Abstract
Activation of the transcription factor STAT3 is thought to potently promote oncogenesis in a variety of tissues, leading to intense efforts to develop STAT3 inhibitors for many tumors, including the highly malignant brain tumor glioblastoma. However, the function of STAT3 in glioblastoma pathogenesis has remained unknown. Here, we report that STAT3 plays a pro-oncogenic or tumor-suppressive role depending on the mutational profile of the tumor. Deficiency of the tumor suppressor PTEN triggers a cascade that inhibits STAT3 signaling in murine astrocytes and human glioblastoma tumors. Specifically, we forge a direct link between the PTEN–Akt–FOXO axis and the leukemia inhibitory factor receptor β (LIFRβ)–STAT3 signaling pathway. Accordingly, PTEN knockdown induces efficient malignant transformation of astrocytes upon knockout of the STAT3 gene. Remarkably, in contrast to the tumor-suppressive function of STAT3 in the PTEN pathway, STAT3 forms a complex with the oncoprotein epidermal growth factor receptor type III variant (EGFRvIII) in the nucleus and thereby mediates EGFRvIII-induced glial transformation. These findings indicate that STAT3 plays opposing roles in glial transformation depending on the genetic background of the tumor, providing the rationale for tailored therapeutic intervention in glioblastoma.
[Keywords: STAT3, astrocyte, glioblastoma, PTEN, EGFRvIII]
Malignant gliomas of the brain, including the devastating tumor glioblastoma, represent a leading cause of cancer-related deaths in the young and middle-aged population (Jemal et al. 2003; Central Brain Tumor Registry of the United States 2002–2003 report, http://www.cbtrus.org). Few tumors carry worse prognosis than glioblastomas (Louis 2006; Furnari et al. 2007). The failed record of clinical trials in the treatment of these tumors may reflect the poorly understood biology of glioblastoma cells and their underlying signal transduction pathways (Stewart 2002; Konopka and Bonni 2003). The majority of glioblastomas bear histologic features of cells along the astrocytic lineage (Louis et al. 2007). These tumors are thus thought to arise from the transformation of astrocytes or their precursors, the neural stem cells (Holland 2001; Bachoo et al. 2002; Uhrbom et al. 2002; Bajenaru et al. 2003). These observations raise the fundamental question of whether deregulation of signaling pathways that promote astrocyte differentiation during normal brain development might contribute to the pathogenesis of these devastating tumors.
During brain development, neural stem cells differentiate into astrocytes (Ware et al. 1995; Bonni et al. 1997; Rajan and McKay 1998). Activation of the cytokine receptor leukemia inhibitory factor receptor β (LIFRβ) and downstream STAT3 signaling pathway stimulates the differentiation of neural stem cells into astrocytes (Ware et al. 1995; Bonni et al. 1997; Rajan and McKay 1998). In addition to promoting astrocyte differentiation, recent studies suggest that STAT3 supports the renewal capacity of neural stem cells (Yoshimatsu et al. 2006). The pleiotropic function of STAT3 in the development of cells along the glial lineage raises the interesting possibility that deregulation of STAT3 signaling might contribute to glial cell transformation.
What is the nature of STAT3 function in the pathogenesis of glioblastomas? STAT3 has been assigned a pro-oncogenic function in several cell types outside the nervous system (Bromberg et al. 1999; Chan et al. 2004; Chiarle et al. 2005; Ling and Arlinghaus 2005; Schlessinger and Levy 2005). STAT3 activation has been described in a subset of glioblastomas (Wang et al. 2004; Weissenberger et al. 2004), and STAT3 has been reported to promote the survival of some glioblastoma cell lines in vitro (Rahaman et al. 2002; Konnikova et al. 2003). These observations have provided the impetus to generate STAT3 inhibitors for a whole host of tumors including glioblastomas (Turkson et al. 2001; Leong et al. 2003; Song et al. 2005). Yet, the functional significance of STAT3 in the pathogenesis of glial malignancy has remained unknown. In addition, how STAT3 function is modulated in the setting of distinct oncogenic lesions known to drive glial cell transformation has been unexplored.
Loss-of-function mutations in the phosphatase PTEN on chromosome 10 are among the most common genetic abnormalities in high-grade gliomas (Li et al. 1997; Steck et al. 1997). PTEN harbors a lipid phosphatase activity that figures prominently in its function as a glioblastoma tumor suppressor (Furnari et al. 1997, 1998). The lipid phosphatase activity of PTEN dephosphorylates phosphatidylinositol 3,4,5 trisphosphate (PIP3) to produce phosphatidylinositol 4,5 bisphosphate (PIP2). PIP3 is a second messenger produced by phosphoinositide 3-kinase (PI3K) and activates its downstream effectors including the protein kinase Akt, a potent inducer of cell proliferation and survival (Cantley 2002). Mutations in PTEN have been linked to tumors outside the brain, including breast, prostate, ovarian, and head and neck tumors (Li et al. 1997; Steck et al. 1997). Thus, PTEN acts as a general tumor suppressor.
In this study, we find an unexpected tumor-suppressive function of STAT3 that is intimately linked to PTEN function. PTEN loss and consequent Akt activation suppresses the LIFRβ–STAT3 signaling pathway in astrocytes. Gene knockout studies reveal that STAT3 plays a key role in the PTEN pathway to suppress malignant transformation of astrocytes. In human glioblastomas, PTEN deficiency tightly correlates with inactivation of STAT3 signaling.
Strikingly, in contrast to STAT3’s tumor-suppressive function in the PTEN pathway, STAT3 associates with the oncoprotein epidermal growth factor receptor type III variant (EGFRvIII) in the nucleus and thereby induces glial transformation. These findings indicate that STAT3 plays distinct roles in cell transformation depending on the oncogenic environment. Our study may thus provide the foundation for tailored therapy of glioblastomas based on their genetic profile.
Results
PTEN loss suppresses the LIFRβ–STAT3 signaling pathway
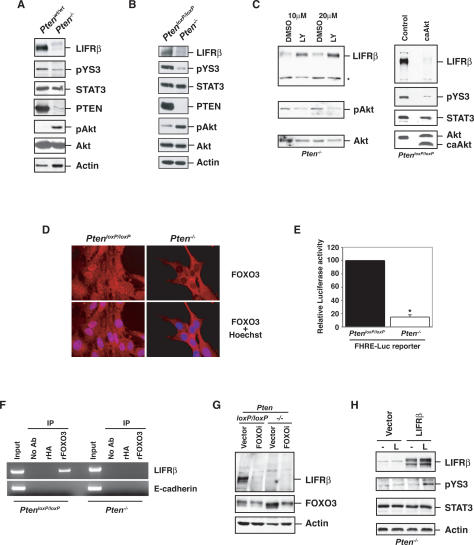
To investigate the role of glial developmental signaling pathways in tumorigenesis, we first determined if PTEN loss impacts on the LIFRβ–STAT3 signaling pathway in glial cells. We measured the levels of LIFRβ in astrocytes in which the PTEN gene was disrupted by Cre-mediated excision of PTEN alleles flanked by loxP (floxed) sites. As predicted, little or no PTEN protein was detected in the Pten−/− astrocytes, and this was associated with enhanced phosphorylation of the serine/threonine kinase Akt on Ser 473, which reflects Akt activation (Fig. 1A,B). The amount of LIFRβ was significantly reduced in astrocytes in which the PTEN gene was knocked out (Pten−/−) when compared with wild-type astrocytes (Ptenwt/wt) or astrocytes with homozygous floxed PTEN allele (PtenloxP/loxP) (Fig. 1A,B). These findings suggest that PTEN loss inhibits LIFRβ expression in astrocytes.
Figure 1.
