Abstract
Can kindreds with tooth agenesis caused by MSX1 or PAX9 mutations be distinguished by their phenotypes? We have identified an MSX1 frameshift mutation (g.62dupG, p.G22RfsX168) that causes non-syndromic autosomal-dominant oligodontia, featuring the absence of multiple permanent teeth, including all second bicuspids and mandibular central incisors. The dominant phenotype is apparently due to haploinsufficiency. We analyzed patterns of partial tooth agenesis in seven kindreds with defined MSX1 mutations and ten kindreds with defined PAX9 mutations. The probability of missing a particular type of tooth is always bilaterally symmetrical, but differences exist between the maxilla and mandible. MSX1-associated oligodontia typically includes missing maxillary and mandibular second bicuspids and maxillary first bicuspids. The most distinguishing feature of MSX1-associated oligodontia is the frequent (75%) absence of maxillary first bicuspids, while the most distinguishing feature of PAX9-associated oligodontia is the frequent (> 80%) absence of the maxillary and mandibular second molars.
Keywords: oligodontia, homeobox, autosomal dominant, MSX1, frameshift, PAX9
INTRODUCTION
MSX1 is a homeobox gene involved in multiple epithelial-mesenchymal interactions during vertebrate embryogenesis, and appears to be most critical during early tooth development. The principle phenotype in Msx1−/Msx1−knockout mice was cleft palate and a failure of tooth development (Satokata and Maas, 1994). A lack of MSX1 expression reduces the expression of downstream signaling molecules, such as Bone Morphogenetic Protein 4 (BMP4), and transcription factors like LEF1 (Chen et al., 1996). MSX1 interacts antagonistically with several transcription factors, such as DLX2 and DLX5 (Zhang et al., 1997), Lhx2 (Bendall et al., 1998), PAX3 (Bendall et al., 1999), PAX9 (Vieira et al., 2004; Ogawa et al., 2005), and binds to a multiprotein transcriptional complex containing TATA-binding protein (TBP), Sp1, or the cAMP-response-element-binding protein (CBP/p300) (Shetty et al., 1999). MSX1 also associates with histone isoform H1b to inhibit gene transcription during myogenesis (Lee et al., 2004). It has been proposed that MSX1 expression maintains cyclin D1 expression, preventing cells from exiting the cell cycle (Hu et al., 2001), and acts as a negative regulator of differentiation (Bendall et al., 1999).
MSX1 is highly expressed in the mesenchyme of developing tooth germs, particularly during the early (bud and cap) stages (Peters et al., 1998). Prior to this report, six different MSX1 defects had been reported that cause multiple congenitally missing teeth, or oligodontia (Vastardis et al., 1996; van den Boogaard et al., 2000; Jumlongras et al., 2001; Lidral and Reising, 2002; Nieminen et al., 2003; De Muynck et al., 2004). An MSX1 nonsense mutation (p.Ser105stop) also produced cleft lip and palate (CL+P) (van den Boogaard et al., 2000; De Muynck et al., 2004). Another MSX1 nonsense mutation, p.Ser202stop, produced oligodontia with nail dysplasia (Jumlongras et al., 2001). The presence of both overlapping (oligodontia) and non-overlapping (CL+P and nail dysplasia) phenotypes suggests that there are contextual influences in the expression and function of MSX1, which presumably relate to the mutations variably interfering with MSX1 interactions with other regulatory molecules, producing unique downstream effects in a tissue-specific manner (Jumlongras et al., 2001). Abnormalities of the short arm of chromosome 4, causing Wolf-Hirschhorn syndrome, exhibited oligodontia as part of the phenotype only in the five families where the defect included deletion of the MSX1 gene (Nieminen et al., 2003).
