Abstract
Mycobacteria can repair DNA double-strand breaks (DSBs) via a nonhomologous end-joining (NHEJ) system that includes a dedicated DNA ligase (LigD) and the DNA end-binding protein Ku. Here we exploit an improved plasmid-based NHEJ assay and a collection of Mycobacterium smegmatis strains bearing deletions or mutations in Ku or the DNA ligases to interrogate the contributions of LigD’s three catalytic activities (polymerase, ligase, and 3′ phosphoesterase) and structural domains (POL, LIG, and PE) to the efficiency and molecular outcomes of NHEJ in vivo. By analyzing in parallel the repair of blunt, 5′ overhang, and 3′ overhang DSBs, we discovered a novel end-joining pathway specific to breaks with 3′ overhangs that is Ku- and LigD-independent and perfectly faithful. This 3′ overhang NHEJ pathway is independent of ligases B and C; we surmise that it relies on NAD+-dependent LigA, the essential replicative ligase. We find that efficient repair of blunt and 5′ overhang DSBs depends stringently on Ku and the LigD POL domain, but not on the LigD polymerase activity, which mainly serves to promote NHEJ infidelity. The lack of an effect of PE-inactivating LigD mutations on NHEJ outcomes, especially the balance between deletions and insertions at blunt or 5′ overhang breaks, argues against LigD being the catalyst of deletion formation. Ligase-inactivating LigD mutations (or deletion of the LIG domain) have a modest impact on the efficiency of blunt and 5′ overhang DSB repair, because the strand sealing activity can be provided in trans by one of the other resident ATP-dependent ligases (likely LigC). Reliance on the backup ligase is accompanied by a drastic loss of fidelity during blunt end and 5′ overhang DSB repair. We conclude that the mechanisms of mycobacterial NHEJ are many and the outcomes depend on the initial structures of the DSBs and the available ensemble of end-processing and end-sealing components, which are not limited to Ku and LigD.
[Keywords: Double-strand breaks, DNA ligase, Ku, end-joining, mutation]
DNA double-strand breaks (DSBs) can be repaired via homologous recombination (HR) or nonhomologous end-joining (NHEJ). Because NHEJ does not rely on a homologous DNA template, it can operate when only one chromosomal copy is available. Thus, NHEJ is a major DSB repair pathway in eukarya during the G1 phase of the cell cycle (Takata et al. 1998; Ferreira and Cooper 2004). Recent studies have illuminated the existence of an NHEJ pathway in bacteria (for review, see Pitcher et al. 2007a; Shuman and Glickman 2007) and its role in repairing DSBs in the bacterial chromosome, especially during quiescent states such as sporulation or late stationary-phase culture (Wang et al. 2006; Moeller et al. 2007; Pitcher et al. 2007b; Stephanou et al. 2007). The common thread in eukaryal and bacterial NHEJ is their reliance on the DNA end-binding protein Ku and a dedicated ATP-dependent DNA ligase (Lig4 in eukarya, LigD in bacteria). Whereas Ku and Lig4 are present in the proteomes of nearly all eukaryal species, only a subset of bacteria have genes encoding Ku and LigD, among which are the human pathogens Mycobacterium tuberculosis, Pseudomonas aeruginosa, and Bacillus anthracis.
Biochemical, structural, and genetic studies of the bacterial NHEJ ligases and Ku proteins have uncovered unique properties of the bacterial NHEJ pathway. For example, LigD differs from all other DNA ligases in that it has multiple catalytic activities within a single polypeptide (Della et al. 2004; Gong et al. 2005; Zhu and Shuman 2007). Mycobacterium, Pseudomonas, and Agrobacterium LigD proteins are composed of three autonomous domains: ligase (LIG), polymerase (POL), and phosphoesterase (PE). The POL and PE domains comprise a suite of DNA end-healing activities that are proposed to remodel the 3′ terminus of the DSB prior to sealing by the LIG component. The POL domain is a minimized member of the archaeal-eukaryal primase-polymerase family that is especially adept at adding a short tract of ribonucleotides to a DNA primer-template and at adding a single nontemplated ribonucleotide at a blunt DNA end (Della et al. 2004; Gong et al. 2005; Zhu and Shuman 2005a; Yakovleva and Shuman 2006; Zhu et al. 2006; Pitcher et al. 2007c). The PE domain, which removes 3′ phosphates and 3′ ribonucleotides from primer-templates, exemplifies a new family of 3′ processing enzymes with a distinctive substrate specificity and active site (Zhu and Shuman 2005b, 2006; Zhu et al. 2005).