PTEN deficiency suppresses the LIFRβ–STAT3 signaling pathway. (A) Lysates of mouse Ptenwt/wt or Pten−/− astrocytes were immunoblotted with antibodies to LIFRβ, Tyr705-phosphorylated STAT3 (pYS3), total STAT3, PTEN, total Akt, Ser473-phosphorylated Akt (pAkt), or actin. (B) Immunoblotting of parental PtenloxP/loxP and Pten−/− astrocytes with antibodies used in A. Loss of PTEN expression was associated with an increase in phospho-Akt levels and with a decrease in LIFRβ and pYS3. Actin served as loading control. (C, left panel) Immunoblotting of lysates of Pten−/− astrocytes treated with the PI3K inhibitor LY294002 or DMSO vehicle control for 48 h. Asterisk indicates a nonspecific band. (Right panel) Immunoblotting of lysates from serum-starved PtenloxP/loxP astrocytes stably expressing a constitutively active form of Akt (caAkt). Activated Akt reduced LIFRβ and pYS3 levels. (D) Immunocytochemical analysis of PtenloxP/loxP and Pten−/− astrocytes with a FOXO3 antibody. Nuclei were stained with a DNA dye (Hoechst). FOXO3 was excluded from the nucleus in Pten−/− astrocytes. (E) PtenloxP/loxP and Pten−/− astrocytes were transfected with a luciferase reporter gene controlled by FOXO-binding sites (FHRE-Luc), together with a renilla expression plasmid to serve as an internal control, and subjected to a dual luciferase assay. FOXO-dependent transcription was reduced in Pten−/− astrocytes. (F) ChIP analysis at the endogenous LIFRβ promoter in PtenloxP/loxP and Pten−/− astrocytes with a FOXO3 antibody. A rabbit anti-HA antibody was used as negative control. The analysis was done with two independent sets of primers for the LIFRβ promoter. Negative controls for the PCR reaction were performed with primers for the E-cadherin promoter. Endogenous FOXO3 occupied the endogenous LIFRβ promoter in PtenloxP/loxP but not in Pten−/− astrocytes. (G) Immunoblotting with the LIFRβ and FOXO3 antibodies of PtenloxP/loxP and Pten−/− astrocytes infected with a FOXO3 RNAi-encoding lentivirus (FOXOi) or an empty vector and selected with puromycin. Actin served as loading control. Knockdown of endogenous FOXO3 reproducibly led to down-regulation of LIFRβ. (H) Immunoblotting of lysates of serum-starved Pten−/− astrocytes transfected with a LIFRβ expression plasmid or a control vector that were left untreated (−) or treated with LIF (L) for 15 min. LIFRβ restored LIF-induced STAT3 phosphorylation in Pten−/− astrocytes.
We next characterized the mechanism by which PTEN loss suppresses LIFRβ expression in astrocytes. PTEN catalyzes the dephosphorylation of the lipid messenger PIP3, which is generated by PI3K (Myers et al. 1997; Maehama and Dixon 1998). Incubation of Pten−/− astrocytes with the PI3K inhibitor LY294002 restored LIFRβ expression in these cells, suggesting that PTEN loss suppresses LIFRβ expression via an increase in the levels of PIP3 (Fig. 1C). In addition, expression of a constitutively active Akt, a critical effector of PIP3 (Cantley 2002), in PtenloxP/loxP cells suppressed LIFRβ expression (Fig. 1C). Together, these results suggest that activated Akt inhibits LIFRβ protein expression in PTEN-deficient astrocytes.
Consistent with the idea that PTEN loss triggers the inhibition of LIFRβ gene expression, the levels of LIFRβ mRNA measured by RT–PCR were significantly reduced in Pten−/− astrocytes (Supplemental Fig. S1). A major substrate of Akt is the transcription factor FOXO3, whose phosphorylation inhibits FOXO-dependent transcription (Burgering and Kops 2002; Van Der Heide et al. 2004). Therefore, we asked whether the LIFRβ gene might represent a direct target of FOXO3 in astrocytes and thus provide the basis for PTEN loss/activated Akt-directed suppression of LIFRβ expression. We confirmed that endogenous FOXO3 was excluded from the nucleus and FOXO-dependent transcription was inhibited in astrocytes upon knockout of PTEN (Fig. 1D,E).
By sequence analysis, we found a conserved FOXO-binding site within the promoters of the human, mouse, and dog LIFRβ gene (data not shown). By chromatin immunoprecipitation (ChIP) analyses, endogenous FOXO3 occupied the endogenous LIFRβ promoter in PtenloxP/loxP astrocytes (Fig. 1F). However, little or no FOXO3 was found at the LIFRβ promoter in Pten−/− astrocytes, suggesting that FOXO3 occupancy at the LIFRβ gene correlates tightly with PTEN status and LIFRβ expression in astrocytes (Fig. 1F).
To determine the role of endogenous FOXO3 in the regulation of LIFRβ expression in astrocytes, we used a lentiviral DNA template-based method of RNAi to knock down FOXO3 in these cells. We found that FOXO3 knockdown in PTEN-expressing (PtenloxP/loxP) astrocytes suppressed LIFRβ expression, mimicking the effect of PTEN knockout (Fig. 1G). Taken together, our findings suggest that LIFRβ is a direct target of FOXO3 in astrocytes and supports the conclusion that PTEN loss and consequent Akt activation inhibit FOXO3-dependent LIFRβ gene expression in astrocytes (see model in Fig. 7A, below).
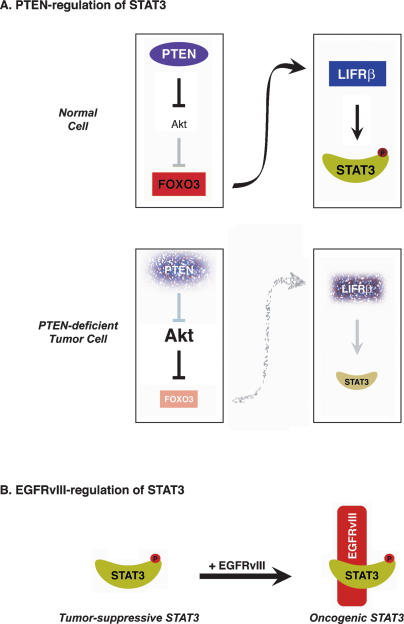
Figure 7.
Dual role of STAT3 in tumorigenesis. (A) A PTEN-regulated STAT3 tumor-suppressive pathway. (B) EGFRvIII induces an oncogenic switch in STAT3 function.
To determine the consequences of the suppression of LIFRβ expression in astrocytes, we measured the level of STAT3 that is phosphorylated at the key regulatory site of Tyr705 (pYS3) in these cells. The Tyr705 phosphorylation triggered by LIFRβ promotes STAT3 dimerization and translocation to the nucleus, where STAT3 regulates transcription of cytokine-responsive genes (Darnell et al. 1994). PTEN loss or expression of a constitutively active Akt in astrocytes significantly inhibited STAT3 phosphorylation concomitantly with LIFRβ down-regulation (Fig. 1A–C). Expression of exogenous LIFRβ in Pten−/− astrocytes restored STAT3 phosphorylation in response to LIF, suggesting that PTEN loss inhibits Tyr705 phosphorylation of STAT3 via down-regulation of LIFRβ (Fig. 1H). In parallel experiments, expression of exogenous PTEN in Pten−/− astrocytes reinstated the expression of endogenous LIFRβ and associated Tyr705 phosphorylation of STAT3 (Supplemental Fig. S2). Taken together, these results suggest that PTEN loss suppresses the STAT3 signaling pathway in astrocytes.
STAT3 suppresses PTEN loss-induced malignant cell transformation
Suppression of STAT3 activation in astrocytes by inactivation of the tumor suppressor PTEN was unanticipated in view of the pro-oncogenic function of STAT3 in nonbrain cell and tumor types (Bromberg et al. 1999; Chan et al. 2004; Chiarle et al. 2005; Ling and Arlinghaus 2005; Schlessinger and Levy 2005). Moreover, STAT3 has been reported to promote the survival of some glioblastoma cells in vitro (Rahaman et al. 2002; Konnikova et al. 2003), and STAT3 activation as reflected by Tyr705 phosphorylation has been described in human gliomas (Weissenberger et al. 2004). At the same time, other studies have provided evidence of the absence of STAT3 activity in a large percentage of these tumors including glioblastomas (Schaefer et al. 2002; Wang et al. 2004), raising questions as to the relevance of STAT3 in glial tumorigenesis.