Familial tooth agenesis can occur as an isolated anomaly or as part of a genetic syndrome (Gorlin et al., 1990). Hypodontia, or the developmental absence of at least 1 tooth, is a common oral finding. Third molars are missing to various degrees in different populations, with most estimates finding that about 20% of the population is missing at least one third molar. Agenesis of at least one third molar was found in 22.5% of the 1000 panoramic radiographs examined from a Czech population (Rozkovcova et al., 2004). Among 4208 orthodontic patients in Germany, 388 patients (9.2%) were missing a total of 826 teeth (excluding wisdom teeth) (Stahl et al., 2003). MSX1 and PAX9 are involved in the etiology of non-syndromic partial tooth agenesis, although many other genes are also believed to play a role (Mostowska et al., 2003b). AXIN2, which encodes a Wnt-signaling regulator, is associated with oligodontia and colorectal neoplasia (Lammi et al., 2004).
The objective of the present study was to identify the mutation responsible for the familial tooth agenesis in our kindred and to identify genotype/phenotype correlations that could improve our understanding of normal and arrested tooth formation and better prioritize candidate genes based upon the pattern of partial tooth agenesis.
MATERIALS & METHODS
Enrollment of Human Subjects
The study protocol and subject consents were reviewed and approved by the Institutional Review Board at the University of Michigan, and appropriate informed consent was obtained from all subjects. Contact information was unavailable for the affected father, who was not recruited for the study.
Primer Design and Polymerase Chain-reaction (PCR)
A 10-mL quantity of peripheral whole blood was obtained from participating family members. Genomic DNA was isolated by means of the QIAamp DNA Blood Maxi Kit (Qiagen Inc., Valencia, CA, USA). Wild-type MSX1 genomic (Acc# AF426432) and mRNA (Acc# NM_002448) sequences were obtained from GenBank. Oligo -nucleotide primers to amplify the two MSX1 coding exons by polymerase chain-reaction (PCR) were designed with the use of Primer3 on the Web (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The PCR primers for exon 1, MSX1x1F (5′-CTGGCCTCGCCTTATTAGC-3′) and MSX1x1R (5′-GCCTGGG TTCTGGCTACTC-3′), used an annealing temperature of 58°C and generated a 766-bp amplification product. The primers for exon 2, MSX1x2F (5′-ACTTGGCGG CACTCAATATC-3′) and MSX1x2R (5′-CAGGGAGCAAAGAGGTGAAA-3′), had an annealing temp of 57°C and generated a 698-bp amplimer. PCR amplifications used the Platinum PCR supermix (Invitrogen, Carlsbad, CA, USA), and 10% DMSO was added for exon 1 amplification. PCR amplification products were purified by the QIAquick PCR Purification Kit and protocol (Qiagen Inc., Valencia, CA, USA). DNA sequencing was performed at the University of Michigan DNA sequencing core.
