Abstract
Regulation of the cell cycle is intimately linked to erythroid differentiation, yet how these processes are coupled is not well understood. To gain insight into this coordinate regulation, we examined the role that the retinoblastoma protein (Rb), a central regulator of the cell cycle, plays in erythropoiesis. We found that Rb serves a cell-intrinsic role and its absence causes ineffective erythropoiesis, with a differentiation block at the transition from early to late erythroblasts. Unexpectedly, in addition to a failure to properly exit the cell cycle, mitochondrial biogenesis fails to be up-regulated concomitantly, contributing to this differentiation block. The link between erythropoiesis and mitochondrial function was validated by inhibition of mitochondrial biogenesis. Erythropoiesis in the absence of Rb resembles the human myelodysplastic syndromes, where defects in cell cycle regulation and mitochondrial function frequently occur. Our work demonstrates how these seemingly disparate pathways play a role in coordinately regulating cellular differentiation.
[Keywords: Rb, cell cycle, cell differentiation, erythropoiesis, hematopoiesis, mitochondrial biogenesis]
The process of erythroid differentiation is intimately coupled with control of the cell cycle. The earliest committed progenitor of the erythroid lineage is the burst-forming unit (BFU-E), which has a slow rate of proliferation (Gregory and Eaves 1977, 1978). BFU-Es then mature into the colony-forming unit erythroid (CFU-E) cells, and in the process undergo a rapid rise in proliferation. A series of three to five cell divisions ensues and further maturation occurs through the proerythroblast, basophilic erythroblast, and polychromatophilic erythroblast stages. Subsequently, cell cycle exit and post-mitotic maturation through the orthochromatophilic erythroblast stage occurs. Enucleation gives rise to reticulocytes, which then loose remaining organelles to become mature erythrocytes that enter the circulation.
While erythroid cell cycle regulation has been extensively studied at a descriptive level, many aspects of the molecular control of this process are not well understood. Studies in cell lines have suggested a role for erythroid transcription factors, such as GATA-1, in modulating the G1-phase arrest that occurs during erythroid maturation (Rylski et al. 2003). However, in vivo studies on the role of cell cycle regulators in erythropoiesis have been limited and are difficult to interpret in light of both cell-type-intrinsic and -extrinsic roles that these genes may have in erythropoiesis (Fero et al. 1996; Ciemerych et al. 2002). Insights into the molecular control of the erythroid cell cycle will further an understanding of how differentiation within this hematopoietic lineage proceeds. Furthermore, modulation of the erythroid cell cycle with S-phase inhibitors is utilized clinically in attempts to increase fetal hemoglobin levels in patients with hemoglobinopathies (Letvin et al. 1985; Stamatoyannopoulos 2005). Knowledge pertaining to how the cell cycle is coupled to erythroid cell maturation may lead to more efficacious therapies for sickle cell anemia and β-thalassemia.
The retinoblastoma protein (Rb) is a central regulator of the G1-to-S-phase transition of the cell cycle (Classon and Harlow 2002). Rb is one member of the pocket protein family of cell cycle regulators that also include p107 and p130. Rb is phosphorylated by activated cyclin-dependent kinases (CDKs), and the resulting hyperphosphorylated form is incapable of binding to its interaction partners, promoting cell cycle progression. The best characterized among these interaction partners are the E2F family of transcription factors that are bound by hypophosphorylated Rb. This interaction represses E2F target genes that are necessary for cell cycle progression.
The role of Rb in erythropoiesis has been the subject of controversy for >15 years. Rb-null mice die at approximately E14.5 with several developmental defects, including a failure of erythroid cell maturation, causing an anemia proposed to be the likely cause of death (Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992). Subsequent work showed that in the context of chimeric mice, Rb-null cells contribute to mature erythrocytes, suggesting that the red cell defects in the original knockout mice were non-cell-autonomous (Maandag et al. 1994; Williams et al. 1994). This conclusion received additional support from the finding that many embryonic defects could be explained by a placental defect that occurs in the absence of Rb with an accompanying deficiency in fetal–maternal nutrient exchange (Wu et al. 2003; Wenzel et al. 2007). Transplantation of fetal liver (FL) erythroid progenitors demonstrated, however, that erythropoiesis is not entirely normal in the absence of Rb (Hu et al. 1997), but the interpretation of this finding is complicated by possible defects in hematopoietic cells resulting from impaired placental nutrient exchange. In sum, prior work has not clarified the intrinsic role, if any, of Rb in erythropoiesis.
Additional efforts to resolve these issues have yielded conflicting results. It was hypothesized that Rb might have a cell-type-intrinsic, but non-cell-autonomous role in erythropoiesis (Whyatt and Grosveld 2002), as appears to be the case in mice that overexpress the GATA-1 protein (Whyatt et al. 2000). Studies using chimeric and transplant models suggested that Rb plays a role in stress erythropoiesis, but the gene appeared to be dispensable for homeostatic erythropoiesis (Spike et al. 2004). Using in vitro culture approaches, one group proposed that Rb serves an intrinsic role in erythroid maturation (Clark et al. 2004), while other investigators suggested that Rb is dispensible in erythroid cells, but necessary in macrophages for the formation of an intact erythroid island in vitro (Iavarone et al. 2004). To date, no direct experiments have been performed to assess an intrinsic role for Rb in erythropoiesis in vivo.
Here, we utilize conditional gene inactivation to delete Rb specifically in the erythroid lineage, as well as in other hematopoietic compartments. We demonstrate that Rb has a cell-type-intrinsic and cell-autonomous role in erythropoiesis. Rb loss in the erythroid compartment leads to a stable anemia that is attributable to ineffective erythropoiesis. Notably, the mild anemia of the mice results from a large degree of in vivo compensation for this ineffective erythropoiesis. Specifically, Rb is necessary for terminal maturation of erythroblasts that occurs concomitantly with cell cycle exit. We demonstrate that the intrinsic role for Rb in erythropoiesis is mediated by coupling the process of mitochondrial biogenesis with cell cycle exit during erythropoiesis. Our findings provide an in vivo demonstration of how cellular differentiation may be efficiently controlled through the coordinate regulation of mitochondrial biogenesis with cellular proliferation. Additionally, this work gives new insight into how ineffective erythropoiesis can occur in human diseases like the myelodysplastic syndromes (MDS) and suggests new therapeutic strategies for these conditions.
Results
Loss of Rb in the erythroid compartment results in a moderate anemia
The Rb gene was deleted specifically in the erythroid compartment by interbreeding pRbfl/fl mice (Sage et al. 2003) with erythropoietin receptor GFPcre knock-in (EpoR-GFPcre) heterozygous mice (Heinrich et al. 2004). The EpoR-GFPcre allele has been shown previously to direct quantitative excision of floxed alleles in the erythroid lineage both during development of the embryo and in the adult (Heinrich et al. 2004; Dumitriu et al. 2006; Maetens et al. 2007). Analysis of RNA and DNA from sorted early erythroblasts (CD71high/Ter-119high bone marrow [BM] cells) demonstrated efficient excision of the floxed Rb allele (Fig. 1A; Supplemental Fig. S1). The expression of the other pocket proteins, p107 and p130, was not appreciably changed. Therefore, increased expression of these proteins does not occur as a compensatory step in response to loss of Rb.