Direct evidence that bacteria can catalyze DSB repair via NHEJ and initial insights to the NHEJ mechanism emerged from studies of the repair of linear plasmid DNAs transfected into Mycobacterium smegmatis (Gong et al. 2005). The plasmid substrate contained a mycobacterial replication origin, a kanamycin resistance gene, and a lacZ gene encoding β-galactosidase. Digestion of the plasmid with a restriction endonuclease that cuts just once within the lacZ gene yields a linear DNA with defined blunt or 5′ overhang ends. For the transfected linear DNA to transform the recipient Mycobacterium to kanamycin resistance, the ends of the plasmid must be joined to form a replication-competent circular DNA. Because the mycobacterial genome has no DNA sequences homologous to the plasmid DNA flanking the DSB, there is no option to effect DSB repair via the HR pathway. This is a critical feature that makes the assay readout specifically a measure of NHEJ. The remarkable finding about mycobacterial NHEJ was that ∼50% of the kanR transformants derived from initially blunt or 5′ overhang DSBs had white colony color on medium containing X-gal, indicating that the lacZ gene had been mutationally inactivated during the repair process (Gong et al. 2005). The efficiency of plasmid NHEJ at blunt or 5′ overhang ends was reduced drastically in M. smegmatis ΔligD or Δku strains (Gong et al. 2005). Analysis of the molecular outcomes of unfaithful NHEJ in vivo in wild-type M. smegmatis highlighted the prevalence of templated insertions at 5′ overhang DSBs and of nontemplated single nucleotide insertions at blunt DSBs, implicating LigD POL as the catalyst of mutagenic insertions (Gong et al. 2005; Zhu et al. 2006). A substantial fraction of unfaithful NHEJ events in vivo at blunt or 5′ overhang DSBs involved deletions at one end, presumably as a consequence of exonucleolytic degradation prior to ligation.
It has been proposed that bacterial Ku and LigD together comprise a two-component NHEJ “machine” possessed of all the activities required for DSB repair (Della et al. 2004). This model is appealing for its simplicity and is supported by studies documenting DSB repair activity in vivo when mycobacterial Ku and LigD were expressed in heterologous species (Della et al. 2004; Malyarchuk et al. 2007). A caveat to assaying NHEJ activity in a heterologous genetic background is that the outcomes might not accurately reflect the mechanism of NHEJ in the naturally NHEJ-proficient bacterium. For example, Malyarchuk et al. (2007) reported that the repair of a linear plasmid with a 3′ overhang DSB in Escherichia coli cells depended on coexpression of mycobacterial Ku and LigD, was predominantly unfaithful, and in virtually all events entailed heterogeneous deletions at the repair junctions. However, as we will show presently, the repair of 3′ overhang DSBs by NHEJ in a mycobacterial cell obeys entirely different rules and elicits dramatically different outcomes. The repair of 5′ overhang DSBs in E. coli expressing mycobacterial Ku and LigD was also reported to be predominantly unfaithful and entailed deletions at all of the repair junctions analyzed (Malyarchuk et al. 2007). This result contrasts starkly with the native mycobacterial pathway of 5′ overhang NHEJ, which is skewed toward insertional frameshift mutations caused by polymerase fill-in of the 5′ overhangs (Gong et al. 2005). Such discrepancies underscore the importance of studying bacterial NHEJ in the native context.
Furthermore, it is not clear that Ku and LigD are the only constituents of the NHEJ pathway in bacterial species that are naturally NHEJ-competent. The two-component model is potentially belied by several factors, including (1) the variability and complexity of the DNA ligase menu in different bacteria taxa, which raises issues of division of labor and functional overlap; (2) the occurrence in vivo of deletional NHEJ events that are not immediately accounted for by the activities of LigD or Ku; and (3) the as yet untested prospect that different types of breaks are handled differently by the bacterial NHEJ system(s).
M. tuberculosis and M. smegmatis have four and five DNA ligases, respectively, including a NAD+-dependent ligase (LigA) plus three different ATP-dependent ligases: LigB, LigC, and LigD (Fig. 1A; Gong et al. 2004, 2005). M. smegmatis has two LigC paralogs (C1 and C2) encoded by adjacent chromosomal genes. Whereas LigA is essential for mycobacterial survival (Korycka-Machala et al. 2007), the genes encoding LigB, LigC, and LigD can be deleted, singly or in pairs, with no apparent effect on cell growth under laboratory conditions (Gong et al. 2005). As noted above, there is ample evidence that LigD is dedicated to NHEJ. Several lines of evidence also point to an NHEJ function for LigC. Genetic studies implicate mycobacterial LigC in a minor Ku-dependent NHEJ pathway operating on blunt DSBs that was revealed when the ligD gene was deleted (Gong et al. 2005). Biochemical studies show that Ku stimulates the repair of DSBs in vitro by LigC (Zhu and Shuman 2007).
