Abstract
Killer cell inhibitory receptors (KIR) protect class I HLAs expressing target cells from natural killer (NK) cell-mediated lysis. To understand the molecular basis of this receptor-ligand recognition, we have crystallized the extracellular ligand-binding domains of KIR2DL2, a member of the Ig superfamily receptors that recognize HLA-Cw1, 3, 7, and 8 allotypes. The structure was determined in two different crystal forms, an orthorhombic P212121 and a trigonal P3221 space group, to resolutions of 3.0 and 2.9 Å, respectively. The overall fold of this structure, like KIR2DL1, exhibits K-type Ig topology with cis-proline residues in both domains that define β-strand switching, which sets KIR apart from the C2-type hematopoietic growth hormone receptor fold. The hinge angle of KIR2DL2 is approximately 80°, 14° larger than that observed in KIR2DL1 despite the existence of conserved hydrophobic residues near the hinge region. There is also a 5° difference in the observed hinge angles in two crystal forms of 2DL2, suggesting that the interdomain hinge angle is not fixed. The putative ligand-binding site is formed by residues from several variable loops with charge distribution apparently complementary to that of HLA-C. The packing of the receptors in the orthorhombic crystal form offers an intriguing model for receptor aggregation on the cell surface.
As part of the innate immune system, natural killer (NK) cells activate their cytolytic killing against certain pathogen-infected or tumor cells. Critical to this function is the ability of NK cells to discriminate between self- and nonself, thereby directing NK-mediated lysis only against appropriate target cells. One mechanism to achieve self-recognition is through regulation by cell surface inhibitory receptors that are capable of interacting with class I MHC molecules expressed on the target cell surface, as proposed by Kärre and coworkers in the “missing-self” hypothesis (1, 2).
To date, two superfamilies of receptors, the Ig superfamily and the C-type lectin superfamily, are known to mediate class I MHC-regulated NK functions (3, 4). The receptors of the Ig superfamily are type I transmembrane glycoproteins, which contain one or more Ig-like extracellular domains (5, 6), whereas the C-type lectin receptors are dimeric type II transmembrane glycoproteins, with each chain containing a single, extracellular C-type lectin domain. The ligand for the human C-type lectin receptor, CD94/NKG2, has been identified recently as the nonclassical class I antigen HLA-E (7–10).
The best-studied Ig superfamily killer cell inhibitory receptors (KIR) consist of the p50, p58, and p70 molecules, now known as KIR2DS, KIR2DL, and KIR3D, respectively, distinguished by their molecular weight. KIR2DS and KIR2DL receptors contain two Ig domains (known as D1 and D2) and differ primarily in the transmembrane and the intracellular regions. KIR2DL molecules are inhibitory receptors possessing intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIM), whereas KIR2DS molecules appear to be noninhibitory receptors with shorter intracellular tails that interact with an immunoreceptor tyrosine-based activation motif (ITAM) containing signaling component DAP12 (11). KIR3D members have three Ig domains (known as D0, D1, and D2) and can be either inhibitory or noninhibitory.
KIR2DL1 and KIR2DL2 represent two distinct classes of p58 molecules that recognize HLA-Cw2, -4, -5, and -6 and HLA-Cw1, -3, -7, and -8 allotypes, respectively (12, 13). Recently, the crystal structure of KIR2DL1 revealed a fold that resembles the hematopoietic growth factor receptor fold (14). In an attempt to understand the molecular basis of HLA allele specificities, we now describe the structure of KIR2DL2.
MATERIALS AND METHODS
Protein corresponding to the extracellular ligand-binding region of 2DL2 was expressed in a bacterial expression system and refolded in vitro by using a procedure similar to that published previously (15). All crystallization trials were performed by using hanging-drop vapor-diffusion method at room temperature. The crystals of 0.05 × 0.04 × 0.1 mm in size were obtained under the conditions of 250 mM calcium acetate, 6% PEG 8000, and 33 mM sodium cacodylate, pH 6.5, using proteins corresponding to amino acids 1–200. These crystals belong to the orthorhombic space group P212121, and they diffracted to 3.0 Å with cell dimensions a = 40.5 Å, b = 71.5 Å, c = 88.8 Å and α = β = γ = 90°. A second trigonal form was obtained in a solution of 400 mM calcium acetate/70 mM sodium cacodylate (pH 6.5). Microseeding of this form yielded crystals 0.1 × 0.1 × 0.4 mm in size that diffracted to 2.9 Å. It belongs to the space group P3221 with cell dimensions of a = b = 91.3 Å, c = 61.4 Å, α = β = 90°, and γ = 120°. Crystals were flash-frozen in a stream of nitrogen gas cooled to −180°C after stepwise equilibration to a cryoprotecting solution consisting of well solution and 30% glycerol.