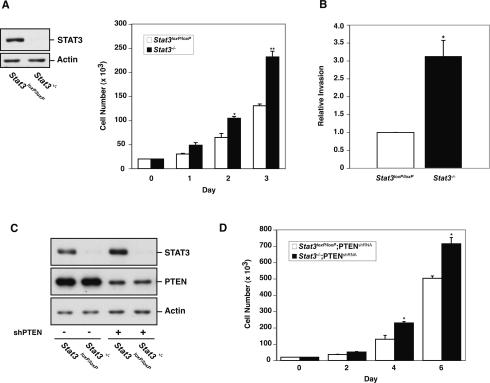
We decided to take a genetic approach to directly determine the role of STAT3 in glial cell transformation by assessing the effect of disruption of the STAT3 gene in astrocytes. We prepared astrocytes from mice carrying a homozygous floxed STAT3 allele (Stat3loxP/loxP) (Raz et al. 1999), and induced knockout of the STAT3 gene by expressing the DNA recombinase Cre. STAT3 loss led to an increase in the number of astrocytes in cell population growth assays (Fig. 2A). In these experiments, STAT3 loss had little or no effect on cell death in astrocytes (data not shown). These results suggest that STAT3 inactivation stimulates astrocyte proliferation. We also examined the effect of knockout of STAT3 on the invasive potential of astrocytes. In matrigel invasion assays, we found that STAT3 knockout (Stat3−/−) astrocytes had significantly higher potential to invade through matrigel (Fig. 2B). Thus, in addition to inducing astrocyte proliferation, STAT3 loss also increases the invasive potential of astrocytes. Together, these results raised the possibility that STAT3 might suppress astrocyte transformation.
Figure 2.
STAT3 knockout promotes astrocyte cell proliferation and invasiveness. (A, left panel) Immunoblotting of Stat3loxP/loxP or Stat3−/− astrocytes with an antibody to STAT3. Actin served as control for loading. (Right panel) Cell population growth of Stat3loxP/loxP and Stat3−/− astrocytes. STAT3 loss significantly increased astrocyte cell population growth rate (representative experiment of three independent experiments performed in triplicate; ANOVA; [*] P < 0.05; [**] P < 0.0001). (B) Quantification of the invasive potential of Stat3−/− astrocytes through a matrigel substrate. Stat3loxP/loxP and Stat3−/− astrocytes were seeded on top of an 8-μm pore size insert coated with matrigel and allowed to invade through the matrigel matrix for 22 h. Knockout of STAT3 significantly increased astrocyte cell invasiveness (n = 3; t-test; [*] P < 0.01). The effect of STAT3 loss on invasiveness was not secondary to a change in cell proliferation, as the invasive potential of these cells was measured at a time (22 h after plating) prior to a significant increase in cell number upon STAT3 knockout. (C) Immunoblotting of STAT3 and PTEN in Stat3loxP/loxP or Stat3−/− astrocytes that were uninfected or infected with a retrovirus encoding a shRNA directed against PTEN (shPTEN). Actin served as control for loading. (D) Cell population growth of PTEN knockdown (PTENshRNA) astrocytes. STAT3 loss significantly increased cell number of PTEN knockdown astrocytes (representative experiment of two independent experiments performed in triplicate; ANOVA; [*] P < 0.0001).
To determine STAT3 function in astrocyte transformation, we tested the ability of Stat3loxP/loxP and Stat3−/− astrocytes to form tumors upon subcutaneous injection in severe combined immunodeficient (SCID) mice. Injection of astrocytes bearing the floxed or recombined STAT3 allele failed to form tumors (data not shown), suggesting that loss of the STAT3 gene on its own is insufficient to transform astrocytes.
Since the inhibition of STAT3 signaling in astrocytes occurred upon loss of PTEN (Fig. 1), we reasoned that STAT3 inactivation might cooperate with other events downstream from PTEN loss to promote glial malignancy. We therefore tested the effect of knockout of STAT3 on the behavior of astrocytes in which expression of the Pten gene was partially reduced by RNAi-mediated knockdown (PTENshRNA) (Fig. 2C). STAT3 knockout significantly increased the number of PTENshRNA astrocytes in cell population growth assays in vitro (Fig. 2D). In addition, STAT3 loss had little or no effect on cell death in PTENshRNA astrocytes (data not shown). Together, these results suggest that STAT3 knockout stimulates the proliferation of PTEN knockdown astrocytes.
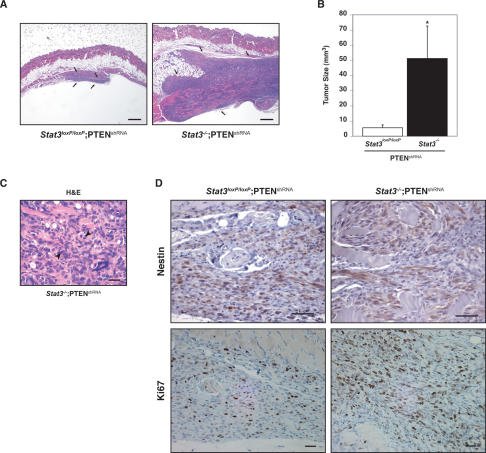
Upon injection of Stat3loxP/loxP;PTENshRNA astrocytes into SCID mice, these cells formed small tumors in a large fraction but not all of the animals examined 8 wk post-injection. In contrast, the Stat3−/−;PTENshRNA astrocytes formed tumors in all injected mice. Importantly, the Stat3−/−;PTENshRNA tumors were significantly greater in size compared with tumors formed by Stat3loxP/loxP;PTENshRNA astrocytes (Fig. 3A,B). Histologic evaluation of the tumors revealed characteristic features of neoplastic transformation including nuclear atypia, pleomorphism, and frequent mitotic activity (Fig. 3C). Tumors also expressed nestin, a marker of gliomas (Fig. 3D; Uhrbom et al. 2002; Dai and Holland 2003). Immunohistochemical analysis using an antibody to the cell proliferation marker Ki67 revealed that the proliferative capacity of Stat3−/−;PTENshRNA tumors was higher than that of Stat3loxP/loxP;PTENshRNA tumors, correlating with the increased size of the Stat3−/−; PTENshRNA tumors (Fig. 3D).
Figure 3.
STAT3 suppresses PTEN deficiency-induced malignant cell transformation. (A,B) Stat3loxP/loxP;PTENshRNA or Stat3−/−;PTENshRNA astrocytes were injected subcutaneously into SCID mice. Eight weeks after injection, tumors were removed, measured, and stained. STAT3 loss induced an increase in tumor size as revealed by hematoxylin and eosin (H&E) staining (A) and tumor size measurements (n = 8; t-test, [*] P < 0.05) (B). Arrows in A show the tumor limits. Bar, 1 mm. (C) Histologic analysis of the Stat3−/−;PTENshRNA tumors by H&E staining. The tumors showed histologic features of neoplastic transformation including nuclear atypia, pleomorphism, and frequent mitotic figures (arrowheads). Bar, 100 μm. (D) Nestin and Ki67 immunostaining of Stat3loxP/loxP;PTENshRNA and Stat3−/−;PTENshRNA tumors. These tumors express nestin, a characteristic marker of glial tumors. Cell proliferation rate, as measured by the percentage of Ki67-positive cells, was higher in Stat3−/−;PTENshRNA tumors as compared with Stat3loxP/loxP;PTENshRNA tumors (64% vs. 25%, respectively; average of two tumors each). Bar, 100 μm.
Collectively, these results show that loss of STAT3 promotes astrocyte proliferation and invasiveness, and strongly potentiates astrocyte tumorigenesis upon knockdown of PTEN. Therefore, our studies strongly support the conclusion that STAT3 suppresses the malignant transformation of astrocytes in the context of the PTEN pathway.
