Abstract
Chromosomes and genes are nonrandomly arranged within the mammalian cell nucleus. However, the functional significance of nuclear positioning in gene expression is unclear. Here we directly probed the relationship between nuclear positioning and gene activity by comparing the location of the active and inactive copies of a monoallelically expressed gene in single cell nuclei. We demonstrate that the astrocyte-specific marker GFAP (glial fibrillary acidic protein) is monoallelically expressed in cortical astrocytes. Selection of the active allele occurs in a stochastic manner and is generally maintained through cell division. Taking advantage of the monoallelic expression of GFAP, we show that the functionally distinct alleles occupy differential radial positions within the cell nucleus and differentially associate with intranuclear compartments. In addition, coordinately regulated astrocyte-specific genes on distinct chromosomes spatially associate in their inactive state and dissociate upon activation. These results provide direct evidence for function-related differential positioning of individual gene alleles within the interphase nucleus.
[Keywords: Monoallelic expression, spatial gene positioning, nuclear architecture, astrocyte]
The genomes of higher eukaryotes are nonrandomly organized within the interphase cell nucleus (Cremer and Cremer 2001; Meaburn and Misteli 2007). Chromosomes and genes occupy preferential radial positions with respect to the nuclear center–periphery axis and relative to other gene loci (Bickmore et al. 2004; Parada et al. 2004; Lanctot et al. 2007). Nuclear positioning has been implicated in gene regulation since some genes change their intranuclear position upon modulation of their transcription status. For example, the IgH and IgK loci move from a peripheral location in hematopoietic precursor cells, where they are inactive, to an internal position upon activation during differentiation into pro-B cells (Skok et al. 2001). Similarly, the β-globin locus takes on a more internal position upon its activation during erythrocyte differentiation, and the CD4 and CD8 loci change their radial position during T-cell differentiation (Kosak et al. 2002; Kim et al. 2004; Ragoczy et al. 2006). However, other observations question the functional relevance of the positional changes of gene loci and of radial gene positioning in general (Misteli 2007). For one, the two alleles of a gene often occupy greatly distinct nuclear positions within the same nucleus despite their apparent functional equivalence, suggesting that the radial position of the allele is unrelated to its activity status. In addition, even for genes that exhibit a nonrandom preferential average position, the location of a gene locus varies greatly in single cells within a population, and all preferential positioning patterns are probabilistic in nature (Parada et al. 2003). Furthermore, it has been difficult to ascertain that the observed repositioning of genes during differentiation was specific and not due to global genome reorganizations that occur during this process. A key experiment that has not been feasible to date is the positional mapping of the two individual alleles of a gene in the same cell nucleus.
Monoallelically expressed genes are an ideal model system to probe the relationship between gene activity and nuclear position. Apart from the well-characterized processes of imprinting and X inactivation, monoallelic expression via allele exclusion has been observed recently for an increasing number of genes, particularly during development and differentiation (Guo et al. 2005b; Yang and Kuroda 2007). Although the detailedmolecular mechanisms for allelic exclusion are unclear, it appears to occur via stochastic selection of the active and inactive allele, and the probabilistic nature of allelic selection has been suggested to be critical for physiological regulation, particularly in the immune system (Guo et al. 2005b). The monoallelic expression of loci has been linked to nuclear organization (Yang and Kuroda 2007). The two X chromosomes physically associated with each other roughly around the time of X inactivation. Furthermore, interchromosomal interactions seem to be essential for imprinting at the Igf2/H-19 locus (Ling et al. 2006; Zhao et al. 2006) and possibly for selective activation of olfactory receptor genes (Lomvardas et al. 2006; Fuss et al. 2007).
Here we report that the astrocyte marker GFAP (glial fibrillary acidic protein) is monoallelically expressed in cortical astrocytes. We took advantage of this expression pattern to directly compare the nuclear position of the active and inactive alleles of a gene locus in single cell nuclei. We demonstrate that the functionally distinct alleles of GFAP are positioned differentially within the cell nucleus, with respect to both nuclear compartments and the nuclear periphery, and that coregulated astrocyte-specific genes are in spatial proximity in their inactive state at a higher than expected frequency.
Results
Monoallelic expression of GFAP during astrocyte differentiation
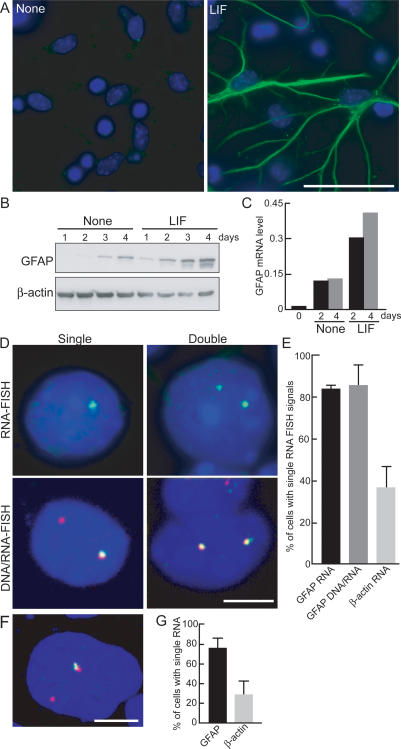
As part of our efforts to characterize changes in nuclear architecture during differentiation processes, we used a previously characterized in vitro astrocyte differentiation system in which neuronal precursor cells (NPCs) prepared from embryonic day 14.5 (E14.5) mouse brain are cultured for 4 d and then stimulated to differentiate into astrocytes in the presence of leukemia inhibitory factor (LIF) (Fig. 1A; Takizawa et al. 2001a; see Materials and Methods). As reported previously, under these conditions, ∼20% of NPCs differentiate into astrocytes as judged by expression of the astrocyte marker GFAP by immunofluorescence staining and Western blotting (Fig. 1A,B; Supplemental Table 1; Takizawa et al. 2001a). As expected, GFAP mRNA increased during the differentiation time course (Fig. 1C). Surprisingly when GFAP mRNA expression was analyzed by fluorescence in situ hybridization (FISH), we found predominant monoallelic expression with >80% of GFAP-expressing cells containing a single RNA FISH signal (Fig. 1D,E; Supplemental Table 2). To exclude the possibility that the absence of signal was due to limited sensitivity of the RNA FISH procedure, we performed simultaneous DNA/RNA FISH. Again, >80% of GFAP-positive cells showed expression from one but not the other allele (Fig. 1D,E). As a control, the biallelically expressed β-actin gene was detected as two FISH signals in the majority of cells (Fig. 1E). To ascertain whether GFAP is also monoallelically expressed in fully differentiated astrocytes and to exclude the possibility that monoallelic expression was an artifact of the in vitro differentiation system or inefficient FISH detection, we prepared primary astrocytes from the cerebral cortex of neonatal mice (Fig. 1F). Simultaneous DNA/RNA FISH revealed monoallelic expression of GFAP in 75% of primary astrocytes (Fig. 1G; Supplemental Table 3). This was not due to differential FISH sensitivity in these cells since two RNA FISH signals were detected in ∼80% of cells for the biallelically expressed β-actin gene (Fig. 1G).