Native data for P212121 and P3221 crystals were collected at Brookhaven National Laboratory X9B beamline by using an MAR345 detector. The KAu(CN)2 anomalous data were collected at the Cornell High Energy Synchrotron Source (CHESS) by using the F2 beamline adjusted to Au absorption edge (Table 1) (16). Data were processed by using denzo, and scalepack (17). The heavy atom positions of the gold derivative were interpreted by shelx-97 and refined by using mlphare (18, 19).
Table 1.
Data collection statistics
| Data set | Space group | Wavelength, Å | Resolution, Å | Unique reflections | Average, I/σ | Rsym,* % | Complete, % | P.P.† | Rcullis |
|---|---|---|---|---|---|---|---|---|---|
| Native 1 | P212121 | 1.54 | 3.4 | 3,832 | 11.2 | 11.9 (28.0)‡ | 99.0 (99.2)‡ | ||
| Native 2 | P212121 | 0.9800 | 3.0 | 4,683 | 18.1 | 7.9 (28.8) | 85.1 (87.9) | ||
| Native 3 | P3221 | 0.9918 | 2.9 | 6,169 | 13.1 | 6.3 (23.6) | 90.3 (75.4) | ||
| KAu(CN)2 1 | P212121 | 1.54 | 3.4 | 5,414* | 8.9 | 12.9 (30.4) | 79.1 (81.1) | 1.41 | 0.67 |
| KAu(CN)2 2 | P212121 | 1.040 | 2.8 | 9,085* | 17.5 | 8.5 (14.8) | 73.5 (70.9) | 1.06 | 0.72 |
Native 1 and 2 are merged to yield a single data set with a total of 5,149 reflections, Rmerge of 11.6% and 92.3% complete to 3.0 Å.
Σ(Ih − 〈I〉h)/ΣIh, Bijvoet pairs are merged separately.
Phasing power. The overall figure of merit is 0.59 for the gold data.
Values in parentheses correspond to the last resolution shell: 3.2–3.0 and 3.0–2.9 Å for P212121 and P3221, respectively.
The structure was solved by using the molecular replacement method combined with heavy atom phasing. The coordinates for the search model were from the KIR2DL1 structure (14). A polyalanine model with the N- and C-terminal domains separated at amino acid 101 to allow for hinge movement was used in the molecular replacement. Several solutions, generated by x-plor and amore with similar correlation coefficients (20, 21), were obtained for each domain, and the correct solution for P212121 was found by visually examining the quality of the electron density maps. It yielded a correlation coefficient of 61.9% and an initial R factor of 41% after the rigid body fitting with amore. The solution for the trigonal P3221 form was obtained by using the P212121 structure as a probe and by varying the hinge angle of the two domains. This search yielded a solution with a correlation coefficient of 63.1% and initial R factor of 42.1% after the rigid body fitting with amore. At this point, most of the β-strands are visible in the density. However, many of the loop regions are not visible. To improve the electron density, the phases obtained from the gold derivative then were combined with those from the molecular replacement solution in the orthorhombic crystal form by using sigmaa (19). Densities from the phase-combined orthorhombic crystal then were averaged with those from the trigonal crystal form. The refinement proceeded with simulated annealing and positional and grouped B factor refinement by using x-plor 3.8 together with alternating model building using the program o (22). Both bulk solvent correction and anisotropic scaling were applied to observed reflections. After initial rounds of refinement, water molecules were added to (2Fo − Fc) electron density peaks that are higher than 1.5 σ (SD of the map) and within hydrogen-bonding distance to protein molecules. The addition of water molecules improved Rfree by 1–2% and Rcryst by 2–3%.
RESULTS AND DISCUSSION
The Overall Structure of KIR2DL2.
The extracellular ligand-recognition portion of the human killer inhibitory receptor 2DL2, corresponding to amino acids 1–200, was expressed in Escherichia coli as inclusion bodies and refolded in vitro. Crystals were obtained in two different space groups, an orthorhombic form of P212121 and a trigonal form of P3221. The structures of the two forms have been determined by using a combination of phases from molecular replacement, a gold heavy atom derivative and crystal averaging to 3.0- and 2.9-Å resolution, respectively (Tables 1 and 2), and were refined independently. In the refined (2Fo − Fc) map of the orthorhombic form, the FG loop region of the N-terminal domain, residues His-85 and Gln-89, and the CC′ loop region of the C-terminal domain, residues Ala-145 to His-146, display discontinuity in electron density when contoured at 1σ (SD) level. Similar regions in the trigonal crystal form, residues Thr-84 to Pro-87 and Glu-142 to Glu-144, also display density breaks in the final (2Fo − Fc) density map at 1σ contour level.