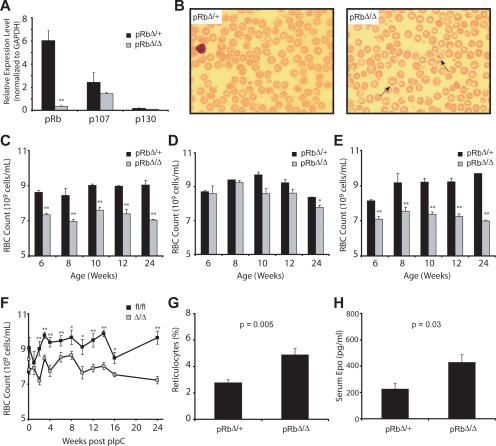
Figure 1.
Deletion of Rb in the erythroid compartment causes a stable anemia. (A) qRT–PCR of RNA from sorted CD71high/Ter-119high BM cells from EpoR-GFPcre/+; pRbfl/+ (pRbΔ/+), and EpoR-GFPcre/+; pRbfl/fl (pRbΔ/Δ) 8- to 12-wk-old animals (n ≥ 3). (**) P < 0.01. (B) Representative PB smears viewed at 600× magnification. Howell-Jolly bodies are seen in adult pRbΔ/Δ mice (arrows). (C–E) RBC counts in EpoR-GFPcre/+ animals (C), LysM-Cre animals (D), and EpoR-GFPcre and LysM-Cre animals (E) at multiple time points during life. (*) P < 0.05; (**) P < 0.01. (F) RBC counts in Mx1-Cre adult animals following somatic deletion. (*) P < 0.05; (**) P < 0.01. (G,H) Reticulocyte percentages (G) and serum erythropoietin levels (H) in EpoR-GFPcre/+ animals (n ≥ 4).
Animals lacking Rb in the erythroid compartment were born with expected Mendelian ratios and did not display visible developmental abnormalities. Peripheral blood (PB) from control (EpoR-GFPcre/+; pRbfl/+, herein pRbΔ/+) and pRbΔ/Δ animals was examined at multiple times. The pRbΔ/Δ animals had occasional Howell-Jolly bodies (nuclear remnants) present in most high-power fields, which were not found in the control animals (Fig. 1B, arrows). This indicates that there may have been either impaired splenic function or a more rapid terminal maturation of erythroid cells. Moderate and stable anemia persisted in adult pRbΔ/Δ animals (Fig. 1C). Of note, the anemia was normocytic and normochromic, as assessed by blood smears and red cell indices (e.g., MCV: pRbΔ/+ 53.8 ± 1.13 fL, pRbΔ/Δ 55.6 ± 0.64 fL; 8 wk of life). Furthermore, peripheral counts of other lineages were normal, indicating that the defect was restricted to the erythroid lineage.
Since prior work posited that macrophages in erythroid islands may require Rb function to support normal erythropoiesis (Iavarone et al. 2004), we deleted Rb in the myeloid lineage (macrophages and granulocytes) using Lysozyme-M-Cre, as has been described previously (Clausen et al. 1999; Walkley et al. 2007). In early adulthood, no anemia was noted, but with age, an exceedingly mild anemia appeared (Fig. 1D). These findings suggest that there either may be an additional age-dependent extrinsic role for Rb in erythropoiesis, or stochastic excision eventually occurs in earlier hematopoietic progenitors, resulting in Rb-null erythroid cells, as has been described previously (Ye et al. 2003). In support of the latter idea, we found excision of Rb in erythroid progenitors from some aged Lys-M-Cre; Rb fl/fl mice (Supplemental Fig. S2). We then combined myeloid and erythroid excision of Rb (using both Lys-M-Cre and EpoR-GFPcre) and found that there was a similar degree of anemia as was found in the EpoR-GFPcre mutant alone (Fig. 1E). Finally, complete excision of Rb in all hematopoietic lineages using the interferon-inducible Mx1-Cre (Kuhn et al. 1995; Walkley et al. 2007) revealed that somatic excision in the adult resulted in a similar degree of anemia (Fig. 1F) as with excision confined to the erythroid lineage. Therefore, these data indicate that Rb has a cell-intrinsic requirement in erythropoiesis.
We further explored the nature of the anemia that was present in erythroid pRbΔ/Δ animals, noting a 1.7-fold increase in the percentage of circulating reticulocytes, which is consistent with a partial physiologic attempt to compensate for the anemia (Fig. 1G). Erythropoietin levels were approximately twofold elevated in the pRbΔ/Δ versus control animals (Fig. 1H), indicating that a secondary response to the anemia was present in these animals and that the compensation is adequate to maintain a stable red cell count throughout life. To evaluate whether the anemia may in part be due to increased red cell destruction, we measured the life span of red blood cells (RBCs) in the circulation by labeling these cells with N-hydroxysuccinimide-biotin, and found that the RBC life span was completely preserved in the pRbΔ/Δ mice (Supplemental Fig. S3). Therefore, these data are consistent with a cell-intrinsic failure in maturation in the course of erythropoiesis.
Late-stage ineffective erythropoiesis occurs upon deletion of Rb
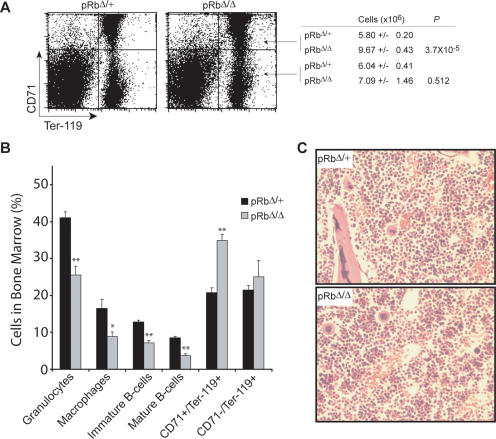
Given the inability of the Rb-null mice to compensate given their degree of anemia, we then examined hematopoiesis in the BM. We first noted that the cellularity of the BM in pRbΔ/Δ and pRbΔ/+ mice was similar (Supplemental Fig. S4A). Phenotypic assessment by FACS analysis revealed a 1.7-fold increase in the absolute numbers of early erythroblasts (CD71high/Ter-119high) in the BM, but near normal numbers of late erythroblasts (CD71low/Ter-119high) (Fig. 2A). The post-mitotic transition between polychromatophilic erythroblasts and orthochromatophilic erythroblasts takes place around the transition point between the CD71high and CD71low fractions of Ter-119high cells (Socolovsky et al. 2001; Zhang et al. 2003). Thus, it appears that a relative block in effective maturation of erythroblasts at this transition is imposed by loss of Rb. Concomitant with the observed expansion of early erythroblasts in the BM, a slight secondary decrease in BM progenitors of the other major lineages was found (Fig. 2B). Histological sections of the marrow confirmed the increase in the number of visually identifiable early erythroblasts present (Fig. 2C). Comparison of early erythroid progenitors using methycellulose colony assays did not reveal differences in progenitor numbers. The frequency of both BFU-E and megakaryocyte-erythroid (MegE) colonies were similar between the pRbΔ/Δ and pRbΔ/+ mice (Supplemental Fig. S4B). Furthermore, CFU-E numbers were comparable in the presence or absence of Rb (Supplemental Fig. S4C). We also examined the number of earlier progenitors (CFU-GEMM) and failed to note differences in Rb-null mice compared with controls (data not shown).