Figure 1.
Monoallelic expression of GFAP. NPC prepared from E14 mice were treated with LIF for 4 d and stained with anti-GFAP antibody (green, GFAP; blue, DAPI) (A) and GFAP protein detected by Western blotting (B). (C) RT–PCR analysis of GFAP mRNA. mRNA was prepared from NPC incubated with or without LIF for 2 or 4 d and subjected to real-time quantitative PCR. GFAP mRNA levels were normalized to β-actin mRNA expression. (D) Monoallelic expression of GFAP. RNA FISH (top panels) or simultaneous DNA/RNA FISH (bottom panels) was performed in NPC treated with LIF. (Green) GFAP RNA; (red) GFAP DNA. (E) Quantitation of monoallelic expression of GFAP. The percentage of cells with a single RNA FISH signal is indicated. Values are averages from two to three experiments ± SEM. N = 41–127. (F) Expression of GFAP in astrocytes. Primary astrocytes were taken from neonatal mice cerebral cortex, and after two passages, simultaneous DNA/RNA FISH for GFAP was performed. (Red) GFAP DNA; (green) GFAP RNA. (G) Quantitation of active GFAP alleles in primary astrocytes. The percentage of cells with a single RNA FISH signal is indicated. Values are averages from three experiments ± SEM. N = 36–248. Bars: A, 50 μm; D,F, 5 μm.
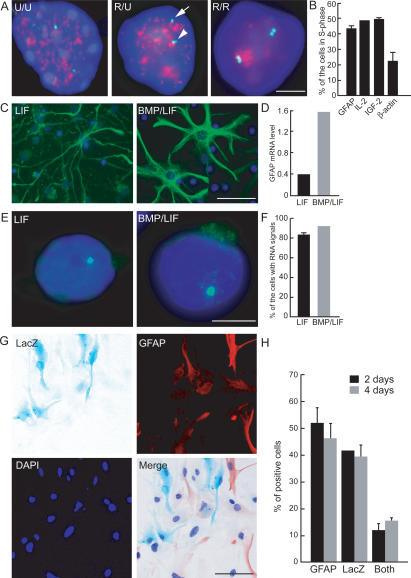
The monoallelic expression of GFAP was confirmed by several functional properties. GFAP was asymmetrically replicated, a hallmark of monoallelically expressed loci (Fig. 2A). Upon BrdU pulse-labeling of primary astrocytes in S phase followed by DNA FISH for GFAP, 40% of cells showed asynchronous replication as indicated by a double FISH signal for one, but not the second, allele (Fig. 2B). As controls, the imprinted gene IGF-2 and IL-2 replicated asynchronously in a similar fraction of cells, whereas the biallelically expressed β-actin locus was only asynchronously replicated in ∼20% of cells (Fig. 2B). Furthermore, monoallelic expression was also indicated by the response of GFAP alleles to stimulation. Astrocyte differentiation and GFAP expression can be stimulated by bone morphogenetic proteins (BMP) in combination with LIF (Nakashima et al. 1999). As expected, BMP2 stimulation in combination with LIF (BMP2/LIF) led to a strong increase in GFAP protein expression in NPC-derived astrocyte cultures (Fig. 2C) and induced an approximately fourfold increase in steady-state GFAP mRNA in the population (Fig. 2D). The increased expression of GFAP in the population was partially due to up-regulation of the single active allele as indicated by the dramatically stronger and larger FISH signals in GFAP-expressing cells (Fig. 2E). In addition, the number of cells expressing GFAP from a single allele increased by about twofold (Supplemental Table 2). In contrast, no significant increase in the number of cells with biallelic expression of GFAP was found (Supplemental Table 2). Specifically, after BMP2/LIF stimulation, >85% of cells expressed GFAP mRNA only from a single allele compared with 83% in LIF-stimulated control cells (Fig. 2F). The absence of activation of the second allele was not an artifact of the in vitro differentiation system, since primary astrocytes behaved identically (Supplemental Table 3). The observed up-regulation of GFAP without activation of the second allele demonstrates the distinct functional properties of the two alleles and supports the monoallelic nature of GFAP expression.
Figure 2.