Table 2.
Refinement statistics
| Protein atoms | Solvent molecules | Rcryst, % | rms
|
|||
|---|---|---|---|---|---|---|
| Bond | Angle | Dihedrals | ||||
| Native 1 & 2 | 1,526 | 29 | 21.9 (30.9)† | 0.010 | 2.00 | 26.5 |
| Native 3 | 1,521 | 10 | 22.7 (33.1) | 0.006 | 1.37 | 26.3 |
Σ|Fo − Fc|/ΣFo.
Values in parentheses are Rfree calculated using randomly chosen 5% reflections.
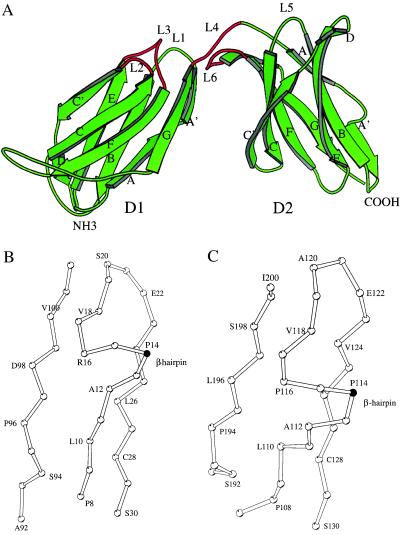
The overall fold of 2DL2 is similar to 2DL1 in that each domain forms a β-sandwich with β-strands ABED and C′CFGA′ (Fig. 1A). The individual D1 and D2 domains of 2DL2 can be superimposed onto the corresponding domains of KIR2DL1, resulting in rms differences of 0.9 Å and 0.6 Å among all their Cα atoms. The N- and C-terminal domains of 2DL2, sharing a 40% sequence identity, can also be superimposed readily, resulting in a rms difference of 1.2 Å among 87 Cα atoms. This fold, designated as K-type (14), is similar but not identical to a subclass of C2-type Ig fold, as found in the hematopoietic receptor family, that includes the human growth hormone receptor (hGHR), tissue factor, the prolactin receptor, interferon γ receptor, and erythropoietin receptor (23–30). The difference is primarily in the pairing of two strands. In KIR structures, the first β-strand splits into A and A′ to hydrogen bond with the B and G strands, resulting in a “strand switching,” but it remains as one strand to hydrogen bond with the B strand in hematopoietic receptors. Second, the strand C′ in the C2 topology pairs only with strand C, whereas the same strand in KIR pairs with both strand C and E, creating a short D strand (Fig. 1). Interestingly, the KIR topology resembles that of E-type Ig fold found in the rel homology domain and the C-terminal domain of galactose oxidase (31).
Figure 1.
(A) Ribbon diagram of 2DL2 structure. The secondary structure assignment for the strands in D1 domain are A (8–12), A′ (16–19), B (23–30), C (36–43), C′ (46–52), D (53–56), E (60–67), F (75–82), and G (90–101). The strands for the D2 domain are: A (108–111), A′ (116–119), B (123–130), C (135–142), C′ (145–152), D (153–156), E (160–167), F (172–180), and G (188–198), respectively. This figure and other structural figures were prepared by using the program molscript 2.1 and raster3d (55, 56). The region around cis-Pro-14 (B) and cis-Pro-114 (C) shows the kink at the proline residue and the β-hairpin.
A cis-Proline Residue in the First Strand Switches Strand A to A′.
Strand switching involving prolines in the first strand has been observed in the structures of V- and I-type Ig fold, such as the V domain of an antibody and the first domain of vascular cell adhesion molecule 1 (32). Similarly, the switch region in KIR involves a sharp kink followed by a four-residue β-hairpin at the junction of strand A and A′ (Fig. 1 B and C). This β-hairpin, which redirects strand A′ to pair with strand G, is found in both domains of 2DL2 as well as 2DL1 and can be attributed to the occurrence of a cis-proline residue in each domain, Pro-14 in D1 and Pro-114 in D2. All other prolines in the structure adopt the more commonly trans-peptide conformation. The presence of a proline residue at this location is found in all human KIR sequences, with Pro-Gly occurring most frequently in D1 domains, Pro-Gly-Pro in D2 domains, and Pro-Ser in D0 domains, suggesting the presence of this strand-switching at similar positions in all KIRs. This cis-proline, not present in hGHR, distinguishes KIR topology apart from that of the hematopoietic receptors. Further structural comparisons between KIR and members of other known Ig families show that this cis-proline is present only in the structures of KIR.