Figure 2.
Erythroid-specific deletion of Rb causes ineffective erythropoiesis in the BM. (A) FACS profile and mean numbers of early (CD71high/Ter-119high) and late (CD71low/Ter-119high) erythroblasts in the BM of EpoR-GFPcre/+ animals (n ≥ 5). (B) Percentages of major lineages in the BM as assessed using phenotypic surface markers (n ≥ 5). (*) P < 0.05; (**) P < 0.01. (C) Representative histological sections of the BM stained with hemotoxylin and eosin (H&E) viewed at 400× magnification.
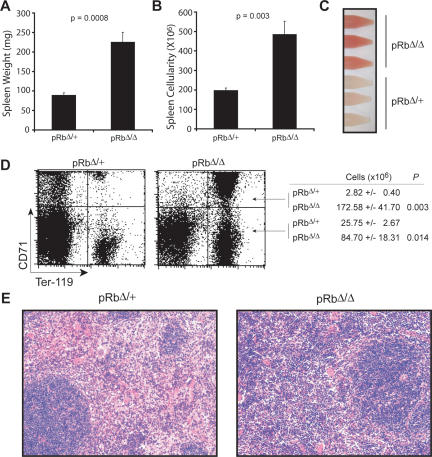
The spleens of the Rb-null mice were considerably enlarged. Spleen weights, as well as total cellularity, were both significantly increased (Fig. 3A,B). Single-cell suspensions prepared from the spleens were markedly different from controls. A greater degree of hemoglobinization was evident in the single-cell suspensions prepared from the pRbΔ/Δ mice (Fig. 3C). This observation indicated that extensive extramedullary erythropoiesis occurs in the spleens of Rb-null animals. Phenotypic analysis revealed a 60-fold increase in the number of early erythroblasts (CD71high/Ter-119high) present in the spleen (Fig. 3D). The numbers of more mature post-mitotic erythroid precursors (CD71low/Ter-119high) was also increased by threefold (Fig. 3D). It is important to note that the relative increase in the early erythroblast fraction in the spleens of the pRbΔ/Δ mice is not nearly accounted for by the smaller increase in terminal erythroblasts. The frequency of BFU-E colonies in the spleen was slightly reduced (1.7-fold) in the pRbΔ/Δ mice compared with littermate controls (Supplemental Fig. S4D); however, when the 2.5-fold increase in cellularity is taken into account, this indicates that the number of BFU-E colonies per spleen is slightly increased. In the case of the CFU-E colonies from the spleen, a threefold expansion in the frequency of colonies was observed (Supplemental Fig. S4E). When the increase in cellularity is also accounted for, it appears that the expansion in erythroblasts in the spleen of the Rb-null mice occurs by increased proliferation at and subsequent to the CFU-E stage. Examination of histological sections of the spleen was consistent with this interpretation and revealed a dramatic expansion of erythroblasts in the red pulp of the spleens of pRbΔ/Δ mice, with preservation of the white pulp architecture (Fig. 3E).
Figure 3.
Extensive extramedullary erythropoiesis in the spleen along with ineffective erythropoiesis from erythroid loss of Rb. (A,B) Spleen weights (A) and cellularities (B) from EpoR-GFPcre/+ animals (n ≥ 5). (C) Representative single-cell suspensions from the spleens. (D) FACS profile and mean numbers of early (CD71high/Ter-119high) and late (CD71low/Ter-119high) erythroblasts in the BM of EpoR-GFPcre/+ animals (n ≥ 5). (E) H&E-stained sections of the spleen viewed at 200× magnification.
The data from the spleen and BM of the pRbΔ/Δ mice is consistent with a phenotype of extensive ineffective erythropoiesis. An additional histological correlate of this finding was seen as splenic siderosis found only in sections of pRbΔ/Δ mice, but not in pRbΔ/+ littermate controls (Supplemental Fig. S5). The pRbΔ/Δ mice also had a reduced response to hemolytic anemia induced by phenylhydrazine (Supplemental Fig. S6), supporting the presence of ineffective erythropoiesis. The Rb-null erythroid cells have features of ineffective erythropoiesis that closely resemble the refractory anemia seen in the context of the MDS (Nathan et al. 2003).
Rb plays a cell-autonomous role in erythropoiesis
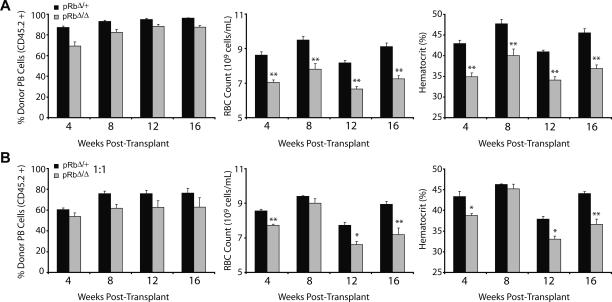
Our data demonstrate that Rb plays an intrinsic role in the erythroid lineage. However, it has been proposed that this intrinsic role may be mediated through cell-nonautonomous functions, such as paracrine signaling (Whyatt and Grosveld 2002). To examine this possibility, we performed a series of transplantation experiments using BM from pRbΔ/Δ mice and heterozygous littermate controls. Lethally irradiated congenic recipients (CD45.1) were transplanted with 2 × 106 BM cells from pRbΔ/Δ mice or heterozygous littermate controls on a C57Bl/6 background. The recipients of pRbΔ/Δ BM consistently showed anemia throughout the course of the transplants (Fig. 4A). The anemia occurred with ∼88% reconstitution from donor cells at 4 mo post-transplantation (Fig. 4A). This observation indicates that wild-type host hematopoietic reconstitution is insufficient to rescue the erythropoietic defect in the absence of Rb.
Figure 4.
Transplantation experiments demonstrate the cell-type-intrinsic and cell-autonomous role of Rb in erythropoiesis. (A,B) Noncompetitive transplants (A) and 1:1 competitive transplants (B) showing the percentage chimerism from the experimental donor marrow, RBC counts, and hematocrit in the PB of recipients over time (n = 5 per group). (*) P < 0.05; (**) P < 0.01.
To investigate these findings further, transplantation assays were performed in the presence of congenic competitor BM (CD45.1/45.2+). We used 106 competitor BM cells mixed with an equal number of pRbΔ/Δ or littermate control BM cells. Wild-type erythroid cells would rescue a putative cell-intrinsic nonautonomous defect if present, in accord with prior hypotheses (Whyatt and Grosveld 2002). The competitive transplants with pRbΔ/Δ cells achieved 62% chimerism of the experimental donor once long-term hematopoiesis was established at 4 mo post-transplantation (Fig. 4B). Interestingly, the anemia was found to be very similar to what is seen at the same time point in the noncompetitive transplantations. This result demonstrates that the defect in the absence of Rb is cell autonomous in nature. Furthermore, the observation that the degree of anemia in the competitive transplantation setting was similar to the noncompetitive setting suggests that the Rb-null erythroid progenitors are able to outcompete similar wild-type progenitors in the context of the microenvironment in which they develop. Alternatively, though less likely, a negative paracrine regulator may be produced by the mutant erythroid cells that blocks maturation of wild-type erythroid progenitors.
