. doi: 10.1128/mcb.13.8.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margottin F, Bour S, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 34.Michel J, Siong Y. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- 35.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piatti S, Bohm T, Cocker J H, Diffley J F, Nasmyth K. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 37.Drury L S, Perkins G, Diffley J F. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kominami K, Toda T. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 39.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert U, Strebel K. J Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul M, Jabbar M A. Virology. 1997;232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]