The Hinge Region.
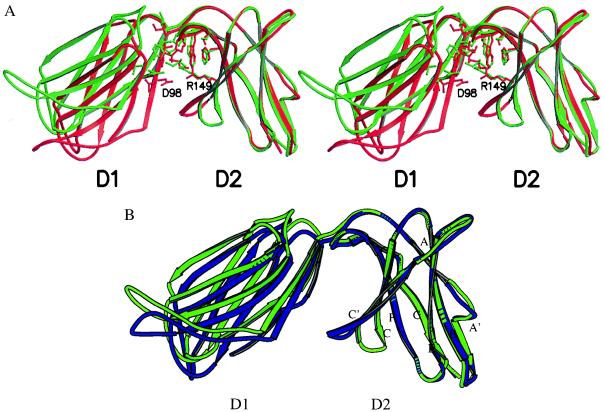
The linker between the N-terminal D1 and the C-terminal D2 domains consists of five residues, Gly-103 to Lys-107, with Leu-104 at the hinge point. The hinge angle between the two domains of KIR2DL2 is approximately 80° in the refined structures of the two crystal forms (calculated using the program hinge). It differs from the 66° angle found in the 2DL1 structure (Fig. 2A) (14). In addition, a 30° rotation with respect to the spindle axis of D1 also is observed between the N-terminal domains of 2DL1 and 2DL2 when their C-terminal domains are superimposed. Such a difference in the hinge angles between the two structures is unexpected because both possess a rather conserved set of hydrophobic residues in the interdomain hinge region that presumably stabilize these angles. Such a hydrophobic core first was observed in the structure of the T cell receptor (TCR) β chain, where it contributed to an acute 76° hinge angle (33). This hydrophobic core in 2DL2 consists of Leu-17, Met-69, Val-100, Ile-101, and Thr-102 from the D1 domain and His-138, Phe-178, Ser-180, Pro-185, Tyr-186, and Trp-188 from the D2 domain. Interestingly, Trp-188 is part of the WSXSS sequence, the KIR family equivalent of WSXWS motif sequence found in the hematopoietic receptor superfamily. This tryptophan packs against the conserved Leu-17 and Val-100 of D1 domain to form the center of the hydrophobic core. A hydrogen bond from Thr-76 to Tyr-186 and a salt bridge from Asp-98 to Arg-149 below the core also stabilize the hinge angle (Fig. 2A). Apart from a Thr-to-Ile change at position 102, the rest of the core is identical to the structure of 2DL1. This suggests that the observed hinge difference most likely is the result of crystal packing rather than the change in core residues. Furthermore, the two crystal forms of 2DL2 also differ by about 5° in their hinge angles (Fig. 2B), indicating that they are not fixed. The total buried solvent-accessible area between the two domains of 2DL2 is 919 Å2 (calculated with the program surface and using a probe radius of 1.4 Å) (19), similar to the 1,076 Å2 observed for 2DL1 and the 833 Å2 between the Vβ and Cβ TCR β chain (14, 33).
Figure 2.
(A) Stereo drawing showing the structure overlay between 2DL1 (in red) and 2DL2 (in green) with their D2 domains superimposed. The hinge angles, calculated with the program hinge, are 66° and 80° for 2DL1 and 2DL2, respectively. Residues involved in the interdomain packing are shown. The coordinates are from the refined orthorhombic crystal form. (B) Overlay of refined 2DL2 structures from the orthorhombic (in green) and the trigonal (in blue) crystal forms.