Erythroid loss of Rb causes defects in cell cycle exit and mitochondrial biogenesis
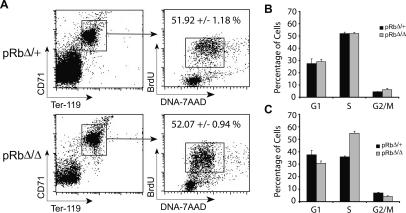
We next investigated whether the CD71high/Ter-119high progenitors that accumulate in the marrow and spleen of the mice in the absence of Rb showed increased rates of proliferation. Adult mice were injected with bromodeoxyuridine (BrdU) for 1 h to assess the cell cycle kinetics of these cells. Interestingly, we found that this population did not demonstrate increased rates of proliferation in the BM (Fig. 5A,B). We obtained similar results when the same population of BM cells was stained with the cell cycle dye Hoescht 33342 (data not shown). We did observe an increased population of proliferating cells in the spleens of the pRbΔ/Δ mice labeled with BrdU (Fig. 5C), but this is likely due to the onset of extramedullary erythropoiesis. Indeed, the rates of proliferation in these splenic erythroblasts are comparable with what is seen in the erythropoietically active BM. Previously, it has been observed that Rb-null erythroid cells appear to show increased proliferation in vitro as a result of defective cell cycle exit (Clark et al. 2004). Our observations of the in vivo behavior of these cells are distinct and are addressed in more detail below.
Figure 5.
Rb-null erythroid progenitors do not show increased rates of proliferation. (A) Representative FACS profiles and mean numbers of CD71high/Ter-119high BM erythroblasts in S phase, labeled in vivo with BrdU and stained with 7-AAD. (B,C) Cell cycle distribution summary of CD71high/Ter-119high fraction of cells from the BM (B) and the spleen (C) (n = 3).
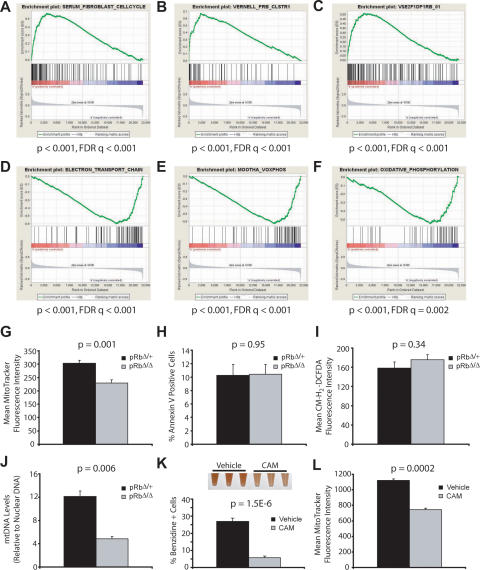
We performed gene expression profiling to gain a global perspective on transcriptional alterations in the absence of Rb. We focused our analysis on the CD71high/Ter-119high fraction of early erythroblasts that accumulates in the mutant animals. Cells were isolated using flow cytometry and RNA was obtained for expression profiling. Since we suspected that there may be subtle, yet important alterations in a variety of pathways that contribute to ineffective erythropoiesis, we used Gene Set Enrichment Analysis (GSEA) to examine >1500 a priori defined gene sets for those that might be deregulated (Mootha et al. 2003; Subramanian et al. 2005). GSEA assesses whether a set of genes involved in a particular biological pathway, or that show common regulation, are significantly enriched in one of two different phenotypes. In our analysis, we compared whether these gene sets were either up- or down-regulated in Rb-null early erythroblasts compared with stage-matched control populations. We noted that a variety of gene sets were up-regulated in the Rb-null cells, which contained numerous S-phase genes that are normally repressed during cell cycle exit (Fig. 6A–C). Indeed, one of the most significantly up-regulated gene sets (Fig. 6A) was experimentally derived by analyzing the subset of genes that increases upon serum stimulation of fibroblasts to enter S phase, which also vary in synchronously dividing cells (Chang et al. 2004). Additionally, many of these up-regulated genes are known or predicted targets of E2F transcription factors (Fig. 6B,C; Vernell et al. 2003), suggesting that the loss of Rb removes the normal repression of these genes in erythroid precursors, and therefore leads to an inability to properly undergo cell cycle exit.
Figure 6.
Rb-null erythroid progenitors display defective cell cycle exit and concomitantly fail to up-regulate the mitochondrial biogenesic program. (A–C) GSEA profiles from microarray data demonstrating up-regulation of S-phase genes (Chang et al. 2004) in early BM erythroblasts from pRbΔ/Δ mice (A), as well as up-regulation of experimental (B) (Vernell et al. 2003) and predicted E2F(C) targets. (D–F) Down-regulation of genes involved in the electron transport chain (D) and OXPHOS pathways (E,F). (G–I) Mean MitoTracker fluorescence intensity (G), Annexin V positivity (H), and mean CM-H2-DCFDA fluorescence intensity (I) in the CD71high/Ter-119high/7-AAD- fraction of BM cells (n ≥ 5 per group). (J) Quantification of mtDNA levels in sorted CD71high/Ter-119high/7-AAD- BM erythroblasts. (K) Percentage of benzidine-positive cells after 48 h of β-estradiol induction of G1E-ER cells, in the presence of 20 μg/mL chloramphenicol (CAM) or vehicle control. Picture of single-cell suspensions at 60 h of induction is shown at the top of the panel. (L) Mean MitoTracker fluorescence in undifferentiated G1E-ER cells in the presence of 20 μg/mL CAM or vehicle control (n = 3).
We then examined other aberrantly regulated gene sets with the intent of better understanding the mechanisms that couple cell cycle exit to differentiation during erythropoiesis. Suprisingly, the most significantly down-regulated gene sets all encompassed components of the mitochondrial electron transport and oxidative phosphorylation (OXPHOS) pathways (Fig. 6D–F; Supplemental Fig. S7). The regulation of these mitochondrial gene sets has been observed previously in other unrelated studies, where modulation of these gene sets and the mitochondrial biogenic program are consistently correlated with activity of the PPARγ coactivator (PGC) family of transcriptional coactivators (Mootha et al. 2003; Cui et al. 2006; Lagouge et al. 2006). We validated the results from our microarray studies using quantitative RT–PCR (qRT–PCR) of independent RNA samples from similar sorted populations and found that the expression of representative OXPHOS genes tested was reduced in Rb-deficient erythroid cells (Supplemental Fig. S8).