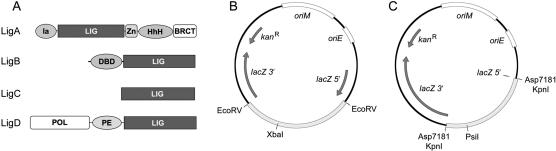
Figure 1.
Mycobacterial ligases and new NHEJ reporter plasmids. (A) The mycobacterial LigA, LigB, LigC, and LigD polypeptides are depicted in cartoon form with the N termini on the left and the C termini on the right. Each enzyme has a core ligase catalytic domain (LIG) composed of a nucleotidyl transferase module and an OB-fold module. Flanking domains of known structure or function are shown. In LigA enzymes, the LIG domain is fused to an N-terminal domain (Ia) and three carboxyl domains: a tetracysteine Zn-finger, a helix–hairpin–helix (HhH) domain, and a BRCT domain. Domain Ia is unique to NAD+-dependent ligases and are required for the reaction of LigA with NAD+ to form the ligase-adenylate intermediate. LigB consists of a C-terminal LIG domain fused to an N-terminal DNA-binding domain (DBD). LigD and LigC are the NHEJ ligases of bacteria. LigC consists only of the catalytic core. LigD is a multifunctional repair protein that has N-terminal polymerase (POL) and central phosphoesterase (PE) catalytic modules fused to the C-terminal LIG domain. (B, C) Maps of the new NHEJ reporter plasmids, which contain a mycobacterial origin of replication (oriM), an E. coli origin of replication (oriE), a kanamycin resistance gene (kanR), and a lacZ gene interrupted by insertion of foreign DNA sequences between restriction sites for EcoRV (B) or the isoschizomers Asp7181/KpnI (C). To prepare the substrate for blunt-end NHEJ, the plasmid in B was digested with EcoRV and XbaI (which cuts uniquely within the foreign insert), and the linear EcoRV plasmid fragment was gel-purified. To prepare the substrates for 5′ overhang and 3′ overhang NHEJ, the plasmid in C was digested with either Asp7181 (5′ overhang) or KpnI (3′ overhang) plus PsiI (which cuts uniquely in the foreign insert), and the linear plasmid fragment was gel-purified.
Here we exploit an improved plasmid-based NHEJ assay and a large collection of M. smegmatis mutant strains to gauge how LigD’s activities and structural domains influence the repair of blunt, 5′ overhang, and 3′ overhang DSBs in vivo. We thereby defined a new Ku- and LigD-independent NHEJ pathway specific to 3′ overhang DSBs that is nonmutagenic and persists in the absence of LigB and LigC. We infer a role for LigA in Ku-independent 3′ overhang NHEJ. We report that repair of blunt and 5′ overhang DSBs requires Ku and the LigD POL domain, but not LigD polymerase catalytic activity, although the polymerase activity is a major determinant of NHEJ infidelity. Phosphoesterase-inactivating LigD mutations had little impact on the prevalence of deletions versus insertions at blunt or 5′ overhang breaks, thus vitiating the hypothesis (Malyarchuk et al. 2007) that LigD is the catalyst of deletion formation in vivo. The strand sealing activity for blunt and 5′ overhang NHEJ is provided by one of the other ATP-dependent ligases (presumably LigC) when the LigD LIG domain is inactivated or deleted. NHEJ catalyzed by the backup ligase is extremely mutagenic. Our results underscore the complexity and nuances of the mycobacterial NHEJ system.
Results
Improved plasmid-based assay for mycobacterial NHEJ
The reporter plasmid used previously to assay mycobacterial NHEJ (Gong et al. 2005) was modified to allow more accurate determinations of repair fidelity in NHEJ-defective strains. Formerly, the reporter plasmid contained a functional lacZ gene and was linearized by digestion with a restriction endonuclease at a single site within lacZ. The linear DNA was electroporated into mycobacteria and the cells were screened simultaneously for acquisition of antibiotic resistance and lacZ expression by plating on agar plates containing kanamycin and X-gal. A caveat to using the original reporter construct is that any residual uncut circular plasmid DNA will, upon electroporation to M. smegmatis, yield blue kanamycin-resistant colonies on X-gal plates that are indistinguishable from the lacZ+ colonies arising from bacteria that repaired the transfected linear plasmid via faithful NHEJ. Whereas the blue colony background from uncut plasmids is typically low and does not affect the assay results in NHEJ-proficient M. smegmatis strains, we found that it is a confounding factor when studying repair fidelity in Δku and ΔligD strains, in which NHEJ efficiency is very weak.