Asynchronous replication timing and monoallelic expression of GFAP after stimulation. (A) Representative images of simultaneous GFAP DNA FISH and BrdU labeling. Astrocytes in S phase were labeled by BrdU 45 min before fixation in methanol/acetic acid. Immunostaining for BrdU was performed after DNA FISH. Arrow and arrowhead indicate the unreplicated and replicated loci, respectively. (Red) BrdU; (green) GFAP DNA; (U) unreplicated locus; (R) replicated locus. (B) Quantitation of replication patterns. The percentage of cells showing asynchronous replication (R/U) of the indicated gene loci in total S-phase cells was determined. DNA FISH for GFAP, IL-2, IGF-2, or β-actin was performed before BrdU staining. Values represent averages from two experiments ± SEM. N = 106–156. (C–F) NPC prepared from E14.5 mice telencephalon were treated with LIF alone or BMP2 plus LIF (BMP/LIF). (C) Immunostaining for GFAP (green) after BMP/LIF stimulation. (D) Quantitative RT–PCR for GFAP mRNA after BMP/LIF stimulation. Data are normalized to β-actin mRNA. (E) GFAP RNA FISH. Monoallelic expression of GFAP mRNA (green) is maintained in BMP/LIF-stimulated cells. (F) Quantitation of cells with a single RNA FISH signal after BMP/LIF stimulation. N = 51–108. (G,H) Astrocytes from GFAP+/LacZ heterozygous mice. (G) X-gal assay and immunostaining for GFAP (red) in astrocytes from GFAP+/LacZ mice. (H) Quantitation of cells expressing LacZ gene product and/or GFAP in astrocytes from GFAP+/LacZ mice. Cells were cultured in the presence of BMP/LIF for 2 or 4 d. Values are averages from three experiments ± SEM. N = 230–937. Bars: A,E, 5 μm; C,G, 50 μm.
Stochastic selection of the active GFAP allele
The monoallelic expression of GFAP might be due to either parental imprinting, random selection of an active allele during development and differentiation, or temporal switching between the two alleles in differentiating astrocytes. To distinguish between these possibilities, we generated heterozygous mice from a GFAP-null strain in which the GFAP locus was disrupted by a lac-insertion cassette expressing LacZ (GFAP+/LacZ mice) (Gomi et al. 1995). If GFAP were parentally imprinted in the germline, astrocytes from GFAP+/LacZ mice would express either the endogenous GFAP or LacZ in the entire cell population; if, on the other hand, the active locus is selected randomly during development, a mixed population of astrocytes expressing either endogenous GFAP or LacZ would be expected. If expression switches rapidly between the two alleles in single cells, all cells would express both markers. Analysis of astrocytes from GFAP+/LacZ animals revealed that >90% of cells expressed one but not the other marker, consistent with stochastic selection of the inactive and active alleles (Fig. 2G,H). In 2-d cultures, 51.7 ± 6.2% of cells expressed GFAP only and 41.4 ± 0.4% expressed LacZ only (values are average from three independent experiments) (Fig. 2H). The slightly lower proportion of LacZ-positive cells is likely a consequence of the shorter half-life of the protein compared with GFAP. In addition, in agreement with the DNA/RNA FISH experiments on wild-type cells, we find 11.8 ± 2.5% of cells simultaneously express GFAP and LacZ. A similar ratio was observed in extended 4-d culture, demonstrating that no slow switching occurs during that period. These results suggest that GFAP is not parentally imprinted, but that the active locus is stochastically selected during development and its expression is then maintained.
To unambiguously demonstrate the monoallelic expression of GFAP, we clonally expanded astrocytes and analyzed the composition of single-cell clones derived from them (Supplemental Fig. 1). In order to generate single-cell-derived clones, we marked GFAP heterozygous astrocytes with retrovirally expressed GFP and cultured them at a low density among an excess of nonlabeled astrocytes (see Materials and Methods). This was necessary since astrocytes do not grow in sparse cultures. Among 103 individual GFP-positive clones, 86 clones (83.5%) expressed either only GFAP (61 out of 64 GFAP-expressing clones) or only LacZ (25 out of 27 LacZ-expressing clones) (Supplemental Fig. 1A,B). Consistent with the FISH and immunostaining results, 10.7% of clones (11 out of 103) were entirely composed of biallelically expressing cells, and six out of 103 (5.8%) clones were mixed (Supplemental Fig. 1B). These results demonstrate the monoallelic nature of GFAP expression, and they indicate that allele selection is generally maintained through cell division but that switching does occur, albeit at very low rates. Similar behavior has been observed for the IL-4 gene in activated T helper cells (Guo et al. 2005a).
Allele-specific nuclear positioning of GFAP
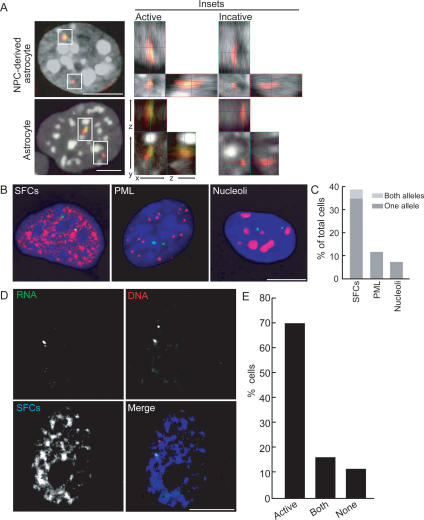
The monoallelic expression of GFAP offered a unique opportunity to directly probe the relationship of gene activity to nuclear positioning by comparing the active and inactive copies of the same gene locus in a single cell nucleus. Since transcriptionally silent loci are often associated with blocks of heterochromatin (Baxter et al. 2002), we first asked whether the active and inactive GFAP alleles localize differently relative to heterochromatin (Fig. 3A). No significant difference in the association frequency of the two alleles relative to heterochromatin domains stained with DAPI was found. Despite their transcriptional repression, inactive GFAP alleles did not associate with centromeric heterochromatin regions (0 out of 28 NPC; one of 48 astrocytes). Furthermore, no difference in the position of the active and inactive alleles with respect to their position in the chromosome territory was observed (data not shown).