Erythroid maturation requires coupling of cell cycle exit and mitochondrial biogenesis
Since a global down-regulation of the mitochondrial biogenic program was seen in the early erythroblasts, we asked whether mitochondrial respiratory function was down-regulated in these cells. MitoTracker dye was used to measure mitochondrial function at the cellular level (Nisoli et al. 2003). We found a significant reduction of 25% in the cellular respiratory function of viable CD71high/Ter-119high BM cells from pRbΔ/Δ mice compared with controls (Fig. 6G). This result indicates that respiratory function was compromised in the early erythroblasts of these mice. Furthermore, we failed to detect an increase in Annexin V staining over controls, suggesting that the phenotype of the mice is not due to a primary apoptotic mechanism (Fig. 6H). High levels of Annexin V staining in the same cell population have been seen in the absence of the survival transcription factor Stat5 (Socolovsky et al. 2001). It should be noted that while there might be an increase in the basal level of apoptosis, it would be difficult to detect in the presence of high reticuloendothelial function in the marrow. Nonetheless, our findings suggest that the absence of Rb is not directly leading to ineffective erythropoiesis via an apoptotic pathway. Using erythroid progenitors from the FLs of germline Rb-null mice, others have suggested that the defects in these cells may be attributable to the presence of high levels of reactive oxygen species (ROS) (Spike and Macleod 2005). Using the dye 5,6-carboxymethyl-2′,7′-dichlorofluorescein diacetate (CM-H2-DCFDA), we failed to detect increased amounts of ROS in the viable fraction of CD71high/Ter-119high BM cells (Fig. 6I). In addition, we isolated DNA from sorted CD71high/Ter-119high BM cells and quantified mtDNA, which serves as another indicator of mitochondrial biogenesis (Lagouge et al. 2006). We found that the mtDNA levels were reduced by almost 60% in the pRbΔ/Δ mice (Fig. 6J). Taken together, this suggests that the ineffective erythropoiesis observed in the absence of Rb is due in part to a primary defect in mitochondrial biogenesis and not to a secondary defect in a pathway that may affect mitochondrial function.
A variety of observations in humans and experimental model organisms have suggested that primary defects in mitochondrial biogenesis result in ineffective erythropoiesis of a similar nature to what we observe in pRbΔ/Δ mice (Firkin 1972; Rotig et al. 1990; Inoue et al. 2000, 2007; Trifunovic et al. 2004; Craven et al. 2005). For example, a specific deletion in mtDNA that causes a reduction in mitochondrial biogenesis, thereby reducing ATP levels in the marrow, results in ineffective erythropoiesis in transplant recipients (Inoue et al. 2000, 2007), which closely resembles the partial differentiation block that we describe here. To ascertain whether mitochondrial biogenesis is indeed necessary for terminal erythroid differentiation, we used chloramphenicol to specifically inhibit mitochondrial biogenesis using a cellular model of late-stage erythropoiesis, the G1E-ER cell line (Welch et al. 2004). At low doses, chloramphenicol is a potent inhibitor of translation by the mitochondrial ribosome and therefore blocks mitochondrial biogenesis. We found that a low dose of chloramphenicol blocks GATA-1-mediated erythroid differentiation of this cell line (Fig. 6K) and chloramphenicol reduced the respiratory function of these cells by 34% in an undifferentiated state (Fig. 6L) with minimal effects on the growth rate of the cells.
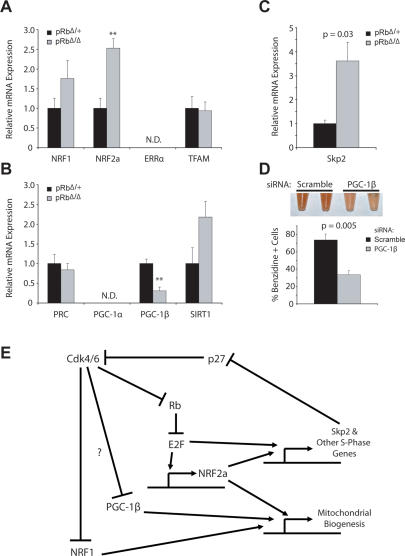
In order to gain a better understanding of the molecular basis for this down-regulation of mitochondrial biogenesis, expression profiling was performed for key regulators of this pathway. We found that the mitochondrial transcription factors, NRF1 and NRF2, were up-regulated (Fig. 7A). Prior work has demonstrated that both genes play a role in cell cycle progression (Cam et al. 2004; Yang et al. 2007) and this up-regulation is likely to reflect this alternative role. Analysis of the PGC family of coactivators revealed that while expression of PRC was unchanged, PGC-1β expression was significantly decreased (Fig. 7B). This finding is likely to at least partially explain the phenotype that we observe, as PGC-1β has been shown to play a critical role in mediating mitochondrial biogenesis (Uldry et al. 2006). It has recently been shown that Cdk4 is capable of phosphorylating NRF1 and inhibiting its mitochondrial biogenic activity (Wang et al. 2006). Additional post-translational modifications of this type are also likely to contribute to the reduction in mitochondrial biogenesis that is seen concomitantly with defective cell cycle exit. In support of this hypothesis, we found that levels of the ubiquitin ligase Skp2 were elevated (Fig. 7C), consistent with the Cdk-inhibitor p27 consequently undergoing proteosomal degradation and thereby allowing Cdk4 to remain active in the Rb-null early erythroblasts. In order to validate the involvement of these pathways for induction of mitochondrial biogenesis during erythropoiesis, we reduced expression of PGC-1β in G1E-ER cells by introduction of a previously characterized siRNA (Uldry et al. 2006). We found that these cells did not differentiate as effectively as cells transfected with scrambled siRNA controls (Fig. 7D; Supplemental Fig. S9), consistent with our interpretation that the global down-regulation in mitochondrial biogenesis seen in pRbΔ/Δ mice may be attributable to decreased activity through the PGC transcriptional axis (Kelly and Scarpulla 2004; Handschin and Spiegelman 2006).
Figure 7.
Defect in PGC pathway may underlie defective mitochondrial biogenesis and differentiation block. (A,B) Relative expression from qRT–PCR of mitochondrial transcription factors (A) and components of the PGC transcriptional axis (B) (n = 3). (C) Relative expression (qRT–PCR) of the ubiquitin ligase Skp2 (n = 3). (D) Percentage of benzidine-positive cells after 60 h of β-estradiol induction of G1E-ER cells stably infected with retrovirus containing a PGC-1β siRNA or a scrambled control. Picture of single-cell suspensions at 60 h of induction is shown at the top of the panel. (E) A model for integration of the Rb–E2F pathway with control of mitochondrial biogenesis through the PGC transcriptional axis. This model is based on the data presented in this study, as well as previously characterized pathways (Classon and Harlow 2002; Kelly and Scarpulla 2004; Bieda et al. 2006; Wang et al. 2006; Yang et al. 2007).
Together, these findings suggest a model for how cell cycle regulation mediated through the Rb/E2F pathway may be linked to control of mitochondrial biogenesis during erythropoiesis (Fig. 7E). All of the molecular changes that are observed in the pRbΔ/Δ mice are entirely consistent with and can be explained through this model. For example, as a result of Rb deletion, the model predicts that S-phase genes, including Skp2, are up-regulated, and that leads to increased Cdk4/6 activity. Concomitantly, it is predicted that mitochondrial gene transcription will be down-regulated, resulting from inhibition of the PGC transcriptional axis. As the components involved in this model are found in a variety of other cell types, our findings may apply more generally and also prove relevant to prior observations in other cell types, such as fibroblasts and mammary epithelium (Sakamaki et al. 2006; Wang et al. 2006). Future studies to extend this model will help to elucidate further molecular links by which cell cycle regulators can have global effects on cellular differentiation.