RESULTS
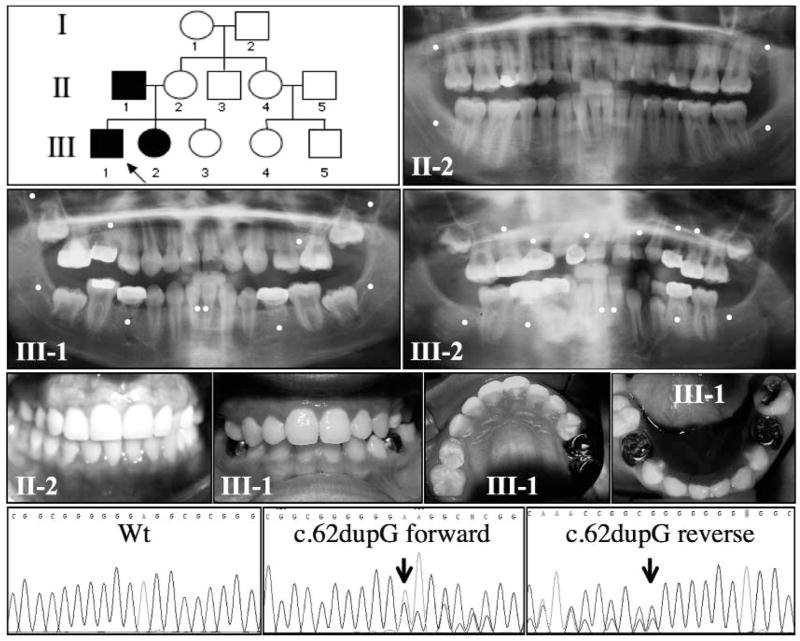
An autosomal-dominant trait of oligodontia was determined in a nuclear family having five members (Fig. 1). Two affected (III-1 and III-2) and two unaffected (II-2 and III-3) persons were recruited for the study. Each participant provided a blood sample for DNA analyses and was given physical, oral, and radiographic examinations. The affection status of the proband's father (II-1) and the maternal side of the family was obtained through an interview with the proband's mother (II-2). Subject II-2 and her family had no history of missing teeth except for third molars. Radiographic examination of the unaffected mother showed that she had no third molars, but the rest of the permanent dentition was present. Therefore, missing third molars was considered to be part of the genetic background in this kindred. Excluding the absence of third molars, the proband (III-1, age 11) was missing 6 permanent teeth, and his affected sister (III-2, age 9) was missing 12 (Fig. 2A). The unaffected youngest sibling (III-3, age 4) did not show any missing permanent teeth, but, owing to the early stage of development, the presence of second premolars and second and third molars could not be confirmed at that time. No one had a history of missing any primary teeth. Tooth size appeared to be similar in unaffected and affected individuals. Despite the lack of mandibular central incisors and second bicuspids, the proband did not display a generalized spacing of the teeth or significant tilting of the mandibular first molars or incisors. Besides the absence of multiple permanent teeth, no other physical symptoms, such as oral clefting or malformation of the finger- or toenails, were observed. These findings supported the diagnosis of autosomal-dominant non-syndromic oligodontia.
Figure 1.
Oral photographs of the unaffected mother (II-2) and the proband (III-1; age 11) show that the MSX1 mutation (g.62dupG) has no apparent effect on tooth size (top). Panorex radiographs demonstrate that the unaffected mother (II-2) has all of her permanent teeth except the third molars, while the proband (III-1) is missing 6 teeth, and his affected sister (III-2; age 9) is missing 12 teeth, not counting the third molars. A white dot indicates the expected location of a congenitally absent tooth. The pedigree indicates that the oligodontia trait in this family was transmitted from the father to the offspring in an autosomal-dominant pattern of inheritance. DNA sequencing chromatograms (bottom) identified the specific MSX1 frameshift mutation (arrows) in both the forward and reverse directions. This mutation was not observed in the wild-type (Wt) MSX1 gene in the unaffected members of the kindred.
Figure 2.