Figure 3.
Relative positioning of GFAP alleles to nuclear bodies. (A) Positioning of GFAP loci relative to heterochromatin. Simultaneous DNA (red) and RNA (green) FISH for GFAP was performed, and confocal images were obtained. Heterochromatin was visualized by DAPI (white). Insets show 3D views of the active or inactive allele for each cell. The association of GFAP alleles with heterochromatin was probed by visual inspection of complete 3D reconstructions. (B,C) Relative positioning of GFAP loci to nuclear bodies. (B) Representative images of immunostaining (red) for SFCs, PML bodies, or nucleoli following DNA FISH for GFAP (green) in astrocytes. (C) Quantitation of percentage of cells with colocalization of GFAP loci to each nuclear body. Values are averages from three experiments for SFCs. N = 73–160. (D,E) Simultaneous DNA (red)/RNA (green) FISH for GFAP in astrocytes followed by immunostaining for SFCs. (E) Quantitation of the number of cells showing colocalization of GFAP active alleles, both active and inactive alleles, and no alleles with SFCs. N = 101. Bars, 5 μm.
We sought to test whether active and inactive GFAP alleles differentially associate with nuclear compartments (Fig. 3B). In DNA FISH experiments, no significant association of GFAP loci with nucleoli (7%, seven out of 106 cells) or with PML bodies (11%, 12 out of 109) was found. In contrast, in 35 ± 11% of cells, one GFAP locus associated with pre-mRNA splicing factor compartments (SFCs) visualized by SF2/ASF, and in an additional 4 ± 2% cells, both GFAP loci associated with SFCs (Fig. 3C). The SFC-associated alleles represented the active GFAP loci, since by RNA FISH 96 ± 1.5% (N = 104, 137) of signals were associated with SFCs. To directly test potential differential association of the GFAP alleles with SFCs, we performed simultaneous RNA/DNA FISH combined with immunostaining. In 70% of cells (70 out of 101), only the active, but not the inactive, allele was associated with SFCs. In an additional 15% of cells, both alleles were found to colocalize with SFCs (Fig. 3E). These results demonstrate activity-dependent allele-specific positioning of GFAP loci to SFCs. This observation is in line with the demonstrated propensity of active, intron-containing genes to associate with SFC, whereas inactive loci do not (Huang and Spector 1996; Meaburn and Misteli 2007).
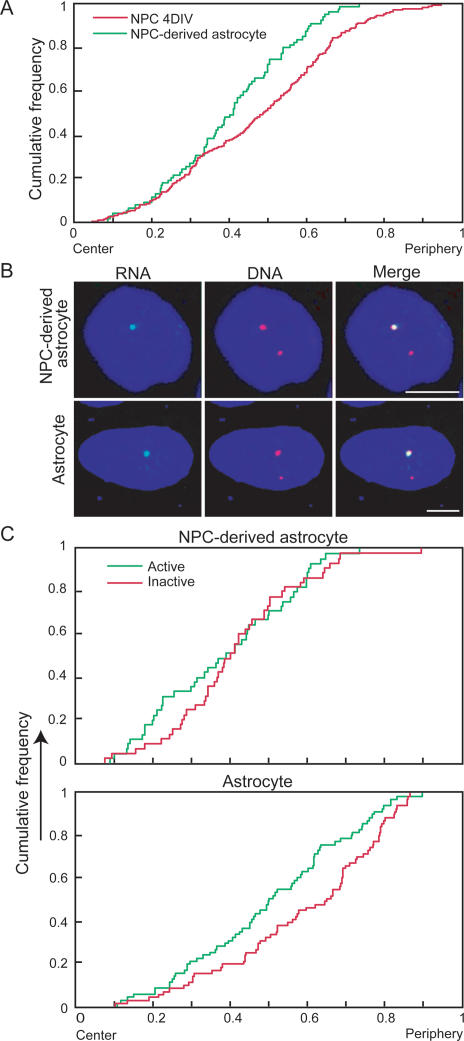
We next examined the radial positioning of the GFAP locus relative to the center of the nucleus in NPC-derived astrocytes and primary astrocytes (Fig. 4). The combined population of active and inactive alleles showed a more internal average localization in NPC-derived astrocytes compared with NPCs (Fig. 4A), suggesting a shift in positioning upon differentiation. The difference was significant at the P < 0.005 level in a Kolmogorov-Smirnov (KS) test. We next asked whether the spatial position of the GFAP locus is related to its transcriptional activity. Using simultaneous DNA/RNA FISH, we found that the localization of active alleles differed significantly from that of the inactive ones in NPC-derived astrocytes (Fig. 4B,C). Direct comparison of radial distances confirmed a significant preference for the transcriptionally active allele to localize toward the nuclear interior (Fig. 4C). In NPC-derived astrocytes, >31% of active alleles localized within the innermost quarter of the nucleus whereas <14% of inactive alleles were found in this region (P < 0.042, χ2 test). Similarly, in primary astrocytes active alleles were consistently more internal than inactive ones (Fig. 4C). These differences were significant at the P < 0.0059 level. Furthermore, in 67% of NPC-derived astrocytes (30 out of 45) and 60% of primary astrocytes (43 out of 72), the active allele was more internally localized than the inactive allele in the same cell nucleus. These values are significantly higher than the 50% expected based on random positioning of the locus (P < 0.0098, P < 0.024, respectively).
Figure 4.