Discussion
We sought to understand the role that Rb plays in erythropoiesis using lineage-specific deletion in the erythroid and other hematopoietic compartments. Prior studies addressing the role of Rb in erythropoiesis have led to conflicting observations and interpretations. The in vivo studies we describe using conditional inactivation of Rb in the erythroid lineage avoid confounding experimental problems and provide novel insights into how Rb loss adversely affects this lineage. We found that Rb plays an intrinsic and cell-autonomous role in erythropoiesis. Unexpectedly, we found that this intrinsic role of Rb in promoting cell cycle exit during late-stage erythropoiesis is coupled to differentiation through mitochondrial biogenesis.
The role of Rb in erythropoiesis
Germline Rb knockout mice are embryonic lethal at around E14.5 and display marked defects in erythropoiesis, as well as in neurogenesis (Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992). Subsequent work showed that these early defects could be largely rescued by the presence of a wild-type placenta (Wu et al. 2003; Wenzel et al. 2007). A major question posed by this work was how a defect in fetal–maternal nutrient exchange might adversely affect particular organ systems, while allowing for relatively normal development of various other cell types. Our findings begin to explain these results. We found that the absence of Rb causes ineffective erythropoiesis, which appears to be due in part to a defect in mitochondrial biogenesis. A cell type, such as the erythroblast, that is already metabolically challenged (from defective mitochondrial biogenesis) is likely to be more adversely affected in the face of compromised nutrient exchange that would result from the Rb-null placenta.
It was hypothesized that the defect in erythropoiesis of Rb-null cells might also be explained by a cell-type-intrinsic but non-cell-autonomous defect, such as through a paracrine effector of erythropoiesis (Whyatt and Grosveld 2002). The results of our competitive transplants demonstrate that is not likely to be the case, as even in the presence of high levels of hematopoietic chimerism by wild-type BM, the anemia resulting from pRbΔ/Δ cells occurs to a similar extent as is seen in the noncompetitive setting. Other work has suggested that Rb may play a role in promoting erythropoiesis in the context of stress erythropoiesis and that Rb promotes erythroid differentiation by promoting enucleation and limiting the production of ROS (Spike et al. 2004; Spike and Macleod 2005). Our findings are distinct from these results, since we show that Rb does indeed play a role in normal erythropoiesis and enucleation defects are not observed in the pRbΔ/Δ mice. Furthermore, we did not detect increased levels of ROS in Rb-null erythroid progenitors. It should be noted that the increased levels of ROS have only been seen in the erythroid cells from the FL of germline Rb-null animals (Spike and Macleod 2005) and may represent the consequence of multiple extrinsic defects.
Studies using ex vivo cell cultures have suggested that Rb may be necessary or dispensible for normal erythropoiesis. One group found that Rb-null FL cells continued to proliferate in a liquid culture system and failed to undergo proper differentiation, as compared with wild-type FL cells (Clark et al. 2004). Work using respiratory chain-deficient cells has demonstrated that metabolically compromised cells can survive in in vitro conditions due to a high level of nutrients, including pyruvate and uridine, that allow the requirement for OXPHOS to be bypassed (King and Attardi 1989). It is likely that in the nutrient-permissive conditions used in the FL cell culture experiments, Rb-null erythroid progenitors that failed to differentiate were able to survive, in contrast to what occurs in vivo (Clark et al. 2004). Other workers used a coculture system that is meant to recapitulate the formation of an erythroid island, involving a supporting macrophage nursing differentiating erythroid progenitors (Iavarone et al. 2004). Under these conditions, Rb activity appeared to be necessary in macrophages, but dispensible in erythroid progenitors, for proper formation of an in vitro erythroid island. Our results contrast with this finding, since animals lacking Rb in macrophages appear to have normal erythropoiesis, with only a slight anemia occurring with age in the animals. It is likely that Rb-null macrophages may behave differently than their wild-type counterparts in the context of the culture conditions used to create ex vivo erythroid islands (Iavarone et al. 2004), but this does not appear to be physiologically relevant. Furthermore, intrinsic defects that can cause ineffective erythropoiesis in vivo may not be detectable in this coculture system.
Cell cycle regulation, mitochondrial biogenesis, and differentiation
The role of cell cycle regulation in erythropoiesis has been carefully studied at a descriptive level. However, the molecular control of this process in vivo and the mechanisms by which this is coupled to differentiation are largely unknown. Our results point to a previously undescribed mechanism that links cell cycle exit to late-stage erythropoiesis. We found that mitochondrial biogenic function is necessary at this transition in erythroid cell maturation, just as commitment to cell cycle exit is initiated. This observation is consistent with the differentiation block seen from primary defects in mitochondrial biogenesis (Firkin 1972; Inoue et al. 2000, 2007; Trifunovic et al. 2004; Craven et al. 2005). In the context of the pRbΔ/Δ erythroid cells, down-regulation of genes involved in cell cycle progression does not occur properly, and concomitantly there is a failure to up-regulate the mitochondrial biogenic program. Late-stage erythropoiesis is critically dependent on mitochondrial function for high-level heme biosynthesis, as well as elevated ATP production for globin gene transcription and translation. In addition to general mitochondrial biogenesis, prior studies have demonstrated that changes in mitochondrial composition occur during these stages of erythropoiesis, including increased transcription of UCP2, ABC-me, and mitoferrin (Shirihai et al. 2000; Shaw et al. 2006). It is interesting to note that cell cycle exit has been decoupled from erythroid differentiation in vitro (Rylski et al. 2003), suggesting that these processes may be separable under some conditions. Mitochondrial biogenesis is an attractive candidate as a mediator of such a process.
The similarity of the ineffective erythropoiesis seen in the pRbΔ/Δ mice to that of human MDS is striking. Not only is the block in erythropoiesis similar to what is seen in MDS, but the finding that this anemia cannot be overcome even in a chimeric situation resembles the refractory anemia of MDS that arises in a clonal manner (Corey et al. 2007). The molecular basis of MDS is not well understood, although the most common known abnormality involves epigenetic silencing of CDKN2B (Boultwood and Wainscoat 2007), a key negative regulator of the cell cycle. It is possible that further studies may find that deregulation of other cell cycle components of the Rb pathway contribute to MDS. At the same time, it is known that deregulated mitochondrial function and structure is frequently seen in MDS, particularly in the erythroid lineage (Greenberg et al. 2002). Our findings show that these seemingly disparate pathways may play key roles in synergistically promoting differentiation. It is likely that the coordination of cell cycle regulation and mitochondrial biogenesis will be important not only in erythropoiesis, but will also play a role in other cases of cellular differentiation. Indeed, a variety of observations in cancer cells have shown that uncontrolled proliferation, which is nearly always driven in part by mutations in the Rb pathway (Classon and Harlow 2002), leads to reduced production of ATP by aerobic respiration and increased dependence upon anaerobic glycolysis (Warburg 1930; Fantin et al. 2006).