STAT3 forms a physical complex with EGFRvIII in the nucleus and mediates EGFRvIII-induced cell transformation
The identification of a PTEN-regulated STAT3 tumor-suppressive function in astrocytes appeared paradoxical in view of the reported oncogenic function of STAT3. These observations led us to ask if STAT3 acts as a tumor suppressor under all circumstances or whether STAT3 function in astrocytes is dictated by the genotype of these cells. Besides PTEN loss, amplifications or rearrangements in the gene encoding the receptor tyrosine kinase (RTK) EGFR are important in the pathogenesis of de novo glioblastomas (Libermann et al. 1985; Wong et al. 1987). A common mutant isoform of EGFR in glioblastomas is the EGFRvIII that lacks exons 2–7 and behaves as a constitutively active form of EGFR (Sugawa et al. 1990; Wong et al. 1992; Moscatello et al. 1996). Expression of EGFRvIII in glial cells drives their malignant transformation (Nishikawa et al. 1994; Holland et al. 1998; Bachoo et al. 2002). We confirmed that expression of EGFRvIII robustly triggers the malignant transformation of astrocytes (Fig. 5A–D, below).
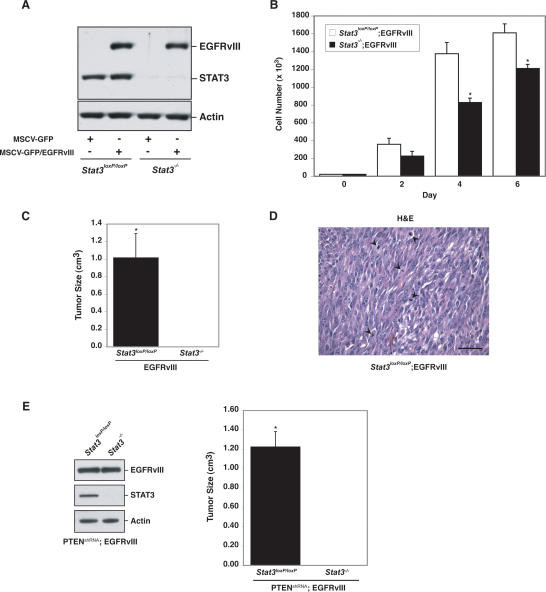
Figure 5.
STAT3 mediates EGFRvIII-induced cell transformation. (A) Immunoblotting of EGFRvIII-expressing or control astrocytes with antibodies against EGFR and STAT3. Actin was used as loading control. (B) Cell population growth curves of Stat3loxP/loxP; EGFRvIII and Stat3−/−;EGFRvIII astrocytes. STAT3 knockout significantly decreased cell number (representative experiment of three independent experiments performed in triplicate; ANOVA; [*] P < 0.0001). (C) Size of EGFRvIII-expressing tumors. Stat3loxP/loxP; EGFRvIII and Stat3−/−;EGFRvIII astrocytes were injected subcutaneously into SCID mice. Four weeks after injection, tumors were excised, measured, and stained. Tumors were only present in Stat3loxP/loxP;EGFRvIII-injected mice (n = 6; t-test; [*] P < 0.05). (D) H&E staining of Stat3loxP/loxP;EGFRvIII tumors confirmed the presence of tumor cells. Mitotic figures are indicated by arrowheads. Bar, 100 μm. (E, left panel) Immunoblotting of PTENshRNA;EGFRvIII astrocytes with EGFR and STAT3 antibodies. Actin served as loading control. (Right panel) Size of PTENshRNA;EGFRvIII-expressing tumors. PTENshRNA;EGFRvIII astrocytes were injected subcutaneously into SCID mice. Four weeks after injection, tumors were excised and measured. Tumors were only present in Stat3loxP/loxP; PTENshRNA;EGFRvIII-injected mice (n = 4; t-test; [*] P < 0.001).
To assess the role of STAT3 in EGFRvIII-induced malignant glial transformation, we characterized the effect of EGFRvIII expression on STAT3 signaling. Activation of RTKs is thought to recruit STAT3 to the RTK at the plasma membrane followed by induction of STAT3 phosphorylation at the key regulatory sites Tyr705 and Ser727 and consequent translocation of STAT3 to the nucleus (Zhong et al. 1994; Leaman et al. 1996; Vignais et al. 1996; Decker and Kovarik 2000). We asked if the constitutively active EGFRvIII recruits STAT3 and regulates its phosphorylation at the two key regulatory sites. We found that EGFRvIII interacted efficiently with STAT3, including endogenous STAT3 in astrocytes (Fig. 4A). However, expression of EGFRvIII surprisingly failed to induce the STAT3 Tyr705 and Ser727 phosphorylation in astrocytes (Fig. 4B). Similarly, EGFRvIII expression in human glioblastoma cells has a modest effect on STAT3 Tyr705 phosphorylation (Huang et al. 2007). Although activated RTKs are generally thought to induce the PI3K–Akt pathway (Cantley 2002; Kapoor and O’Rourke 2003), EGFRvIII expression also had little effect on Akt activation in astrocytes (Fig. 4B). Consistent with these results, whereas PTEN deficiency significantly reduced the STAT3 Tyr705 phosphorylation, EGFRvIII failed to reduce the STAT3 Tyr705 phosphorylation in astrocytes (Fig. 4B).
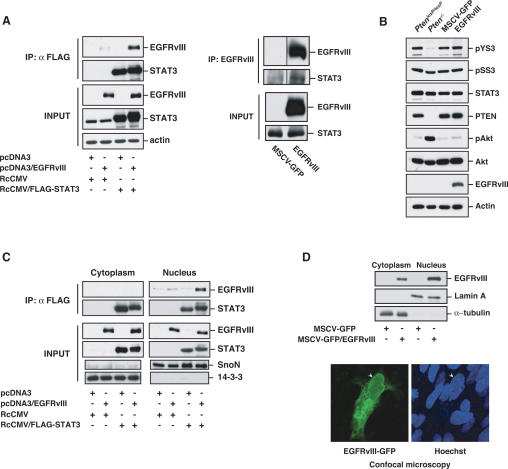
Figure 4.
STAT3 interacts with EGFRvIII in the nucleus. (A, left panel) Lysates of 293T cells transfected with expression plasmids encoding Flag-STAT3, EGFRvIII, or both were immunprecipitated with a Flag antibody followed by immunoblotting with an EGFR or STAT3 antibody. Total lysates (Input) were also immunoblotted with these antibodies. Actin served as loading control. EGFRvIII interacted efficiently with STAT3. (Right panel) Lysates of stable MSCV-GFP and MSCV-GFP/EGFRvIII astrocytes were immunoprecipitated with an EGFRvIII antibody followed by immunoblotting with an EGFR or STAT3 antibody. Total lysates (Input) were also immunoblotted with these antibodies. EGFRvIII interacted with endogenous STAT3 in astrocytes. (B) Immunoblotting of PtenloxP/loxP and Pten−/− astrocytes, or stable MSCV-GFP and MSCV-GFP/EGFRvIII astrocytes with antibodies that recognize Tyr705-phosphorylated STAT3 (pYS3), Ser727-phosphorylated STAT3 (pSS3), total STAT3, PTEN, Ser473-phosphorylated Akt (pAkt), total Akt, and EGFR. Actin was used as loading control. EGFRvIII expression had little effect on STAT3 Tyr705 and Ser727 phosphorylation. (C) Lysates of 293T cells transfected with Flag-STAT3, EGFRvIII, or both were fractionated into cytoplasmic and nuclear fractions and then subjected to immunoprecipitation and immunoblotting analyses as in A. SnoN and 14–3–3 were used as nuclear or cytosolic markers, respectively. EGFRvIII interacted with STAT3 more efficiently in the nuclear than in the cytoplasmic fraction. (D) EGFRvIII is present in the nucleus of astrocytes. (Top panel) Lysates of EGFRvIII-expressing and control astrocytes were subjected to subcellular fractionation and immunoblotting with the EGFR antibody. Lamin A and α-tubulin were used as nuclear or cytosolic markers, respectively. (Bottom panel) Immortalized astrocytes transfected with a plasmid encoding an EGFRvIII-GFP fusion protein were fixed and imaged by confocal microscopy to detect GFP fluorescence. Hoechst was used to visualize nuclei. Arrowheads indicate the position of the nucleus.