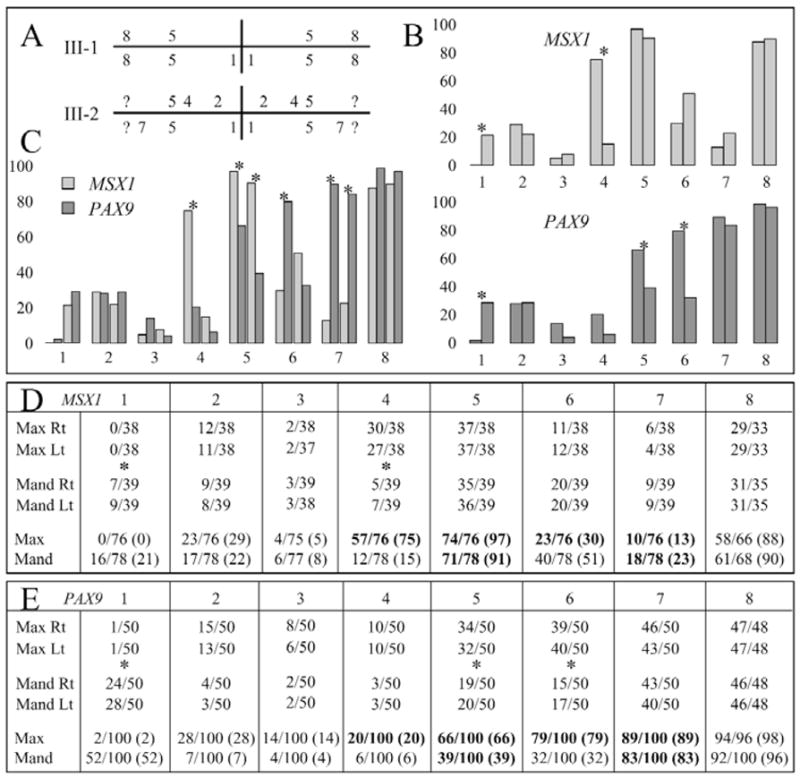
Quadrant diagram (Palmer notation system) indicating the teeth missing in the two affected members (III-1 and III-2) of the MSX1 kindred (A). The number of missing teeth in people with defined MSX1 mutations was compiled for each position in the human dentition (32 teeth) based on the data from this report, as well as from the six previous reports of MSX1 mutations. The MSX1 mutations are summarized in the chart (D) as fractions, with the numerators being the number of missing teeth and the denominators being the number of individuals. There were no statistically significant differences between the numbers of teeth missing on the left and right, so the data for equivalent teeth on the left and right were combined (bottom of D). We plotted the percentage of teeth missing for each position in the maxillary and mandibular arches (B top), and indicated statistically significant differences (p < 0.001) between the maxillary and mandibular arches with asterisks. The number of missing teeth at each position in people with defined PAX9 mutations was compiled from the 8 previous reports of 10 PAX9 mutations (E). As with MSX1, there were no statistically significant differences between the numbers of teeth missing on the left and right, so the data for equivalent teeth on the left and right were combined (bottom of E). In subsequent analyses, we averaged the values for the mandibular central and lateral incisors, since which tooth is missing is often ambiguous. We plotted the percentage of teeth missing for each position in the maxillary and mandibular arches (B bottom), with statistically significant differences (p < 0.001) between the maxillary and mandibular arches marked with asterisks. We plotted the percentage of missing teeth at each maxillary and mandibular position for people with defined MSX1 and PAX9 mutations (C). For each tooth position in the bar graphs (B,C), the bars for the maxillary teeth are on the right, and bars for the mandibular teeth are on the left. Statistically significant differences (p < 0.001) between MSX1 and PAX9 are marked with asterisks in C, and by bold type in D and E. Note that the absence of maxillary first bicuspids (#4) is the most distinguishing feature of an MSX1 mutation, while the absence of second molars (#7) is the most distinguishing feature of PAX9 mutations. Key: 1, central incisor; 2, lateral incisor; 3, cuspid; 4, first bicuspid; 5, second bicuspid; 6, first molar; 7, second molar 8, third molar; ?, not able to be determined; x axis = tooth position; y axis = percent missing.
Mutational Analyses
A G duplication (g.62dupG) was identified in exon 1 of the MSX1 gene (4p16.1) in both the proband and his affected sister (III-2), but not in the unaffected participants (II-2 and III-3). This extra G shifts the translation reading frame after Gly21, so that 146 novel amino acids are substituted for the rest of the protein (p.G22RfsX168). The mutant protein would have only 167 amino acids (there are 297 in the native protein), and only the first 21 amino acids would be the same as in the native protein.