Spatial nuclear positioning of GFAP alleles. (A) Cumulative distribution graphs of the radial position of GFAP alleles in NPC cultured for 4 d in vitro (4DIV) and NPC-derived astrocytes. N = 45, 138. (B) Representative images of NPC-derived astrocytes and primary astrocytes with only a single active GFAP allele. (Green) RNA; (red) DNA FISH. The active allele is preferentially internally located compared with the inactive allele. (C) Cumulative distribution graphs of the radial position of active and inactive alleles in NPC-derived astrocytes and primary astrocytes. N = 45, 72. Bars, 5 μm.
Activity-dependent radial positioning of the GFAP allele was further confirmed by simultaneous RNA FISH for endogenous GFAP and the LacZ insertion in astrocytes from GFAP+/LacZ mice. In the population of biallelically expressing cells (∼25%) (Fig. 1G; Supplemental Table 3), the radial position of both alleles was similar to that of the active allele in wild-type cells (Supplemental Fig. 2), indicating that the internal position of an allele is related to its activity. Interestingly, differential radial positioning of monoallelically expressed genes does not seem to be limited to GFAP, since we find that the active allele of the well-characterized, stochastically monoallelically expressed gene IL-4 (Guo et al. 2005b) is similarly more internally positioned than the inactive allele (Supplemental Fig. 3A,B). Taken together, these observations strongly point to allele-specific radial positioning in monoallelically expressed loci.
Spatial association of coregulated astrocyte marker genes
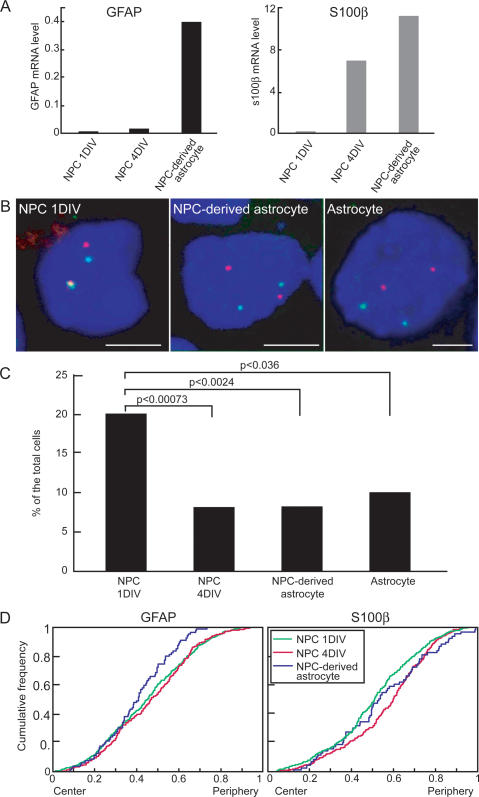
One possible reason for differential positioning of active and inactive alleles is the clustering of multiple genes with similar functional status to form nuclear microenvironments (Chakalova et al. 2005; Fraser and Bickmore 2007). To begin to address this possibility, we determined the localization of the GFAP locus that is located on chromosome 11 relative to the calcium-binding protein, S100β, an astrocyte marker that, like GFAP, is regulated by Jak–STAT pathways and DNA demethylation but is located on chromosome 10 (Namihira et al. 2004; He et al. 2005). As expected, GFAP and S100β were not expressed in freshly isolated NPCs as judged by quantitative RT–PCR (Fig. 5A). After 4 d in culture, S100β was active whereas no GFAP mRNA could be detected yet. In contrast, in NPC-derived mature astrocytes, both genes showed robust expression (Fig. 5A). Upon double-labeling by DNA FISH for GFAP and S100β in NPC, the two genes were closely positioned in 20% of cells (close positioning defined as a distance of <500 nm) (Fig. 5B,C). This value is comparable with previously characterized pairs of coregulated genes and is significantly above the value expected for two randomly positioned genes, suggesting a propensity of these loci to colocalize (Osborne et al. 2004; Spilianakis et al. 2005). As observed for other genes, in most cells, only one allele of each gene colocalized. The spatial proximity of these two genes was lost upon differentiation (Fig. 5B,C). In 4-d NPC cultures, where S100β but not GFAP is active, no colocalization was observed (P < 0.00073, χ2 test) (Fig. 5C). Similarly, in NPC-derived astrocytes where both genes are active, significant proximity was found in only 8% of cells (P < 0.0024, χ2 test) (Fig. 5B,C). As a control, two unrelated genes, IL-2 and GFAP, were in proximity in 6.2% of NPC-derived astrocytes (P < 0.0032).
Figure 5.
Relative positioning of GFAP and S100β in NPC during differentiation. (A) Quantitative RT–PCR of GFAP and S100β in NPC cultured for 1 or 4 d in vitro (DIV) and differentiated into astrocytes by LIF stimulation for another 4 d (NPC-derived astrocyte). Data are normalized to β-actin mRNA level. (B) Relative positioning of GFAP and S100β. Confocal images of DNA FISH for GFAP (red) and S100β (green). Relative distances between GFAP and S100β were determined for each focal plane. Bar, 5 μm. (C) Quantitation of percentage of cells showing close proximity of GFAP and S100β signals. N = 113–327. P values were obtained by a χ2 test. (D) Cumulative distribution graphs of the radial position of GFAP and S100β alleles in NPC cultured for 1 or 4 d and NPC-derived astrocytes. N = 45–245.
To determine whether the change in colocalization of GFAP and S100β loci was due to a change in the position of both genes or only one, we analyzed the radial position of GFAP and S100β at various time points during differentiation. The radial position of both genes changed at the time of their respective activation (Fig. 4D). While GFAP loci did not show any change in radial position during the first 4 d in culture, the average position of the gene shifted toward the interior during the transition from 4-d cultures to mature astrocytes (Fig. 4D). Similarly, while almost 50% of S100β alleles were within the inner half of the nuclear diameter in freshly isolated NPC, <35% were after 4 d in culture (P < 0.000035, KS-test) (Fig. 4D). These positional changes coincide precisely with the time point of activation of each gene, further supporting a link between the radial position of GFAP and S100β and their functional status. These findings suggest that GFAP and S100β pair in their inactive state and their association is lost upon dissociation of S100β during its activation during differentiation.