Since EGFRvIII associated efficiently with STAT3 but failed to significantly alter STAT3 phosphorylation at Tyr705 or Ser727, we further characterized the interaction of EGFRvIII with STAT3. Several activated RTKs may translocate to the nucleus, raising the possibility of a direct signaling mechanism from the cell surface to the nucleus (Wells and Marti 2002; Carpenter 2003). Subcellular fractionation analyses revealed that EGFRvIII, like STAT3, was present in both the nuclear and cytoplasmic fractions (Fig. 4C,D). In addition, EGFRvIII staining was present in the nucleus as well as the cytoplasm in astrocytes in confocal fluorescence microscopy experiments (Fig. 4D). Strikingly, EGFRvIII formed a physical complex with STAT3 more efficiently in the nuclear than cytoplasmic fraction (Fig. 4C). These results suggest that the constitutively active EGFRvIII localizes in the nucleus where it associates with STAT3. Corroborating our findings, activated wild-type EGFR has been reported to interact with STAT3 in the nucleus leading to direct transcriptional activation of the pro-oncogenic genes VEGF and iNOS in breast cancer cells (Lo et al. 2005). Expression of EGFRvIII in astrocytes stimulated iNOS expression in a STAT3-dependent manner as monitored by RT–PCR assays (data not shown). Taken together, these observations support the conclusion that EGFRvIII associates with STAT3 in the nucleus and raised the possibility that STAT3 might directly couple EGFRvIII signals to oncogenic responses in glial cells.
To determine STAT3 function in EGFRvIII-induced malignant astrocyte transformation, we assessed the effect of STAT3 knockout on the behavior of astrocytes expressing EGFRvIII (Fig. 5A). In cell population growth experiments, STAT3 knockout led to a significant reduction in the number of EGFRvIII-expressing astrocytes (Fig. 5B). In these assays, STAT3 loss had little or no effect on cell death (data not shown). Together, these results suggest that STAT3 promotes cell proliferation of astrocytes upon EGFRvIII expression.
We next measured the ability of Stat3loxP/loxP;EGFRvIII and Stat3−/−;EGFRvIII astrocytes to undergo malignant transformation upon injection into SCID mice (Fig. 5C,D). We found that Stat3loxP/loxP;EGFRvIII astrocytes formed large tumors with histological features typical of malignancy, including nuclear atypia and frequent mitotic figures (Fig. 5C,D). Remarkably, STAT3 knockout blocked the ability of EGFRvIII-expressing astrocytes to undergo malignant transformation (Fig. 5C). Thus, in contrast to STAT3-mediated suppression of transformation in PTEN knockdown astrocytes, STAT3 is essential for EGFRvIII-mediated astrocyte transformation. Collectively, our results indicate that STAT3 has opposing functions in cell transformation depending on the genetic environment. While STAT3 has an oncogenic function in the context of EGFRvIII expression, STAT3 behaves as a tumor suppressor specifically in the PTEN pathway.
Identification of the opposing functions of STAT3 in glial transformation in the distinct setting of PTEN deficiency or EGFRvIII expression raised the intriguing question of the behavior of STAT3 in cells subjected to both oncogenic stimuli of PTEN loss and EGFRvIII expression. To address this question, we generated Stat3loxP/loxP and Stat3−/− astrocytes that are both deficient in PTEN (PTENshRNA) and express EGFRvIII (Fig. 5E). We found that STAT3 knockout blocked malignant transformation of PTENshRNA;EGFRvIII astrocytes (Fig. 5E). These results support the conclusion that nuclear EGFRvIII acts as a switch to convert STAT3 from a tumor-suppressive to pro-oncogenic protein.
STAT3 signaling in human glioblastomas
The elucidation of opposing STAT3 functions in PTEN loss- and EGFRvIII-induced transformation in genetic studies in mouse astrocytes prompted us to consider the role of STAT3 in human glial tumors. We addressed this question by measuring the levels of PTEN, EGFRvIII, and STAT3 Tyr705 phosphorylation in a panel of human glioblastoma specimens (Fig. 6A). We monitored STAT3 phosphorylation in the human glioblastoma specimens by immunoblotting using a well-established phosphoTyr705-STAT3 antibody (Bonni et al. 1997). Since STAT3 was inhibited in Pten−/− mouse astrocytes due to down-regulation of LIFRβ, we also assessed the levels of LIFRβ in the glioblastoma specimens (Fig. 6A). The levels of phosphorylated STAT3, LIFRβ, PTEN, and EGFRvIII were measured as continuous variables and normalized to GAPDH.
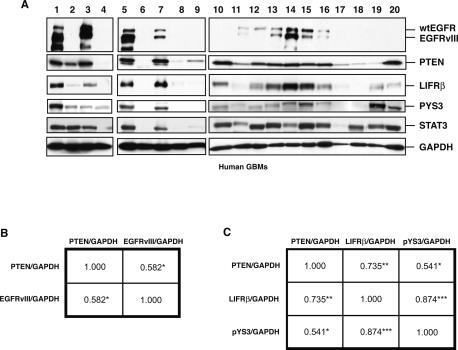
Figure 6.
STAT3 signaling in human glioblastoma specimens. (A) Immunoblotting of lysates of human glioblastoma samples (GBM) with antibodies to EGFR that recognize both wild-type and EGFRvIII forms, PTEN, LIFRβ, Tyr705-phosphorylated STAT3 (pYS3), or total STAT3. GAPDH was used as loading control. (B,C) Spearman correlation matrix of PTEN and EGFRvIII levels (B) or PTEN, LIFRβ, and pYS3 levels (C) measured as continuous variables in the immunoblots shown in A. The Spearman rank correlation coefficients, rs ([*] P < 0.05; [**] P < 0.005; [***] P < 0.0001), are shown.
We found that EGFRvIII expression and low PTEN levels stratified into distinct tumors (Fig. 6A). Upon subjecting the immunoblotting data to regression analysis, a positive correlation was found between PTEN expression and EGFRvIII expression with a Spearman rank correlation coefficient of rs = 0.582 (P < 0.05) (Fig. 6B). These results are corroborated by other studies demonstrating that PTEN loss and EGFRvIII mark distinct molecular subsets of human glioblastoma, with a small subset of tumors displaying both genetic alterations (Choe et al. 2003; Mellinghoff et al. 2005). Immunohistochemical analysis of human glioblastoma specimens expressing high levels of EGFRvIII revealed EGFRvIII immunoreactivity predominantly in the cytoplasm in tumor cells. Remarkably, EGFRvIII staining was also found in the nucleus in some tumor cells (Supplemental Fig. S3). In coimmunoprecipitation studies, EGFRvIII and STAT3 interacted in the nuclear fraction of glioblastomas expressing high EGFRvIII levels (Supplemental Fig. S4). These results corroborate our findings in mouse astrocytes, and suggest that EGFRvIII and STAT3 may form a physical nuclear complex in human glioblastomas.
We also found that low levels of PTEN were associated with down-regulation of LIFRβ and phosphorylated STAT3 in human glioblastomas (Fig. 6A). Regression analysis revealed a strong positive correlation between PTEN expression and LIFRβ expression with a Spearman rank correlation coefficient of rs = 0.735 (P < 0.005) (Fig. 6C). Similarly, levels of PTEN and phosphorylated STAT3 correlated tightly with a Spearman rank correlation value of rs = 0.541 (P < 0.05) (Fig. 6C). Consistent with the interpretation that STAT3 and LIFRβ act in a common signaling pathway, the levels of LIFRβ and phosphorylated STAT3 also correlated in a highly significant manner, displaying a Spearman rank correlation coefficient of rs = 0.874 (P < 0.0001) (Fig. 6C). Collectively, these findings demonstrate that PTEN loss and EGFRvIII mark distinct subsets of glioblastoma, and PTEN loss, but not EGFRvIII, is associated with inhibition of the LIFRβ–STAT3 signaling pathway in human glioblastomas.