Frequency of Tooth Loss with MSX1 Mutations
The normal human permanent dentition is comprised of 32 teeth, which are divided into 4 quadrants, each having 8 teeth. The 4 quadrants are the maxillary right and left and the mandibular right and left. The numbering of the teeth in each quadrant starts with the central incisor (numbered 1) and works back to the third molars (numbered 8). The teeth missing in the two affected individuals in our kindred are shown in Fig. 2A. With this report, there are now 39 individuals with defined MSX1 mutations who have known patterns of tooth agenesis in their permanent dentitions. For a few of these 39 individuals, data are not available for every tooth position, because of the early age of the subject or, in one case, because of the presence of a maxillary complete denture. We compiled the number of missing teeth at each of the 8 positions in the 4 quadrants (Fig. 2D) and performed chi-square tests to determine if there were statistically significant differences between the number of missing teeth in the 8 sites in the maxillary right quadrant relative to the same sites in the maxillary left quadrant, and between mandibular right and mandibular left quadrants. No statistically significant differences were observed, meaning that for a particular type of tooth (e.g., a maxillary second bicuspid), there is an equal probability of that tooth being missing on the left side as on the right side.
We combined the data for the left and right sides of each arch and performed chi-square tests to determine if there were statistically significant differences between the number of missing teeth in the 8 sites of the combined maxillary right and left quadrants (maxillary arch) relative to the same sites in the combined mandibular right and mandibular left quadrants (mandibular arch). Statistically significant differences (p < 0.001) were found at positions 1 (central incisor) and 4 (first bicuspid) (Figs. 2B top, 2D). An individual with an MSX1 mutation is more likely to miss a mandibular central incisor than a maxillary central incisor. Among individuals with partial tooth agenesis associated with a defined MSX1 mutation, about 1 out of every 5 mandibular central incisors is missing, while no instances of a missing maxillary central incisor have yet been observed. In addition, persons with an MSX1 deficit miss, on average, 75% of their maxillary first bicuspids, while only about 15% of their mandibular first bicuspids fail to develop.
Frequency of Tooth Loss with PAX9 Mutations
We also compiled the number of missing teeth at each of the 8 positions in the 4 quadrants for individuals with defined PAX9 mutations (Fig. 2E) and performed analyses similar to those described above (Stockton et al., 2000; Nieminen et al., 2001; Das et al., 2002, 2003; Frazier-Bowers et al., 2002; Lammi et al., 2003; Mostowska et al., 2003a; Jumlongras et al., 2004). As with MSX1, there is an equal probability of a given tooth being missing on the left and right sides, so data for the left and right were combined. Statistically significant differences (p < 0.001) were observed between the maxillary and mandibular arches at positions 1 (central incisor), 5 (second bicuspid), and 6 (first molar) (Figs. 2B bottom, 2E). Persons with PAX9 deficits are more likely to be missing their mandibular central incisors, maxillary second bicuspids, and maxillary first molars relative to the same teeth in the opposing arch.
Comparing Frequency of Tooth Loss with MSX1 and PAX9 Mutations
We compared the frequency with which each type of tooth in the maxillary and mandibular arches was missing in patients with MSX1 mutations relative to those with PAX9 mutations (Fig. 2C). Statistically significant differences were observed at 6 positions. Patients with MSX1 defects were more likely to be missing maxillary first bicuspids, maxillary second bicuspids, and mandibular second bicuspids. Patients with PAX9 defects were more likely to be missing maxillary first molars, maxillary second molars, and mandibular second molars.
DISCUSSION
We have identified an MSX1 frameshift mutation in the affected members of a kindred with autosomal-dominant oligodontia without clefting or nail dysplasia. In this family, a G duplication at nucleotide position 62 in exon 1 of MSX1 introduced 146 novel amino acids following the first 21 amino acids, while deleting the normal protein sequence from Gly22 through Thr297. This MSX1 mutation is associated with the absence of multiple permanent teeth, including all second bicuspids and mandibular central incisors. The mutation must be considered a rare MSX1 sequence variation, since it was not observed during the complete sequencing of MSX1 from over 1000 individuals with cleft lip and/or cleft palate, and from over 500 controls of unaffected persons (Jezewski et al., 2003). Since so little of the native MSX1 protein sequence (21 of 297 amino acids) is synthesized from the mutant allele, the mutant translation product is presumed to be inactive and should not interfere with the DNA- and protein-protein interactions of MSX1 expressed from the wild-type allele. Therefore, we propose that the dominant phenotype of partial tooth agenesis in our kindred is due to haploinsufficiency, rather than a dominant-negative mechanism.