Discussion
In this study, we identified GFAP as a monoallelically expressed gene during astrocyte differentiation and demonstrated the allele-specific radial position of the functionally distinct alleles. The monoallelic expression of GFAP is likely a regulatory mechanism to control the physiological level of GFAP in vivo. Maintenance of low GFAP levels appears of key physiological relevance since overexpression of GFAP caused by knockout of its regulator N-CoR results in an increase in binucleated cells, suggesting that excess GFAP inhibits proper cytokinesis (Hermanson et al. 2002). Furthermore, overexpression of human GFAP in mouse recapitulates one of the characteristic pathological features of Alexander disease—the Rosenthal fibers, which are intracellular aggregates containing GFAP—and mice overexpressing GFAP die within a few weeks of birth (Messing et al. 1998). Monoallelic expression of GFAP appears to be tightly controlled since astrocytes of hippocampal origin express both alleles (T. Takizawa, unpubl.), whereas expression in cortical astrocytes is monoallelic, suggesting distinct requirements for GFAP levels among astrocyte populations and indicating a significant degree of heterogeneity in the global astrocyte population in the brain. Our analysis of GFAP+/LacZ mice indicates that the selection of the active allele appears to occur in a stochastic fashion and is largely maintained over long periods of time and during cell division. Similar stochastic monoallelic expression has been reported for cytokine genes such as IL-4 and IL-2 (Guo et al. 2005b). The molecular mechanism for the probabilistic selection and maintenance of the active allele for these genes is unknown.
The monoallelic nature of GFAP expression allowed us for the first time to directly compare within the same cell nucleus the position of gene alleles that differ in their activity status. We found a significant difference in the positioning of the active and inactive alleles. This conclusion is supported by population analysis where the average position of active alleles is more internal compared with inactive ones, and is corroborated by single-cell analysis where in a majority of cells the active allele is more internal than the inactive one in the same cell nucleus. In addition, in a subpopulation of cells with two active alleles, both occupy an internal location.
A potential driving force for the relocalization of gene loci is their association with coregulated genes (Fraser and Bickmore 2007; Misteli 2007). In support, we find that GFAP colocalizes with the astrocyte-specific marker gene S100β at a higher than expected frequency, both of which are activated in a STAT-dependent manner during astrocytogenesis (He et al. 2005). As the genes are activated, their physical proximity is lost due to the repositioning of the S100β gene. Again, this situation is reminiscent of that for IL-4 or IFN-γ, which colocalize in naïve T cells where they are not expressed but become physically separated upon their activation during differentiation (Spilianakis et al. 2005). Indeed, we show here that, similar to GFAP, the active and inactive alleles of IL-4 also occupy distinct radial positions in activated T cells. For some genes, which colocalize upon activation, it has been suggested that their pairing is driven by their association with shared nuclear transcription sites (Osborne et al. 2004). In analogy, the colocalization of GFAP and S100β in their inactive state may be related to their shared silencing, and the differential localization of functionally distinct GFAP alleles serves to stabilize and maintain their respective transcriptional states.
Our results suggest that functionally distinct gene loci occupy different radial positions. These results extend studies on biallelically expressed genes, some of which have been shown to undergo repositioning upon activation during differentiation (Skok et al. 2001; Kosak et al. 2002; Parada et al. 2003; Kim et al. 2004). However, the functional relevance of radial gene positioning remains unclear. The strongest argument that radial positioning does not determine activity is the observation that, for many genes, functionally identical alleles often occupy vastly different locations within the same nucleus (Roix et al. 2003; Ragoczy et al. 2006). Similarly, most gene loci can be found at any radial position within a population. This behavior is confirmed for GFAP, whose active allele could be found at any radial position within a population although it was preferentially found in the nuclear interior. Furthermore, while in a majority of cells the active GFAP allele occupies a more internal position, in other cells it does not; nevertheless, the allele appears fully active. Although we cannot distinguish in our experiments whether the GFAP locus changes its position prior to or after activation, observations on the β-globin gene during erythrocyte differentiation indicate that repositioning of alleles occurs after transcriptional activation. The silent β-globin locus localizes to the nuclear periphery, becomes activated at the periphery, and only then begins its relocalization toward the interior as differentiation proceeds (Ragoczy et al. 2006). These observations suggest that the radial position of an allele is largely a consequence of its activity and not a prerequisite for its proper expression. A key question that now remains is whether, in analogy to our observations on two monoallelically expressed genes, the frequently observed differential radial position of the two copies of biallelically expressed genes in single cell nuclei may reflect subtle differences in the expression behavior of the two alleles and, if so, what these differences are.
Materials and methods
Cell culture
NPC cultures were grown as described previously (Takizawa et al. 2001b). The telencephalons from E14.5 cd-1 mice were triturated and cultured in N2-supplemented (Invitrogen) DMEM/F12 with basic FGF (10 ng/mL; R&D Systems). For astrocyte differentiation, the cells were replated on laminin/poly-D-Lysine-coated glass coverslips (BD) after 4 d of culture, and were stimulated for 4 d in the presence of LIF (80 ng/mL; Chemicon) and, in some cases, in a combination of LIF and BMP2 (80 ng/mL; R&D Systems). For primary astrocyte cultures, the cerebral cortices of postnatal day 0 or 1 cd-1 mice were dissociated using papain (Sigma), cultured in N2-supplemented DMEM/F12 containing 10% fetal calf serum, and passaged twice before assays. For cytokine stimulation of astrocytes, media were changed to G5-supplemented (Invitrogen) DMEM/F12 and cultured with LIF and BMP2 for 2 or 4 d.