Discussion
We discovered an unexpected tumor-suppressive function for STAT3 that is regulated by the tumor suppressor PTEN. The major findings in this study are as follows: (1) Knockout experiments in mouse astrocytes demonstrate that STAT3 inhibits astrocyte proliferation and invasiveness in vitro and suppresses the malignant transformation of PTEN knockdown astrocytes in vivo. (2) PTEN loss, acting via the Akt–FOXO signaling pathway, triggers the down-regulation of the cytokine receptor LIFRβ in astrocytes and consequently inhibits the phosphorylation of STAT3 at the key regulatory site Tyr705. (3) In human glioblastoma specimens, PTEN loss correlates tightly with low levels of LIFRβ expression and inactivation of STAT3. (4) In contrast to the tumor-suppressive function of STAT3 in the PTEN pathway, STAT3 forms a physical complex with the pro-oncogenic protein EGFRvIII in the nucleus and mediates EGFRvIII-induced astrocyte transformation. Collectively, these findings reveal that STAT3 harbors distinct pro-oncogenic and tumor-suppressive functions that are dictated by key genetic alterations associated with glioblastoma (Fig. 7). These results provide the foundation for the development of selective treatments in glioblastoma that are based on specific tumor-associated signaling events.
The identification of a STAT3 tumor-suppressive function may have significant ramifications in the management of glioblastoma. The prevailing view is that STAT3 acts as a general positive regulator of cell transformation and oncogenesis in diverse tissues including the brain (Bromberg et al. 1999; Turkson and Jove 2000; Chan et al. 2004; Chiarle et al. 2005; Schlessinger and Levy 2005). This has led to intense efforts to develop inhibitors of STAT3 using small molecule, peptide, and decoy oligonucleotide approaches with the hope of treating many tumor types including brain tumors (Turkson et al. 2001; Leong et al. 2003; Song et al. 2005). Our finding that STAT3 may operate in either a tumor suppressor or tumor-promoting capacity suggests that careful consideration must be given to STAT3 function and the associated genetic profile in each tumor type in order to determine whether inhibition of STAT3 activity might be ultimately beneficial or potentially harmful as a treatment strategy.
Analysis of the regulatory mechanisms underlying the unexpected tumor suppressor function of STAT3 in glial cells illuminates how loss of the tumor suppressor PTEN promotes the pathogenesis of glial tumors. Identification of the cytokine receptor LIFRβ as a direct target of the transcription factor FOXO3 establishes a mechanism by which the PTEN–Akt–FOXO cascade modulates the glial developmental LIFRβ–STAT3 signaling pathway. The intersection of these two signaling networks allows PTEN loss to down-regulate LIFRβ expression and inhibit STAT3 activity, thereby relieving STAT3’s suppression of glial cell proliferation, invasiveness, and transformation. The inhibition of STAT3 signaling may thus constitute a mechanism by which PTEN loss induces a dedifferentiation program in astrocytes leading to their malignant transformation. Akt inhibition of the DNA-binding activity of STAT3 may also occur in glioblastoma cell lines, although the underlying mechanism is unclear (Ghosh et al. 2005). An important question for future investigation is whether the PTEN-regulated STAT3 tumor suppressor pathway operates specifically in glial cells or whether STAT3 might also function in a tumor-suppressive capacity in other cell types in which PTEN is a critical tumor suppressor. Interestingly, the PI3K–Akt signaling pathway may also inhibit STAT3 in melanoma cells, although the functional significance of STAT3 inhibition in these cells remains to be investigated (Krasilnikov et al. 2003).
In stark contrast to the tumor-suppressive role of STAT3 in the PTEN pathway, STAT3 forms a physical complex with the pro-oncogenic protein EGFRvIII in the nucleus and thus mediates the ability of EGFRvIII to induce glial cell transformation. In the absence of either PTEN loss or EGFRvIII overexpression, STAT3 suppresses astrocyte cell proliferation and invasiveness. This is consistent with the view that the predominant function of STAT3 in astrocytes is to prevent malignant transformation. Thus, nuclear EGFRvIII may trigger a switch in the function of STAT3 from tumor suppressing to tumor promoting. A prediction of this hypothesis is that STAT3 might exert a tumor-promoting function in glioblastomas that are both PTEN-deficient and express EGFRvIII. Consistent with this conclusion, we found that STAT3 is required for the malignant transformation of astrocytes that are both PTEN-deficient and express EGFRvIII. In future studies, it will be interesting to determine the extent of nuclear EGFRvIII function in other tumors. The presence of EGFRvIII in the nucleus has been described in breast and prostate cancer (Ge et al. 2002; Edwards et al. 2006). A nuclear protein complex comprising of activated wild-type EGFR and STAT3 activates transcription of pro-oncogenic genes in breast cancer cells (Lo et al. 2005). Since EGFRvIII and STAT3 are expressed in many tumors (Bowman et al. 2000; Bromberg 2001; Pedersen et al. 2001; Lorimer 2002; Mizoguchi et al. 2006), a complex of nuclear EGFRvIII and STAT3 may thus act widely to promote malignant cell transformation.
Beyond EGFRvIII, the nuclear localization of membrane-bound RTKs, such as FGFR (fibroblast growth factor receptor) and wild-type EGFR, has been reported in diverse cell types (Maher 1996; Lin et al. 2001; Offterdinger et al. 2002). Although the mechanism of nuclear localization of RTKs is unknown, several hypotheses have been proposed (Wells and Marti 2002; Carpenter 2003). RTKs might reach the endoplasmic reticulum via the endocytic pathway, then might be extracted into the cytoplasm by the ERAD (endoplasmic reticulum-associated degradation) system (Tsai et al. 2002), and transferred into the nucleoplasm by the nuclear import machinery (Carpenter 2003). Alternatively, binding of chaperone-like factors to the receptor might mask the hydrophobicity of the membrane-spanning domain, allowing it to become soluble in the cytoplasm and translocate into the nucleus (Wells and Marti 2002). The requirement of importin-β for nuclear translocation of FGFR and HER2 (Reilly and Maher 2001; Giri et al. 2005) suggests that the nuclear import machinery might participate in nuclear targeting of RTKs. Once in the nucleus, RTKs might regulate transcription, phosphorylate nuclear proteins, alter chromatin conformation, and/or modulate mRNA processing (Wells and Marti 2002).
The elucidation of STAT3 as a dual regulator of PTEN loss- and EGFRvIII-induced glial malignancy in this study may provide clues for the design of patient-tailored therapies aimed at treating patients with glioblastomas according to their underlying molecular pathogenesis. STAT3 inhibitors may be useful in the treatment of glioblastomas in which EGFRvIII is amplified, while STAT3 activators may have therapeutic value in PTEN-deficient brain tumors.
Materials and methods
Cell culture
PtenloxP/loxP and Pten−/− astrocytes were cultured from mice in which the PTEN gene contained loxP sites flanking exon 5 (M.J. You and R.A. DePinho, unpubl.). Stat3loxP/loxP and Stat3−/− astrocytes were obtained from mice with floxed STAT3 alleles (Stat3loxP/loxP) (Raz et al. 1999) and immortalized by retroviral-mediated expression of the SV40 large T antigen protein. The genes flanked by loxP sites were excised in vitro using adenovirus encoding the recombinase Cre (University of Iowa). To obtain constitutively active Akt-expressing cells, PtenloxP/loxP astrocytes were infected with pWZL/MyrAkt or empty vector retroviruses, and infected cells were selected with hygromycin (250 μg/mL). FOXO3 knockdown cells were obtained by infection of PtenloxP/loxP and Pten−/− astrocytes with pLL3.7-Puro/FOXOi lentiviruses and selection with puromycin (4 μg/mL). PTEN knockdown and EGFRvIII-expressing astrocytes were generated by infection of Stat3loxP/loxP astrocytes with pSUPER-Puro retoviruses encoding an shRNA directed against PTEN or MSCV-IRES-GFP/EGFRvIII retroviruses, respectively.
Plasmids
The pcDNA3/EGFRvIII and pEGFP-N1/EGFRvIII constructs were obtained by subcloning from the MSCV-IRES-GFP/ EGFRvIII plasmid (kind gift from Dr. Alonzo Ross, University of Massachusetts Medical School) into pcDNA3 and pEGFP-N1, respectively.