In our MSX1 kindred, missing third molars were observed in affected and unaffected individuals and were considered to be part of the genetic background. Our analysis shows that MSX1 and PAX9 kindreds have a high, but equal, probability of missing third molars, so the absence of third molars is not a useful indicator of which gene (MSX1 or PAX9) is likely to be affected in a given kindred. Analysis of the data for the seven MSX1-deficient kindreds shows that the teeth most likely to be absent were the maxillary and mandibular second bicuspids (91–97%), and the maxillary first bicuspids (75%). All teeth were equally likely to be missing from the left and right sides, which also appears to be true of hypodontia in general (Stahl et al., 2003).
The characterization of additional genes and the mutations that cause oligodontia should improve our ability to prioritize the list of candidate genes based upon the specific pattern of partial tooth agenesis in a kindred. Currently, the developmental absence of maxillary and mandibular second bicuspids and maxillary first bicuspids, while most mandibular first bicuspids are retained, appears to be the pattern of tooth agenesis that best indicates the presence of an MSX1 mutation. In contrast, patients with PAX9 mutations typically show agenesis of almost all of their molars, with the absence of second molars best distinguishing them from persons with MSX1 defects. Furthermore, a history of missing primary teeth has been reported in some PAX9 kindreds (Nieminen et al., 2001; Das et al., 2002), but never in MSX1 kindreds.
Very recently, a mutation in the PAX9 translation initiation codon has been reported (Klein et al., 2005). In the two affected individuals for whom data was provided, there was a history of missing primary teeth, while all of the molars (as well as many other teeth) were absent. This further supports these criteria as being able to distinguish accurately between MSX1 and PAX9 mutations, based upon the dental phenotype.
Acknowledgments
We thank all the family members for their cooperation. This investigation was supported by the Foundation of the American Academy of Pediatric Dentistry, and by USPHS Research Grants DE15846 and DE11301 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 29892, USA.
References
- Bendall AJ, Rincon-Limas DE, Botas J, Abate-Shen C. Protein complex formation between Msx1 and Lhx2 homeoproteins is incompatible with DNA binding activity. Differentiation. 1998;63:151–157. doi: 10.1046/j.1432-0436.1998.6330151.x. [DOI] [PubMed] [Google Scholar]
- Bendall AJ, Ding J, Hu G, Shen MM, Abate-Shen C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Das P, Stockton DW, Bauer C, Shaffer LG, D'Souza RN, Wright T, et al. Haploinsufficiency of PAX9 is associated with autosomal dominant hypodontia. Hum Genet. 2002;110:371–376. doi: 10.1007/s00439-002-0699-1. [DOI] [PubMed] [Google Scholar]
- Das P, Hai M, Elcock C, Leal SM, Brown DT, Brook AH, et al. Novel missense mutations and a 288-bp exonic insertion in PAX9 in families with autosomal dominant hypodontia. Am J Med Genet A. 2003;118:35–42. doi: 10.1002/ajmg.a.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muynck S, Schollen E, Matthijs G, Verdonck A, Devriendt K, Carels C. A novel MSX1 mutation in hypodontia. Am J Med Genet A. 2004;128:401–403. doi: 10.1002/ajmg.a.30181. [DOI] [PubMed] [Google Scholar]
- Frazier-Bowers SA, Guo DC, Cavender A, Xue L, Evans B, King T, et al. A novel mutation in human PAX9 causes molar oligodontia. J Dent Res. 2002;81:129–133. [PubMed] [Google Scholar]
- Gorlin RJ, Cohen M, Jr, Leven L. Syndromes of the head and neck. 3. New York: Oxford Universtiy Press; 1990. [Google Scholar]
- Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001;128:2373–2384. doi: 10.1242/dev.128.12.2373. [DOI] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, et al. A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet. 2001;69:67–74. doi: 10.1086/321271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumlongras D, Lin JY, Chapra A, Seidman CE, Seidman JG, Maas RL, et al. A novel missense mutation in the paired domain of PAX9 causes non-syndromic oligodontia. Hum Genet. 2004;114:242–249. doi: 10.1007/s00439-003-1066-6. [DOI] [PubMed] [Google Scholar]
- Klein ML, Nieminen P, Lammi L, Niebuhr E, Kreiborg S. Novel mutation of the initiation codon of PAX9 causes oligodontia. J Dent Res. 2005;84:43–47. doi: 10.1177/154405910508400107. [DOI] [PubMed] [Google Scholar]
- Lammi L, Halonen K, Pirinen S, Thesleff I, Arte S, Nieminen P. A missense mutation in PAX9 in a family with distinct phenotype of oligodontia. Eur J Hum Genet. 2003;11:866–871. doi: 10.1038/sj.ejhg.5201060. [DOI] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- Lidral AC, Reising BC. The role of MSX1 in human tooth agenesis. J Dent Res. 2002;81:274–278. doi: 10.1177/154405910208100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowska A, Kobielak A, Biedziak B, Trzeciak WH. Novel mutation in the paired box sequence of PAX9 gene in a sporadic form of oligodontia. Eur J Oral Sci. 2003a;111:272–276. doi: 10.1034/j.1600-0722.2003.00036.x. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Kobielak A, Trzeciak WH. Molecular basis of non-syndromic tooth agenesis: mutations of MSX1 and PAX9 reflect their role in patterning human dentition. Eur J Oral Sci. 2003b;111:365–370. doi: 10.1034/j.1600-0722.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Arte S, Tanner D, Paulin L, Alaluusua S, Thesleff I, et al. Identification of a nonsense mutation in the PAX9 gene in molar oligodontia. Eur J Hum Genet. 2001;9:743–746. doi: 10.1038/sj.ejhg.5200715. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Kotilainen J, Aalto Y, Knuutila S, Pirinen S, Thesleff I. MSX1 gene is deleted in Wolf-Hirschhorn syndrome patients with oligodontia. J Dent Res. 2003;82:1013–1017. doi: 10.1177/154405910308201215. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kapadia H, Wang B, D'Souza RN. Studies on Pax9-Msx1 protein interactions. Arch Oral Biol. 2005;50:141–145. doi: 10.1016/j.archoralbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozkovcova E, Markova M, Lanik J, Zvarova J. Agenesis of third molars in young Czech population. Prague Med Rep. 2004;105:35–52. [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Shetty S, Takahashi T, Matsui H, Ayengar R, Raghow R. Transcriptional autorepression of Msx1 gene is mediated by interactions of Msx1 protein with a multi-protein transcriptional complex containing TATA-binding protein, Sp1 and cAMP-response-element-binding protein-binding protein (CBP/p300) Biochem J. 1999;339:751–758. [PMC free article] [PubMed] [Google Scholar]
- Stahl F, Grabowski R, Wigger K. Epidemiological significance of Hoffmeister's “genetically determined predisposition to disturbed development of the dentition”. J Orofac Orthop. 2003;64:243–255. doi: 10.1007/s00056-003-0220-z. [DOI] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D'Souza RN, Patel PI. Mutation of PAX9 is associated with oligodontia. Nat Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Meira R, Modesto A, Murray JC. MSX1, PAX9, and TGFA contribute to tooth agenesis in humans. J Dent Res. 2004;83:723–727. doi: 10.1177/154405910408300913. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]