To avoid this problem, we inserted foreign DNA fragments into the EcoRV or Asp7181/KpnI sites, thereby inactivating the lacZ gene (Fig. 1B,C). To prepare the linear NHEJ substrate, the new plasmids were digested with EcoRV or Asp7181 (or its isoschizomer KpnI) to excise the foreign insert, and then digested with a second restriction endonuclease that cuts within the foreign insert. The linear plasmids were gel-purified prior to use in NHEJ assays. In the modified regime, the only way that a blue kanR colony can arise is by faithful NHEJ. Although a white colony could, in principle, be generated by residual uncut circular plasmid, in practice we recovered the original vector only three times in a collection of 664 individual white kanR colonies for which the structures of the repair junctions were determined. The assay improvement was instantly apparent from a comparison of the relative efficiency and absolute fidelity of blunt end NHEJ in wild-type M. smegmatis versus the Δku mutant (Fig. 2). Loss of Ku reduced blunt NHEJ efficiency by a factor of 500, in agreement with prior results (Gong et al. 2005). The key finding was that <1% of the blunt NHEJ events in the Δku strain were faithful, compared with 57% fidelity in wild-type mycobacteria. Previously, using the original reporter assay, we noted that 100% of the kanR Δku colonies in the blunt NHEJ experiment were blue (Gong et al. 2005). In light of the new results, we conclude that the exclusively blue Δku colonies recovered in the prior experiment resulted mainly from uptake of intact circular plasmid, not NHEJ.
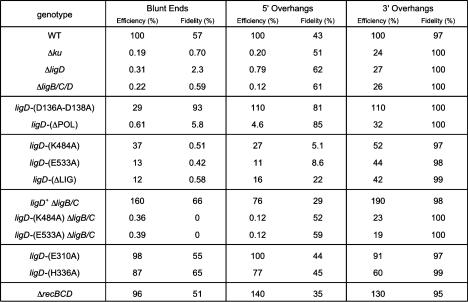
Figure 2.
NHEJ efficiency and fidelity in wild-type and mutant M. smegmatis strains. Genotypes are indicated in the left column. NHEJ efficiencies for repair of transfected linear plasmids with blunt, 5′ overhang, and 3′ overhang DSBs in the M. smegmatis mutants were normalized to the wild-type values (100%). NHEJ fidelity is the fraction (percentage) of kanR transformants that were lacZ+. The fidelity values for wild-type M. smegmatis for repair of blunt, 5′ overhang, and 3′ overhang ends were calculated from counts of 4128, 2259, and 2626 kanR colonies, respectively. The fidelity values in the Δku mutant for blunt, 5′ overhang, and 3′ overhang NHEJ were calculated from counts of 143, 79, and 2169 kanR colonies. The fidelity values in the ΔligD strain for blunt, 5′ overhang, and 3′ overhang NHEJ were calculated from counts of 523, 1539, and 4953 kanR colonies. The fidelity values in the ΔligB/C/D strain for blunt, 5′ overhang, and 3′ overhang NHEJ were calculated from counts of 169, 126, and 1613 kanR colonies.
Outcome of NHEJ depends on the structure of the DSB ends
The influence of DSB end configuration on NHEJ fidelity in wild-type mycobacteria was profound. Whereas the repair of linear plasmid DNA with blunt ends or complementary 5′ overhangs ends was highly mutagenic (57% and 43% fidelity, respectively), the repair of 3′ overhang DSBs was remarkably error-free, correctly restoring the lacZ ORF in 97% of the kanR transformants (Fig. 2). The faithful outcome of 3′ overhang NHEJ cannot be ascribed to a particular “sticky” overhang sequence, or to a positional effect within the lacZ reporter gene, insofar as the unfaithfully repaired 5′ overhang DSBs have an identical sticky-end sequence and are located at the same site in the plasmid as the 3′ overhang DSBs, because these DSBs are generated by incision with the isoschizomers Asp7181 (5′ overhang) and KpnI (3′ overhang), respectively (Fig. 1C).