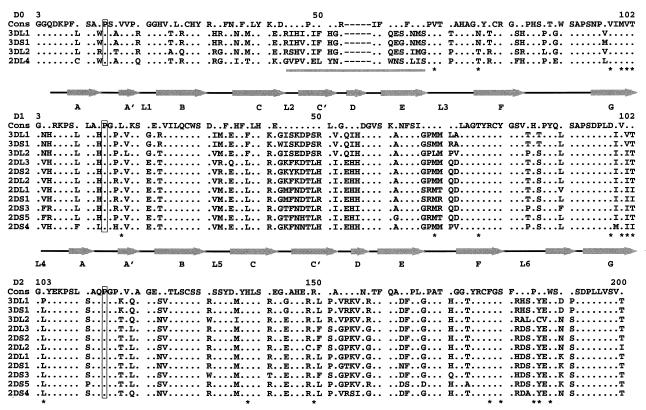
Sequence alignment shows that most of the residues in this hydrophobic core are well conserved among the known KIR sequences (Fig. 4), indicating the existence of a conserved interdomain packing in this receptor family to restrict their hinge to more acute angles. On the other hand, this core does not appear to impose absolute rigidity on the hinge angle. The ability of hinge angle to vary may reflect the at-the-hinge location of the hydrophobic core, which makes it less sensitive to angle changes. The phenomenon of a ligand-induced change in hinge angle first was observed in the structure of hGHR, where two identical receptor subunits bind to separate regions of growth hormone using the same sets of residues (23). The asymmetry of the hormone induces different angles in the two receptor subunits, 95° in subunit I and 85° in the subunit II. It can be argued that smaller hinge angles enable KIR molecules to adopt a preshaped conformation with an exposed upward-situated binding epitope to facilitate their scanning through a large number of HLA class I molecules with different binding specificity, whereas the flexibility may be necessary to allow induced fit upon binding to their ligand class I HLA molecules.
Figure 4.
Sequence alignment for members of the KIR family. The sequences are shown for the D0 domain (Top), D1 domain (Middle), and D2 domain (Bottom). The secondary structure of 2DL2 is shown above the sequences for D0, D1, and D2 domains. Conserved positions are indicated by dots. The respective cis-proline residues are boxed. Residues involved in the hinge hydrophobic core are marked with an ∗. The variable region in the D0 domain is underlined. Dashes indicate deletions in a sequence. The numbering is consistent with the mature amino acid sequence numbering.
The Charges in the KIR Putative-Binding Region Correlate with Those in the HLA Putative Receptor-Binding Site.
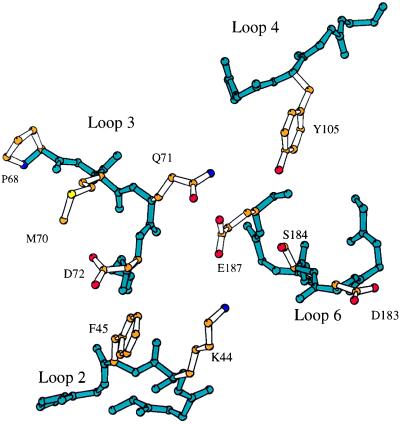
Several residues near the C-terminal end of the α1 helix of HLA-C molecules, including the dimorphic residues 77 and 80 (Asn and Lys in Cw 2, 4, 5, and 6 versus Ser and Asn in Cw 1, 3, 7, and 8) and residues 73, 76, and 90, have been shown to affect receptor-binding specificity and affinity (34). In addition, mutational analysis of the KIR has identified several residues that are important for the recognition of HLA class I ligands. Position 44, a lysine residue in 2DL2, appears particularly important in determining the receptor specificity (35). Other residues, including phenylalanine 45 and methionine 70, also influence the ligand affinity (36, 37). Four loops (L2, L3, L4, and L6) are situated in this region defined by mutational analysis (Figs. 1A and 3). Loop 2, residues 43–46, between strands C and C′, contains both lysine 44 and phenylalanine 45. Loop 3, residues 69–75, between the strands E and F, is found adjacent to L2 and contains Met-70 (Fig. 3). Several other residues, including Gln-71, Asp-72 from L3, Leu-104 and Tyr-105 from L4, and Asp-183, Ser-184, and Glu-187 from the C-terminal domain L6, also are found near this region. The overall appearance of this putative ligand-binding site is quite polar, with three acidic residues conferring a net negative charge.
Figure 3.
The residues observed in the putative ligand-binding region. The backbones of loop 2 (residues 43–46), loop 3 (69–75), loop 4 (103–107), and loop 6 (181–187) are shown in blue, and the side chains of residues in this region are shown in color according to atom type: oxygen, red; nitrogen, blue; carbon, orange; sulfur, yellow.