Virus production and infection
Recombinant retroviruses were made by transfecting 293T cells with pMD.MLV gag.pol, pHDM.G (VSVG pseudotype), and the transfer plasmid (e.g., MSCV-IRES-GFP). Cells were infected with equal amounts of retroviruses and selected with the appropriate resistance drug or FACS-sorted for GFP expression.
Cloning of recombinant lentiviruses coding for a shRNA directed against FOXO was carried out using a modified pLL3.7 vector that encodes resistance to puromycin (pLL3.7 Puro). The following complementary oligonucleotides were inserted into pLL3.7 Puro: FOXOi fw, 5′-TGCGTGCCCTACTTCAAGGA TTCAAGAGATCCTTGAAGTAGGGCACGCTTTTTGGA AAC-3′; FOXOi rev, 5′-TCGAGTTTCCAAAAAGCGTGCC CTACTTCAAGGATCTCTTGAATCCTTGAAGTAGGGC ACGCA-3′. Hairpin structures containing the stem sequences (underlined) and the loops (bold italics) are indicated. Lentiviruses were generated by cotransfecting pLL3.7 and packaging vectors (VSVG, RSV-REV, and pMDL g/p RRE) into 293T cells. Cells were infected with equal amounts of lentiviruses and selected with puromycin.
The PTEN-encoding adenovirus was a kind gift from Dr. Christopher D. Kontos (Duke University School of Medicine).
ChIP
ChIP analyses were done as described (Shi et al. 2003). Following immunoprecipitation, a PCR reaction was used to amplify the LIFRβ promoter with the following primers: first PCR reaction, mouse LIFRβ fw, 5′-TGTGGGAAAGAATGGGGATA-3′; mouse LIFRβ rev, 5′-AACCGCTGTCATTGCACTTT-3′; nested PCR reaction, mouse LIFRβ nest fw, 5′-CCAGAAAC AGTCATGGACAGC-3′; mouse LIFRβ nest rev, 5′-GCGGAG GAGGAAACTCGT-3′. The following primers designed to amplify the E-cadherin promoter were used as negative controls for the PCR reaction: mouse E-cad fw, 5′-ACCGTCGGAGAA ATAGCTCA-3′; mouse E-cad rev, 5′-AACTTCCTCCACCCC TGTCT-3′.
Matrigel invasion assays
Matrigel precoated invasion chambers (BD) with an 8-μm pore size membrane were utilized according to the manufacturer’s instructions. Cells (2.5 × 104) in 500 μL of serum-free DMEM were added to each of the inserts and incubated for 22 h at 37°C. Cells on the lower surface of the membrane that had migrated through the matrigel were fixed, stained with crystal violet, and counted. Noninvasive NIH3T3 cells were used as a negative control. Equivalent numbers of NIH3T3 failed to invade the matrigel.
Mouse injections
Cells (1 × 106) were resuspended in serum-free, antibiotic-free media and injected subcutaneously into 4- to 6-wk-old SCID mice. Approximately 8 wk (PTEN knockdown astrocytes) or 4 wk (EGFRvIII astrocytes) after injection, the mice were sacrificed and the tumors were removed, measured, and fixed for histologic analysis.
Human tumor tissue samples
Discarded excess human glioblastoma tissue from neurosurgical specimens was collected and used in accordance with local IRB policies. Flash-frozen tumor tissue was processed for Western blotting analysis.
Statistical analysis
Correlation analysis of immunoblotting of human glioblastoma specimens was done by the nonparametric Spearman rank correlation test using the StatView statistics package.
Immunohistochemistry
Staining was performed on 5-μm paraffin sections. Sections were deparaffinized through xylenes and graded ethanol, and then rehydrated in PBS. Antigen retrieval was performed by microwaving for 20 min in 1 mM EDTA/5 mM Tris buffer (pH 8.0). The antibodies used were rabbit polyclonal against Ki-67 (Vector Laboratories), mouse monoclonal anti-Nestin (BD Biosciences), and mouse monoclonal anti-EGFRvIII (Skybio). Detection was performed with the Envision+ system (Dako) per the manufacturer’s protocol.
Acknowledgments
We thank Rod Bronson for histologic analysis of tumors, Jennifer Zheng for help in the statistical analysis of the human glioblastomas data, Cathy Nutt and David Louis for providing glioblastoma tumor specimens, and John P. Carroll for help in the maintenance of mouse colonies. We also thank James A. DeCaprio, Alonzo H. Ross, Jeng-Shin Lee, Joan Brugge, and Heinrich Baumann for providing plasmids. This work was supported by awards from the Stewart Trust of Washington, D.C. (to A.B.), the Armenise-Harvard Foundation (to A.B.), and the Carolyn and Peter Lynch Research Fund (to A.B.); NIH grants to A.B. (NS051255, NS41021, and NS047188), D.E.L., and R.A.D.; a post-doctoral fellowship from the Fundación Ramón Areces, Spain (N.d.I.); and a Taplin Post-doctoral fellowship (N.d.I.). R.A.D. is an American Cancer Society Research Professor and is supported by the Robert A. and Renee E. Belfer Institute for Innovative Cancer Science. A.B. is the recipient of a fellowship from the Alfred P. Sloan Foundation, a Robert H. Ebert Clinical Scholar Award from the Esther A. and Joseph Klingenstein Fund, an EJLB Foundation award, and a Sidney Kimmel Foundation Award.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1606508
References
- Bachoo R.M., Maher E.A., Ligon K.L., Sharpless N.E., Chan S.S., You M.J., Tang Y., DeFrances J., Stover E., Weissleder R., et al. Epidermal growth factor receptor and Ink4a/Arf. Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Bajenaru M.L., Hernandez M.R., Perry A., Zhu Y., Parada L.F., Garbow J.R., Gutmann D.H. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63:8573–8577. [PubMed] [Google Scholar]
- Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D.A., Rozovsky I., Stahl N., Yancopoulos G.D., Greenberg M.E. Regulation of gliogenesis in the central nervous system by the JAK–STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Bowman T., Garcia R., Turkson J., Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- Bromberg J.F. Activation of STAT proteins and growth control. Bioessays. 2001;23:161–169. doi: 10.1002/1521-1878(200102)23:2<161::AID-BIES1023>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C., Darnell J.E. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Burgering B.M., Kops G.J. Cell cycle and death control: Long live Forkheads. Trends Biochem. Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- Cantley L.C. The phosphoinositide 3′-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr. Opin. Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- Chan K.S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., DiGiovanni J. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R., Simmons W.J., Cai H., Dhall G., Zamo A., Raz R., Karras J.G., Levy D.E., Inghirami G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- Choe G., Horvath S., Cloughesy T.F., Crosby K., Seligson D., Palotie A., Inge L., Smith B.L., Sawyers C.L., Mischel P.S. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- Dai C., Holland E.C. Astrocyte differentiation states and glioma formation. Cancer J. 2003;9:72–81. doi: 10.1097/00130404-200303000-00002. [DOI] [PubMed] [Google Scholar]
- Darnell J.E., Kerr I.M., Stark G.R. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Decker T., Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- Edwards J., Traynor P., Munro A.F., Pirret C.F., Dunne B., Bartlett J.M. The role of HER1–HER4 and EGFRvIII in hormone-refractory prostate cancer. Clin. Cancer Res. 2006;12:123–130. doi: 10.1158/1078-0432.CCR-05-1445. [DOI] [PubMed] [Google Scholar]
- Furnari F.B., Lin H., Huang H.S., Cavenee W.K. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc. Natl. Acad. Sci. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari F.B., Huang H.J., Cavenee W.K. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C., et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes & Dev. 212683–2710. [DOI] [PubMed] [Google Scholar]
- Ge H., Gong X., Tang C.K. Evidence of high incidence of EGFRvIII expression and coexpression with EGFR in human invasive breast cancer by laser capture microdissection and immunohistochemical analysis. Int. J. Cancer. 2002;98:357–361. doi: 10.1002/ijc.10224. [DOI] [PubMed] [Google Scholar]
- Ghosh M.K., Sharma P., Harbor P.C., Rahaman S.O., Haque S.J. PI3K–AKT pathway negatively controls EGFR-dependent DNA-binding activity of Stat3 in glioblastoma multiforme cells. Oncogene. 2005;24:7290–7300. doi: 10.1038/sj.onc.1208894. [DOI] [PubMed] [Google Scholar]