Clonal culture of GFAP heterozygous astrocytes
Cortical astrocytes from GFAP heterozygotes (GFAP+/LacZ) were labeled with GFP-expressing retrovirus and cultured for 2 d. Cells were mixed with nonlabeled GFAP heterozygous astrocytes at a ratio of 1:1000 and plated onto glass coverslips. After 4–6 d, cells were stimulated with BMP2/LIF for 4 d. Single clonal colonies identified by their GFP signal were used for analysis.
Immunoblotting
Cells were lysed with NP40 lysis buffer (0.5% NP40, 10 mM Tris-Cl at pH 7.4, 150 mM NaCl, 5 mM EDTA, Protease inhibitor cocktail [Calbiochem]). Lysates were separated by SDS-PAGE and subsequently immunoblotted with antibodies against GFAP (DAKO) or β-actin (Sigma). Detection was performed with an ECL system (Amersham).
Immunocytochemistry and X-Gal staining
Cells were fixed with 4% paraformaldehyde (PFA) in PBS and treated as described previously (Takizawa et al. 2001a). Rabbit polyclonal antibodies against PML and GFAP (DAKO), and mouse monoclonal antibodies against GFAP (Sigma), SF2/ASF, and Nucleophosmin (Zymed) were used as primary antibodies. Alexa488- or Alexa568-conjugated secondary antibodies (Invitrogen) were used. For simultaneous FISH and immunostaining, immunostaining was performed after FISH. To detect LacZ gene expression, astrocytes were stained with X-Gal staining solution (Genlantis) immediately after being fixed with 2% PFA.
Quantitative RT–PCR
Total RNAs were extracted with RNeasy and DNase kit (Qiagen) and reverse-transcribed with SuperScript III (Invitrogen). Quantitative PCR was performed with iQ SYBR Green Supermix (Bio-Rad) using the MyiQ Real-Time PCR system (Bio-Rad). The following primers were used: 5′-ATCGAGATCGCCACCT ACAG-3′ and 5′-CTCACATCACCACGTCCTTG-3′ (for the GFAP); 5′-TTCCGATGCCCTGAGGCTC-3′ and 5′-TGCGCT CAGGAGGAGCAATG-3′ (for the β-actin).
FISH
FISH was essentially carried out as described in Roix et al. (2003). For three-dimensional FISH (3D-FISH), cells were fixed with 4% PFA/0.5% Triton X-100 and kept in 70% ethanol at −20°C until use. The cells were permeabilized with 0.5% saponin/0.5% Triton X-100 and treated with 0.1 N HCl for 10 min. Cells were denatured for 5 min at 75°C in 70% formamide with 2× SSC. Hybridization was performed overnight at 37°C with biotin- or digoxigenin (DIG)-labeled probes and detected with Texas Red-conjugated streptavidin (Vector) or FITC-conjugated anti-DIG antibody (Roche). Probes for DNA FISH were generated by nick translation of BAC clones (BACPAC Resources). The following BAC clones were used: RP24-155G1 (GFAP), RP23-476J7 (β-actin), RP23-42P16 (IL-2), RP23-51J21 (IGF-2), RP23-213P2 (S100β), and RP23-449L19 (IL-4). For RNA FISH probes, plasmids containing cDNAs pcDNA3-mGF (GFAP, from M. Brenner) and pUC-β-actin (β-actin, from C. Smith) were nick-translated using standard protocols. For GFAP RNA/DNA FISH, cells were simultaneously hybridized with the RNA- and DNA-specific probes after denaturation. Specificity of FISH probes was verified with RNase and DNase treatments. For IL-4 RNA/DNA FISH, Th2 cells were fixed with 4% PFA containing 10% acetic acid and hybridized with an ssDNA probe against IL-4 cDNA (Open Biosystems) after dehydration through ethanol treatment. RNA was detected with Alexa598-conjugated tyramide using the TSA kit (Invitrogen), and DNA FISH was performed after RNase treatment. For replication timing assays, cells were labeled with BrdU (Amersham) for 45 min prior to methanol/acetic acid fixation and treated with RNase before hybridization. Anti-BrdU (Serotc AbD) was used to detect incorporated BrdU.
Microscopy and image analysis
For immunocytochemistry, a Nikon Eclipse E800 microscope was used. For RNA signal quantification, between 100 and 107 nuclei containing RNA FISH signals were counted. For replication analysis, 136–150 nuclei were counted. For spatial positioning analysis of FISH signals, we used a Zeiss 510 META confocal microscope. Projected images were obtained from stacked serial Z-sections of 0.9-μm thickness in 0.3-μm intervals. Two-dimensional (2D) maximal projections were generated and analyzed for radial distribution of FISH spots as described below. For colocalization analysis, distances between two FISH spots were examined in each Z-section by a Zeiss LSM Image Browser.
Quantitative analysis of FISH signal distributions
For automated nucleus segmentation and detection of FISH signals, we used a three-stage process involving (1) noise reduction, (2) segmentation, and (3) post-processing. For multicolor FISH analysis, background noise was removed in each channel by applying an adaptive nonlinear noise-reduction technique (Smith and Brady 1997). We applied a fuzzy C-means clustering algorithm (Castleman 1996; Duda et al. 2001) on the noise-reduced gray channels to delineate objects within them. We converted the fuzzy images from the segmentation process into binary images (hard segmentation) in order to obtain a collection of potential segmented objects (both FISH spots and nucleus). We used gray-weighted thresholds to select only high-intensity FISH signals and used a sequence of 2D morphological operations (opening and closing) for filling holes and smoothing boundaries of the segmented nucleus. Manual comparison demonstrated successful identification of >99% of FISH signals and a false-positive rate of <1%.