Sequence comparison indicates that three of the four loops, L2, L3, and L6, display considerably more sequence variation than the rest of the molecule (Fig. 4). This is particularly obvious when L2 and L3 are compared with their topological equivalent loops in D2, residues 143–146 and 167–171, and when L6 is compared with its topological equivalent loop, residues 85–90 in D1, which shows much less sequence variation. This is consistent with the notion that these three loops are involved functionally in the recognition of HLA molecules. Another loop, residues 153–158 between the strands C′ and E of the D2 domain, displays similar variability to L3. It is located near the hinge region but away from L2, L3, L4, and L6. It is unclear whether this loop is also involved in HLA binding.
The influence of charge on KIR recognition of HLA has been reported in a number of studies involving both KIR3D and KIR2D receptors (38–40). Although the molecular interactions between killer cell receptors and HLA molecules are yet to be established, the structure of the receptor, particularly the charge distribution in the putative ligand-binding region, does offer some insight into the apparent importance of charge in recognition. For example, both 2DL1 and 2DL2 carry several negatively charged residues in their ligand-binding region, which may explain the need for a lysine residue either from HLA molecules (Lys-80 of HLA-Cw2, 4, and 6) or from the receptor (Lys-44 of 2DL2), but not both, for recognition. When methionine 44 of 2DL1 is replaced with lysine, it can no longer recognize the HLA-C allotypes possessing Asn-77 and Lys-80 but, instead, recognizes the uncharged Ser-77 and Asn-80 (35). Similarly, substitution of Thr-70 with Lys in 2DL1 may have disrupted the existing charge balance, thus resulting in decreased binding (37). Furthermore, when negatively charged residues were substituted into the binding peptide of HLA-Cw4, a decreased receptor binding was observed, again consistent with the disruption of the receptor–ligand charge balance (39).
The D0 Domain.
Among the three Ig domains of KIR3D, the D1 and D2 domains share 70% or higher sequence identity with the respective domains of 2DL2. The D0 domain of KIR3D, however, shares less than 40% sequence identity with either D1 or D2. Nevertheless, the presence of a proline residue at the corresponding cis-proline location of 2DL2 in its sequence suggests that D0 possesses a KIR topology. The sequence alignment shows that there is a five-residue deletion near the C′ and E strands in D0 as compared with D1 and D2 (Fig. 4). This likely results in a shorter loop between the two strands. When the hydrophobic core residues at the hinge are compared, the D0 domain appears to be D1-like as opposed to D2-like. An interesting example is 2DL4, which consists of D0 and D2 without D1 (41). Because D0 possesses a D1-like hydrophobic core at the hinge position, it can be argued that 2DL4 retains the structure similar to that of 2DL2. However, the corresponding aspartate 98 of D1 is replaced with a valine in D0, precluding a salt bridge between Asp-98 and Arg-149 in 2DL4. In addition, the D0 domain of 2DL4 contains a variable loop in the same region as the L2 loop of 2DL2, suggesting that the loop is part of the ligand-recognition site. In contrast to 2DL4, the loops in the D0 domain of KIR3D molecules show little sequence variation, suggesting that they are not involved directly in ligand recognition. Instead, the requirement for the D0 domain in the function of KIR3D molecules may be for structural or for general affinity (42).
Other KIR-Like Molecules.
Two new families of KIR-like receptors, the Ig-like transcripts (ILTs, also known as leukocyte Ig-like receptors or LIRs) and the leukocyte-associated Ig-like receptors (LAIR-1 and -2) have been identified recently and appeared to recognize MHC class I molecules (43–46). Both LAIR and ILT families contain the proline residues at the corresponding cis-proline location of 2DL2 in their Ig domains. Among the four Ig domain ILTs, the third and fourth domains share 44% sequence identity with the D1 and D2 domains of 2DL2, and the regions of conserved sequence align well with the regions of regular secondary structural elements. In addition, the murine Gp49A and B are found homologous to KIR with approximately 35% sequence identity (47). Both are predicted to contain the characteristic cis-proline residues, making them undoubtedly part of the KIR fold.
KIR2DL2 Forms an Orderly Aggregate in Crystal.