- Giri D.K., Ali-Seyed M., Li L.Y., Lee D.F., Ling P., Bartholomeusz G., Wang S.C., Hung M.C. Endosomal transport of ErbB-2: Mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland E.C. Gliomagenesis: Genetic alterations and mouse models. Nat. Rev. Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- Holland E.C., Hively W.P., DePinho R.A., Varmus H.E. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.H., Mukasa A., Bonavia R., Flynn R.A., Brewer Z.E., Cavenee W.K., Furnari F.B., White F.M. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Murray T., Samuels A., Ghafoor A., Ward E., Thun M.J. Cancer statistics, 2003. CA Cancer J. Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- Kapoor G.S., O’Rourke D.M. Receptor tyrosine kinase signaling in gliomagenesis: Pathobiology and therapeutic approaches. Cancer Biol. Ther. 2003;2:330–342. doi: 10.4161/cbt.2.4.507. [DOI] [PubMed] [Google Scholar]
- Konnikova L., Kotecki M., Kruger M.M., Cochran B.H. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G., Bonni A. Signaling pathways regulating gliomagenesis. Curr. Mol. Med. 2003;3:73–84. doi: 10.2174/1566524033361609. [DOI] [PubMed] [Google Scholar]
- Krasilnikov M., Ivanov V.N., Dong J., Ronai Z. ERK and PI3K negatively regulate STAT-transcriptional activities in human melanoma cells: Implications towards sensitization to apoptosis. Oncogene. 2003;22:4092–4101. doi: 10.1038/sj.onc.1206598. [DOI] [PubMed] [Google Scholar]
- Leaman D.W., Pisharody S., Flickinger T.W., Commane M.A., Schlessinger J., Kerr I.M., Levy D.E., Stark G.R. Roles of JAKs in activation of STATs and stimulation of c-fos gene expression by epidermal growth factor. Mol. Cell. Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong P.L., Andrews G.A., Johnson D.E., Dyer K.F., Xi S., Mai J.C., Robbins P.D., Gadiparthi S., Burke N.A., Watkins S.F., et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc. Natl. Acad. Sci. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Libermann T.A., Nusbaum H.R., Razon N., Kris R., Lax I., Soreq H., Whittle N., Waterfield M.D., Ullrich A., Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y., Bourguignon L., Hung M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Ling X., Arlinghaus R.B. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- Lo H.W., Hsu S.C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J.Y., Hung M.C. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Lorimer I.A. Mutant epidermal growth factor receptors as targets for cancer therapy. Curr. Cancer Drug Targets. 2002;2:91–102. doi: 10.2174/1568009023333926. [DOI] [PubMed] [Google Scholar]
- Louis D.N. Molecular pathology of malignant gliomas. Ann. Rev. Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K. World Health Organization histological classification of tumours of the central nervous system. IARC Press; International Agency for Research on Cancer (IARC), Lyon, France: 2007. [Google Scholar]
- Maehama T., Dixon J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maher P.A. Nuclear translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J. Cell Biol. 1996;134:529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff I.K., Wang M.Y., Vivanco I., Haas-Kogan D.A., Zhu S., Dia E.Q., Lu K.V., Yoshimoto K., Huang J.H., Chute D.J., et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Mizoguchi M., Betensky R.A., Batchelor T.T., Bernay D.C., Louis D.N., Nutt C.L. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: Correlation with EGFR status, tumor grade, and survival. J. Neuropathol. Exp. Neurol. 2006;65:1181–1188. doi: 10.1097/01.jnen.0000248549.14962.b2. [DOI] [PubMed] [Google Scholar]
- Moscatello D.K., Montgomery R.B., Sundareshan P., McDanel H., Wong M.Y., Wong A.J. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- Myers M.P., Stolarov J.P., Eng C., Li J., Wang S.I., Wigler M.H., Parsons R., Tonks N.K. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl. Acad. Sci. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R., Ji X.D., Harmon R.C., Lazar C.S., Gill G.N., Cavenee W.K., Huang H.J. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl. Acad. Sci. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offterdinger M., Schofer C., Weipoltshammer K., Grunt T.W. c-erbB-3: A nuclear protein in mammary epithelial cells. J. Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M.W., Meltorn M., Damstrup L., Poulsen H.S. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann. Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Rahaman S.O., Harbor P.C., Chernova O., Barnett G.H., Vogelbaum M.A., Haque S.J. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- Rajan P., McKay R.D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R., Lee C.K., Cannizzaro L.A., d’Eustachio P., Levy D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly J.F., Maher P.A. Importin β-mediated nuclear import of fibroblast growth factor receptor: Role in cell proliferation. J. Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L.K., Ren Z., Fuller G.N., Schaefer T.S. Constitutive activation of Stat3α in brain tumors: Localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;21:2058–2065. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- Schlessinger K., Levy D.E. Malignant transformation but not normal cell growth depends on signal transducer and activator of transcription 3. Cancer Res. 2005;65:5828–5834. doi: 10.1158/0008-5472.CAN-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sawada J., Sui G., Affar el B., Whetstine J.R., Lan F., Ogawa H., Luke M.P., Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Song H., Wang R., Wang S., Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc. Natl. Acad. Sci. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck P.A., Pershouse M.A., Jasser S.A., Yung W.K., Lin H., Ligon A.H., Langford L.A., Baumgard M.L., Hattier T., Davis T., et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stewart L.A. Chemotherapy in adult high-grade glioma: A systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet. 2002;359:1011–1018. doi: 10.1016/s0140-6736(02)08091-1. [DOI] [PubMed] [Google Scholar]
- Sugawa N., Ekstrand A.J., James C.D., Collins V.P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl. Acad. Sci. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T.A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Turkson J., Jove R. STAT proteins: Novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- Turkson J., Ryan D., Kim J.S., Zhang Y., Chen Z., Haura E., Laudano A., Sebti S., Hamilton A.D., Jove R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J. Biol. Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- Uhrbom L., Dai C., Celestino J.C., Rosenblum M.K., Fuller G.N., Holland E.C. Ink4a–Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- Van Der Heide L.P., Hoekman M.F., Smidt M.P. The ins and outs of FoxO shuttling: Mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M.L., Sadowski H.B., Watling D., Rogers N.C., Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 1996;16:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang W., Huang H.J., Liao W.S., Fuller G.N. Lab Invest. Vol. 84. 2004. Analysis of the activation status of Akt, NFκB, and Stat3 in human diffuse gliomas; pp. 941–951. [DOI] [PubMed] [Google Scholar]
- Ware C.B., Horowitz M.C., Renshaw B.R., Hunt J.S., Liggitt D., Koblar S.A., Gliniak B.C., McKenna H.J., Papayannopoulou T., Thoma B., et al. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- Weissenberger J., Loeffler S., Kappeler A., Kopf M., Lukes A., Afanasieva T.A., Aguzzi A., Weis J. IL-6 is required for glioma development in a mouse model. Oncogene. 2004;23:3308–3316. doi: 10.1038/sj.onc.1207455. [DOI] [PubMed] [Google Scholar]
- Wells A., Marti U. Signalling shortcuts: Cell-surface receptors in the nucleus? Nat. Rev. 2002;3:697–702. doi: 10.1038/nrm905. [DOI] [PubMed] [Google Scholar]
- Wong A.J., Bigner S.H., Bigner D.D., Kinzler K.W., Hamilton S.R., Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc. Natl. Acad. Sci. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.J., Ruppert J.M., Bigner S.H., Grzeschik C.H., Humphrey P.A., Bigner D.S., Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Natl. Acad. Sci. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T., Kawaguchi D., Oishi K., Takeda K., Akira S., Masuyama N., Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133:2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Wen Z., Darnell J.E. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]