Importantly, our findings provide insight into the pathophysiology underlying ineffective erythropoiesis and suggest novel therapeutic modalities. Of note, we implicated PGC-1β in the mitochondrial dysfunction seen in erythroid precursors lacking Rb. Of particular interest, PGC-1β is haploinsufficient in the 5q− subtype of MDS (Boultwood et al. 2002). Modulation of mitochondrial function is already being attempted in neurodegenerative diseases. Similar approaches may benefit hematologic diseases like MDS. Indeed, initial research in this area has shown some promise (Greenberg et al. 2002; Galili et al. 2007). While modulation of the cell cycle is a common therapeutic target in MDS (Corey et al. 2007), addition of agents that strike at the dysfunctional mitochondria of cells arising from surviving dysplastic clones might be beneficial. This may explain in part the demonstrated clinical efficacy of arsenic trioxide, a potent inhibitor of the OXPHOS component succinate dehydrogenase, in MDS (Schiller et al. 2006; Vey et al. 2006). Our findings on the molecular control of erythroid differentiation, through the coordination of cell cycle exit and mitochondrial biogenesis, may have important implications in attempting to develop therapies for a set of largely incurable diseases involving ineffective erythropoiesis.
Materials and methods
A detailed version of the Materials and Methods can be found in the Supplemental Material.
Experimental animals
EpoR-GFPcre mice (Heinrich et al. 2004) were kindly provided by Drs. Ursula Klingmüller and Ben Neel. pRbfl/fl mutant mice (Sage et al. 2003), Mx1-Cre transgenic mice (Kuhn et al. 1995; Walkley and Orkin 2006), and Lys-M-Cre animals (Clausen et al. 1999) have been described previously (Walkley et al. 2007). All experiments were performed with the approval of the CHB Institute Animal Ethics Committee.
Flow cytometry analysis
All antibodies and clone numbers are listed in the Supplemental Material. Flow cytometry was performed on a FACSCalibur, and sorting was performed on a FACS Aria; all data were analyzed using Cell Quest Pro software (Becton Dickinson).
Cell culture
G1E-ER cells were cultured and synchronously differentiated as described previously (Rylski et al. 2003). Details of these procedures can be found in the Supplemental Material.
Gene expression analysis
A detailed description of the methods used for gene expression analysis using qRT–PCR and microarray can be found in the Supplemental Material. The microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE9717.
Statistical analyses
Statistical analyses were performed using the paired and unpaired Student’s t-test, with a P value ≤0.05 being considered significant. Results are depicted as mean ± standard error of the mean (SEM) for n given samples.
Acknowledgments
We are grateful to B. Spiegelman and W. Yang for providing reagents and helpful suggestions, and to U. Klingmüller, B. Neel, T. Jacks, B. Lowell, and M. Huntgeburth for providing mouse strains. We thank D. Nathan, L. Purton, J. Hirschhorn, M. Weiss, L. Zon, V. Mootha, and C. Sieff for valuable discussion and critical comments; the Children’s Animal Facility staff for care of experimental animals; G. Losyev, J. Daley, and S. Lazo-Kallanian for assistance with FACS sorting; the DFCI Microarray Core; and the DFCI/HCC Rodent Histology Core. This work was supported in part by a Center of Excellence Award in Molecular Hematology from the NIH-NIDDK (S.H.O.). V.G.S. was supported by MSTP and NRSA awards from the NIH. C.R.W. is a Special Fellow of the Leukemia and Lymphoma Society and S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1627208
References
- Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J., Wainscoat J.S. Gene silencing by DNA methylation in haematological malignancies. Br. J. Haematol. 2007;138:3–11. doi: 10.1111/j.1365-2141.2007.06604.x. [DOI] [PubMed] [Google Scholar]
- Boultwood J., Fidler C., Strickson A.J., Watkins F., Gama S., Kearney L., Tosi S., Kasprzyk A., Cheng J.F., Jaju R.J., et al. Narrowing and genomic annotation of the commonly deleted region of the 5q− syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Chang H.Y., Sneddon J.B., Alizadeh A.A., Sood R., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych M.A., Kenney A.M., Sicinska E., Kalaszczynska I., Bronson R.T., Rowitch D.H., Gardner H., Sicinski P. Development of mice expressing a single D-type cyclin. Genes & Dev. 2002;16:3277–3289. doi: 10.1101/gad.1023602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.J., Doyle K.M., Humbert P.O. Cell-intrinsic requirement for pRb in erythropoiesis. Blood. 2004;104:1324–1326. doi: 10.1182/blood-2004-02-0618. [DOI] [PubMed] [Google Scholar]
- Clarke A.R., Maandag E.R., van Roon M., van der Lugt N.M., van der Valk M., Hooper M.L., Berns A., te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Clausen B.E., Burkhardt C., Reith W., Renkawitz R., Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Corey S.J., Minden M.D., Barber D.L., Kantarjian H., Wang J.C., Schimmer A.D. Myelodysplastic syndromes: The complexity of stem-cell diseases. Nat. Rev. Cancer. 2007;7:118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- Craven S.E., French D., Ye W., de Sauvage F., Rosenthal A. Loss of Hspa9b in zebrafish recapitulates the ineffective hematopoiesis of the myelodysplastic syndrome. Blood. 2005;105:3528–3534. doi: 10.1182/blood-2004-03-1089. [DOI] [PubMed] [Google Scholar]
- Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Dumitriu B., Patrick M.R., Petschek J.P., Cherukuri S., Klingmuller U., Fox P.L., Lefebvre V. Sox6 cell-autonomously stimulates erythroid cell survival, proliferation, and terminal maturation and is thereby an important enhancer of definitive erythropoiesis during mouse development. Blood. 2006;108:1198–1207. doi: 10.1182/blood-2006-02-004184. [DOI] [PubMed] [Google Scholar]
- Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Fero M.L., Rivkin M., Tasch M., Porter P., Carow C.E., Firpo E., Polyak K., Tsai L.H., Broudy V., Perlmutter R.M., et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Firkin F.C. Mitochondrial lesions in reversible erythropoietic depression due to chloramphenicol. J. Clin. Invest. 1972;51:2085–2092. doi: 10.1172/JCI107015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N., Sechman E.V., Cerny J., Mehdi M., Mumtaz M., Westervelt P., Maguire J., Raza A. Clinical response of myelodysplastic syndromes patients to treatment with coenzyme Q10. Leuk. Res. 2007;31:19–26. doi: 10.1016/j.leukres.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Greenberg P.L., Young N.S., Gattermann N. Myelodysplastic syndromes. Hematology Am. Soc. Hematol. Educ. Program. 2002;2002:136–161. doi: 10.1182/asheducation-2002.1.136. [DOI] [PubMed] [Google Scholar]
- Gregory C.J., Eaves A.C. Human marrow cells capable of erythropoietic differentiation in vitro: Definition of three erythroid colony responses. Blood. 1977;49:855–864. [PubMed] [Google Scholar]
- Gregory C.J., Eaves A.C. Three stages of erythropoietic progenitor cell differentiation distinguished by a number of physical and biologic properties. Blood. 1978;51:527–537. [PubMed] [Google Scholar]
- Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Heinrich A.C., Pelanda R., Klingmuller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104:659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- Hu N., Gulley M.L., Kung J.T., Lee E.Y. Retinoblastoma gene deficiency has mitogenic but not tumorigenic effects on erythropoiesis. Cancer Res. 1997;57:4123–4129. [PubMed] [Google Scholar]
- Iavarone A., King E.R., Dai X.M., Leone G., Stanley E.R., Lasorella A. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature. 2004;432:1040–1045. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
- Inoue K., Nakada K., Ogura A., Isobe K., Goto Y., Nonaka I., Hayashi J.I. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat. Genet. 2000;26:176–181. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- Inoue S., Yokota M., Nakada K., Miyoshi H., Hayashi J. Pathogenic mitochondrial DNA-induced respiration defects in hematopoietic cells result in anemia by suppressing erythroid differentiation. FEBS Lett. 2007;581:1910–1916. doi: 10.1016/j.febslet.2007.03.092. [DOI] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Kelly D.P., Scarpulla R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes & Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- King M.P., Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lee E.Y., Chang C.Y., Hu N., Wang Y.C., Lai C.C., Herrup K., Lee W.H., Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Letvin N.L., Linch D.C., Beardsley G.P., McIntyre K.W., Miller B.A., Nathan D.G. Influence of cell cycle phase-specific agents on simian fetal hemoglobin synthesis. J. Clin. Invest. 1985;75:1999–2005. doi: 10.1172/JCI111918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maandag E.C., van der Valk M., Vlaar M., Feltkamp C., O’Brien J., van Roon M., van der Lugt N., Berns A., te Riele H. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 1994;13:4260–4268. doi: 10.1002/j.1460-2075.1994.tb06746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M., Doumont G., Clercq S.D., Francoz S., Froment P., Bellefroid E., Klingmuller U., Lozano G., Marine J.C. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nathan D.G., Orkin S.H., Look A.T., Ginsburg D. Nathan and Oski’s hematology of infancy and childhood. Saunders; Philadelphia, PA: 2003. [Google Scholar]
- Nisoli E., Clementi E., Paolucci C., Cozzi V., Tonello C., Sciorati C., Bracale R., Valerio A., Francolini M., Moncada S., et al. Mitochondrial biogenesis in mammals: The role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Rotig A., Cormier V., Blanche S., Bonnefont J.P., Ledeist F., Romero N., Schmitz J., Rustin P., Fischer A., Saudubray J.M., et al. Pearson’s marrow-pancreas syndrome. A multisystem mitochondrial disorder in infancy. J. Clin. Invest. 1990;86:1601–1608. doi: 10.1172/JCI114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylski M., Welch J.J., Chen Y.Y., Letting D.L., Diehl J.A., Chodosh L.A., Blobel G.A., Weiss M.J. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sakamaki T., Casimiro M.C., Ju X., Quong A.A., Katiyar S., Liu M., Jiao X., Li A., Zhang X., Lu Y., et al. Cyclin D1 determines mitochondrial function in vivo. Mol. Cell. Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller G.J., Slack J., Hainsworth J.D., Mason J., Saleh M., Rizzieri D., Douer D., List A.F. Phase II multicenter study of arsenic trioxide in patients with myelodysplastic syndromes. J. Clin. Oncol. 2006;24:2456–2464. doi: 10.1200/JCO.2005.03.7903. [DOI] [PubMed] [Google Scholar]
- Shaw G.C., Cope J.J., Li L., Corson K., Hersey C., Ackermann G.E., Gwynn B., Lambert A.J., Wingert R.A., Traver D., et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- Shirihai O.S., Gregory T., Yu C., Orkin S.H., Weiss M.J. ABC-me: A novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000;19:2492–2502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Spike B.T., Macleod K.F. The Rb tumor suppressor in stress responses and hematopoietic homeostasis. Cell cycle. 2005;4:42–45. doi: 10.4161/cc.4.1.1337. [DOI] [PubMed] [Google Scholar]
- Spike B.T., Dirlam A., Dibling B.C., Marvin J., Williams B.O., Jacks T., Macleod K.F. The Rb tumor suppressor is required for stress erythropoiesis. EMBO J. 2004;23:4319–4329. doi: 10.1038/sj.emboj.7600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlof S., Oldfors A., Wibom R., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Vernell R., Helin K., Muller H. Identification of target genes of the p16INK4A-pRB–E2F pathway. J. Biol. Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- Vey N., Bosly A., Guerci A., Feremans W., Dombret H., Dreyfus F., Bowen D., Burnett A., Dennis M., Ribrag V., et al. Arsenic trioxide in patients with myelodysplastic syndromes: A phase II multicenter study. J. Clin. Oncol. 2006;24:2465–2471. doi: 10.1200/JCO.2005.03.9503. [DOI] [PubMed] [Google Scholar]
- Walkley C.R., Orkin S.H. Rb is dispensable for self-renewal and multilineage differentiation of adult hematopoietic stem cells. Proc. Natl. Acad. Sci. 2006;103:9057–9062. doi: 10.1073/pnas.0603389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley C.R., Shea J.M., Sims N.A., Purton L.E., Orkin S.H. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li Z., Lu Y., Du R., Katiyar S., Yang J., Fu M., Leader J.E., Quong A., Novikoff P.M., et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc. Natl. Acad. Sci. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. The metabolism of tumors. Arnold Constable; London, UK: 1930. [Google Scholar]
- Welch J.J., Watts J.A., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wenzel P.L., Wu L., de Bruin A., Chong J.L., Chen W.Y., Dureska G., Sites E., Pan T., Sharma A., Huang K., et al. Rb is critical in a mammalian tissue stem cell population. Genes & Dev. 2007;21:85–97. doi: 10.1101/gad.1485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt D., Grosveld F. Cell-nonautonomous function of the retinoblastoma tumour suppressor protein: New interpretations of old phenotypes. EMBO Rep. 2002;3:130–135. doi: 10.1093/embo-reports/kvf033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt D., Lindeboom F., Karis A., Ferreira R., Milot E., Hendriks R., de Bruijn M., Langeveld A., Gribnau J., Grosveld F., et al. An intrinsic but cell-nonautonomous defect in GATA-1-overexpressing mouse erythroid cells. Nature. 2000;406:519–524. doi: 10.1038/35020086. [DOI] [PubMed] [Google Scholar]
- Williams B.O., Schmitt E.M., Remington L., Bronson R.T., Albert D.M., Weinberg R.A., Jacks T. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 1994;13:4251–4259. doi: 10.1002/j.1460-2075.1994.tb06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., de Bruin A., Saavedra H.I., Starovic M., Trimboli A., Yang Y., Opavska J., Wilson P., Thompson J.C., Ostrowski M.C., et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Yang Z.F., Mott S., Rosmarin A.G. The Ets transcription factor GABP is required for cell-cycle progression. Nat. Cell Biol. 2007;9:339–346. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- Ye M., Iwasaki H., Laiosa C.V., Stadtfeld M., Xie H., Heck S., Clausen B., Akashi K., Graf T. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- Zhang J., Socolovsky M., Gross A.W., Lodish H.F. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]