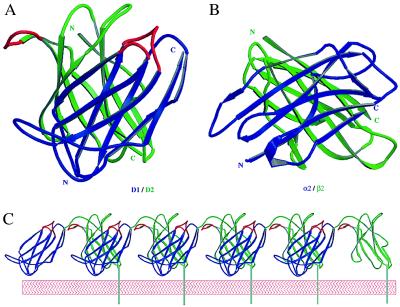
Ligand-induced receptor aggregation is presumed to be a common mechanism for initiating receptor-mediated signaling (48, 49). Evidence of MHC peptide-mediated T cell receptor oligomerization has been found both in solution and on the cell surface (50, 51). To date, several different structure models have been proposed as possible mechanisms for receptor aggregation and signal transduction (23, 52, 53). We observed an intriguing form of receptor dimer in the orthorhombic crystals of 2DL2 (Fig. 5A). Two receptor molecules with the same orientation dimerize in such a way that the amino-terminal D1 domain of one receptor packs against the carboxyl-terminal D2 domain of the other molecule with their strands in an approximate orthogonal orientation, creating a D1/D2 heterodimer reminiscent of a handshake. The interface is away from the ligand-binding region, and it involves portions of the C′, D, and E strands from both domains with a total buried solvent-accessible area of 480 Å2, approximately half the size of hGHR dimer interface (23). This handshake packing between D1 and D2, interestingly, resembles that between the α2 and β2 domains of class II MHC molecules, which buries a total of 680 Å2 solvent-accessible area. Both use the ABED β-sheet as the common interfaces, with D strand mediating the primary contact (Fig. 5B). It is necessary to point out, however, that this is only the crystallographic packing, and efforts to isolate dimeric receptors in solution has not been successful. This kind of packing permits receptor molecules to form oligomeric aggregates through alternating dimerization of their D1 and D2 domains (Fig. 5C), as seen in the crystal lattice of 2DL2. The formation of an orderly aggregate is consistent with the observation that receptor activation often is achieved through cross-linking by antibody. As a result of the D1/D2 packing, a D2-domain loop of the second receptor, residues 150–156, is brought into close proximity with the putative ligand-binding region of the first receptor. Previously, a zipping model involving the adhesion domain of an N-cadherin has been proposed to be important to cell adhesion (54). It is possible that upon recognizing HLA class I molecules, the killer cell receptors form these D1/D2 cross dimers, which, in turn, lead to the formation of an orderly aggregation. The extent of receptor aggregation will depend on the concentration of the ligand HLA molecules and the affinity of the binding, because both influence the stability of the aggregate.
Figure 5.
Dimerization and aggregation of receptors. (A) Crystal lattice interaction results in two molecules of 2DL2 forming a D1/D2 packing. The N-terminal D1 domains are shown in blue and the C-terminal D2 domains are in green. The putative ligand-binding loops are shown in red. (B) The packing between the α2 (blue) and β2 (green) domains of HLA-DR1. The coordinates are from Protein Data Bank reference 1dlh (57). (C) Receptors form an orderly aggregate in crystal. The N-terminal domains are in blue and C-terminal domains are in green. The C-terminal transmembrane tail and membrane bilayer are modeled in to illustrate the orientation on the cell surface.
Acknowledgments
We thank Dr. C. Titlow for assistance in molecular cloning, Drs. Y. Zhang and J. Boyington for assistance in crystallographic data collection and insightful suggestions throughout the project, Mr. S. Motyka for providing all the sequence-alignment results, Dr. C. Hammer for providing the mass spectrometry measurements, and Dr. M. Garfield for amino acid sequencing. We also thank Dr. E. Long and C. Winter for providing valuable opinions on the manuscript. We thank Dr. Q. Fan for providing the coordinates of the refined KIR2DL1 structure. This work is supported by intramural research funding of National Institute of Allergy and Infectious Diseases.
ABBREVIATIONS
- KIR
killer Ig receptor
- NK
natural killer
- 2DL2
two domains and long-form number two
- 2DL1
two domains and long-form number one
- hGHR
human growth hormone receptor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 2dl2).