For quantitation of signal distributions, we computed intensity gravity center (IGCN), area of nucleus (AN), theoretical ellipse fitted to the nucleus boundary (Fitzgibbon et al. 1999), orientation of principal axis (θN), and exclusive-or of the area enclosed by segmented nucleus boundary and theoretical ellipse (AN,T). For each FISH signal, we computed its intensity gravity center (IGCF) and the nearest pixels on the actual segmented nucleus boundary and on the theoretical ellipse. Additionally, each DNA-based FISH signal within the nucleus was also automatically marked active based on its spatial proximity to the RNA FISH signal. These values were used to determine the radial position of each signal.
For statistical analysis, to compare the distances of FISH signals from the IGCN across multiple nuclei, we transformed each FISH signal into a template ellipse by applying a two-stage affine transform on each segmented nucleus. This was done such that its principal axis was horizontally oriented and then projecting each FISH signal into a user-defined, common template ellipse via homographic projection between the template ellipse, and the ellipse was fitted to the segmented nucleus. In order to test for underlying spatial randomness of the FISH signals, we generated a population of random points within the user-defined template ellipse and statistically compared these two distributions using the two-sample 1D KS test. We constrained our spatial analysis of FISH signals to only those nuclei that had the ratio of AN,T to AN <0.1. All analysis tools were implemented using custom software written in Matlab with the DIPImage toolbox.
Acknowledgments
We thank M. Brenner and C. Smith for plasmids, P.G. McQueen for statistical analysis, and T. Karpova and C. Combs for imaging support. Imaging was performed at the National Cancer Institute Fluorescence Imaging Facility and National Heart, Lung, and Blood Institute Light Microscopy Facility. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. T.T. was supported by a Human Frontier Science Program Long-term Fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1634608
References
- Baxter J., Merkenschlager M., Fisher A.G. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 2002;14:372–376. doi: 10.1016/s0955-0674(02)00339-3. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A., Mahy N.L., Chambeyron S. Do higher-order chromatin structure and nuclear reorganization play a role in regulating Hox gene expression during development? Cold Spring Harb. Symp. Quant. Biol. 2004;69:251–257. doi: 10.1101/sqb.2004.69.251. [DOI] [PubMed] [Google Scholar]
- Castleman K.R. Digital image processing. Prentice Hall; Englewood Cliffs, NJ: 1996. [Google Scholar]
- Chakalova L., Debrand E., Mitchell J.A., Osborne C.S., Fraser P. Replication and transcription: Shaping the landscape of the genome. Nat. Rev. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Duda R.O., Hart P.E., Stork D.G. Pattern classification. Wiley; New York: 2001. [Google Scholar]
- Fitzgibbon A., Pilu M., Fisher R.B. Direct least square fitting of ellipses. IEEE Trans. Pattern Anal. Mach. Intell. 1999;21:476–480. [Google Scholar]
- Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Fuss S.H., Omura M., Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Gomi H., Yokoyama T., Fujimoto K., Ikeda T., Katoh A., Itoh T., Itohara S. Mice devoid of the glial fibrillary acidic protein develop normally and are susceptible to scrapie prions. Neuron. 1995;14:29–41. doi: 10.1016/0896-6273(95)90238-4. [DOI] [PubMed] [Google Scholar]
- Guo L., Hu-Li J., Paul W.E. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity. 2005a;23:89–99. doi: 10.1016/j.immuni.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Guo L., Hu-Li J., Paul W.E. Probabilistic regulation of IL-4 production. J. Clin. Immunol. 2005b;25:573–581. doi: 10.1007/s10875-005-8218-5. [DOI] [PubMed] [Google Scholar]
- He F., Ge W., Martinowich K., Becker-Catania S., Coskun V., Zhu W., Wu H., Castro D., Guillemot F., Fan G., et al. A positive autoregulatory loop of Jak–STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O., Jepsen K., Rosenfeld M.G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Huang S., Spector D.L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., McQueen P.G., Lichtman M.K., Shevach E.M., Parada L.A., Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet. Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Lanctot C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D.J., Mendelsohn M., Kirkland J., Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Meaburn K.J., Misteli T. Cell biology: Chromosome territories. Nature. 2007;445:379–381. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Messing A., Head M.W., Galles K., Galbreath E.J., Goldman J.E., Brenner M. Fatal encephalopathy with astrocyte inclusions in GFAP transgenic mice. Am. J. Pathol. 1998;152:391–398. [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Namihira M., Nakashima K., Taga T. Developmental stage dependent regulation of DNA methylation and chromatin modification in a immature astrocyte specific gene promoter. FEBS Lett. 2004;572:184–188. doi: 10.1016/j.febslet.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parada L.A., Roix J.J., Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13:393–396. doi: 10.1016/s0962-8924(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Parada L.A., Sotiriou S., Misteli T. Spatial genome organization. Exp. Cell Res. 2004;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes & Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix J.J., McQueen P.G., Munson P.J., Parada L.A., Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Skok J.A., Brown K.E., Azuara V., Caparros M.L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Brady J.M. SUSAN—New approach to low level image processing. Int. J. Comput. Vis. 1997;23:45–78. [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001a;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Yanagisawa M., Ochiai W., Yasukawa K., Ishiguro T., Nakashima K., Taga T. Directly linked soluble IL-6 receptor–IL-6 fusion protein induces astrocyte differentiation from neuroepithelial cells via activation of STAT3. Cytokine. 2001b;13:272–279. doi: 10.1006/cyto.2000.0831. [DOI] [PubMed] [Google Scholar]
- Yang P.K., Kuroda M.I. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjolinder M., Gondor A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]