References
- 1.Kärre K, Ljunggren H G, Piontek G, Kiessling R. Nature (London) 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren H G, Kärre K. Immunol Today. 1990;11:237–243. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Long E O, Wagtmann N. Curr Opin Immunol. 1997;9:344–350. doi: 10.1016/s0952-7915(97)80080-5. [DOI] [PubMed] [Google Scholar]
- 4.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 5.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 6.Colonna M, Samaridis J. Nature (London) 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 7.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J. Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braud V M, Allan D S, O’Callaghan C A, Söderström K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, et al. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 9.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carretero M, Palmieri G, Llano M, Tullio V, Santoni A, Geraghty D E, Lopez-Botet M. Eur J Immunol. 1998;28:1280–1291. doi: 10.1002/(SICI)1521-4141(199804)28:04<1280::AID-IMMU1280>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 11.Lanier L L, Corliss B C, Wu J, Leong C, Phillips J H. Nature (London) 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 12.Moretta A, Vitale M, Bottino C, Orengo A M, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 14.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 15.Fan Q R, Garboczi D N, Winter C C, Peruzzi M, Long E. Proc Natl. Acad Sci USA. 1996;93:7178–7183. doi: 10.1073/pnas.93.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick G. Acta Cryst A. 1990;46:467–473. [Google Scholar]
- 19.Collaborative Computational Project No. 4. Acta Crystallogr D. 1995;50:760–763. [Google Scholar]
- 20.Brünger A T. X-PLOR: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 21.Navaza J. Acta Cryst A. 1994;50:157–163. [Google Scholar]
- 22.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.DeVos A M, Ultsch M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 24.Harlos K, Martin D M, O’Brien D P, Jones E Y, Stuart D I, Polikarpov I, Miller A, Tuddenham E G, Boys C W. Nature (London) 1994;370:662–666. doi: 10.1038/370662a0. [DOI] [PubMed] [Google Scholar]
- 25.Muller Y A, Ultsch M H, Kelley R F, de Vos A M. Biochemistry. 1994;33:10864–10870. doi: 10.1021/bi00202a003. [DOI] [PubMed] [Google Scholar]
- 26.Banner D W, D’Arcy A, Chene C, Winkler F K, Guha A, Konigsberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 27.Somers W, Ultsch M, de Vos A M, Kossiakoff A A. Nature (London) 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 28.Walter M R, Windsor W T, Nagahhushan T L, Lundell D J, Lunn C A, Zauodny P J, Narula S K. Nature (London) 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 29.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 30.Syed R S, Reid S W, Li C, Cheetham J C, Aoki K H, Liu B, Zhan H, Osslund T D, Chlrino A J, Zhang J, et al. Nature (London) 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 31.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;47:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 32.Sun P D, Boyington J C. Current Protocols in Protein Science. New York: Wiley; 1997. pp. 1–100. [Google Scholar]
- 33.Bentley G A, Boulot G, Karjalainen K, Mariuzza R A. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 34.Mandelboim O, Reyburn H T, Sheu E G, Valés-Gómez M, Davis D M, Pazmany L, Strominger J L. Immunity. 1997;6:341–350. doi: 10.1016/s1074-7613(00)80336-2. [DOI] [PubMed] [Google Scholar]
- 35.Winter C C, Long E O. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 36.Winter C C, Gumperz J E, Parham P, Long E O, Wagtmann N. J. Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 37.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 38.Peruzzi M, Parker K C, Long E O, Malnati M S. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 39.Rajagopalan S, Long E. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gumperz J E, Barber L D, Valiante N M, Percival L, Phillips J H, Lanier L L, Parham P. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 41.Selvakumar A, Steffens U, Dupont B. Tissue Antigens. 1996;48:285–295. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 42.Rojo S, Wagtmann N, Long E. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 43.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 44.Borges L, Hsu M-L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 45.Colonna M, Navarro F, Bellón T, Llano M, Garcia P, Samaridis S, Angman L, Cella M, López-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyaard L, Adema G J, Chang C, Woollatt E, Sutherland G R, Lanier L L, Phillips J H. Immunity. 1997;7:283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- 47.Arm J P, Gurish M F, Reynolds D S, Scott H C, Gartner C S, Austen K F, Katz H R. J Biol Chem. 1991;266:15966–15973. [PubMed] [Google Scholar]
- 48.Germain R N. Curr Biol. 1997;7:R640–R644. doi: 10.1016/s0960-9822(06)00323-x. [DOI] [PubMed] [Google Scholar]
- 49.Qian D, Weiss A. Curr Opin Cell Biol. 1997;9:205–212. doi: 10.1016/s0955-0674(97)80064-6. [DOI] [PubMed] [Google Scholar]
- 50.Reich Z, Boniface J J, Lyons D S, Borochov N, Wachtel E J, Davis M M. Nature (London) 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 51.Konig R, Shen X, Germain R N. J Exp Med. 1995;182:779–787. doi: 10.1084/jem.182.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 53.Fields B A, Ober B, Malchiodi E L, Lebedeva M I, Braden B C, Ysern X, Kim J-K, Shao X, Ward E S, Mariuzza R A. Science. 1995;270:1821–1824. doi: 10.1126/science.270.5243.1821. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro L, Fannon A M, Kwong P D, Thompson A, Lehmann M S, Grübel G, Legrand J-F, Als-Nielsen J, Colman D R, Hendrickson W A. Nature (London) 1995;374:327–336. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 55.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 56.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 57.Stern L J, Brown J H, Jardetzky T J, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]