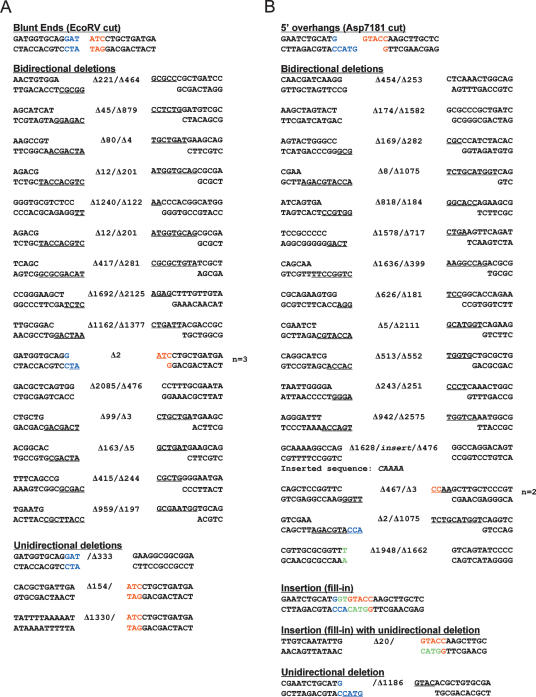
To gain further insight to whether the mutagenic NHEJ mechanism differs according to DSB end configuration, we determined the junction sequences of the repaired blunt, 5′ overhang, and 3′ overhang reporter plasmids derived from at least 20 independent lacZ− kanR transformants of wild-type M. smegmatis. The individual NHEJ outcomes at blunt and 5′ overhang ends are shown in Figure 3 and categorized in Figure 4. The results agree with previous data (Gong et al. 2005) and are summarized as follows: The predominant mechanisms of blunt NHEJ entailed either (1) the insertion of a single nontemplated nucleotide at the otherwise unperturbed DSB ends, thereby creating a +1 frameshift mutation (eight of 21 events), or (2) a unidirectional deletion entailing resection of a variably sized tract from one end followed by ligation to the other intact blunt DSB (nine of 21 events). Various other permutations, involving nontemplated single nucleotide addition at one intact end and unidirectional deletion at the other end (n = 3) or a bidirectional deletion (n = 1), were also noted (Fig. 3A). In sum, 11 of 21 unfaithful blunt DSB repairs involved nucleotide addition and 13 involved a deletion. A striking aspect of the deletional repairs is that none appeared to exploit a microhomology at the junction to assist end joining. The single most prevalent outcome of 5′ overhang NHEJ in wild-type mycobacteria involved templated addition of 4 nucleotides (nt) at both single-stranded tails to form a blunt DSB that was then sealed to create a +4 frameshift (six of 22 events) (Fig. 3B). One junction was completely filled in and then received a nontemplated single nucleotide addition prior to sealing (+5 frameshift) (shown in Fig. 3B with the nontemplated junction nucleotide colored magenta). Seven of 22 repair events entailed partial fill-in of the 5′ overhangs to generate +2 frameshifts. Another subset of junctions was formed by templated fill-in at one 5′ overhang end and unidirectional deletion of the other end (n = 7). Only one event involved bidirectional deletion. In sum, 21 of 22 of the 5′ overhang DSB repairs involved templated nucleotide addition, and eight of 22 involved a deletion. None of the deletions had a microhomology at the junction.
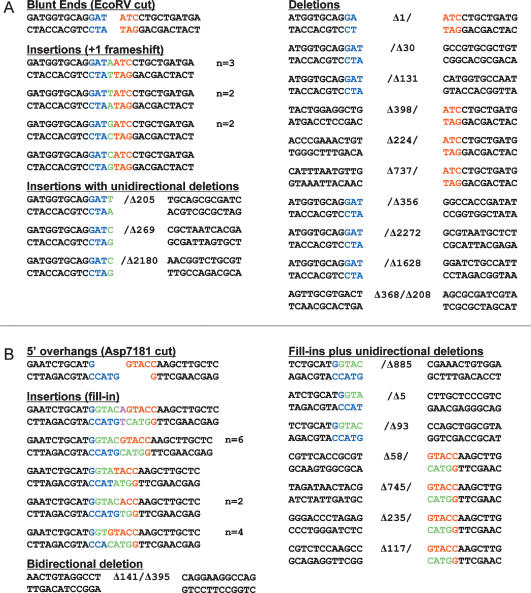
Figure 3.
Outcomes of unfaithful blunt and 5′ overhang DSB repair in wild-type M. smegmatis. The nucleotide sequences of NHEJ junctions from independent lacZ− kanR colonies that had been transformed with EcoRV-cut (A) or Asp7181-cut (B) linear plasmid DNAs are shown. The two halves of the original restriction sites are shown in blue and red. Inserted nucleotides are colored green (or magenta in B when a nontemplated nucleotide is added after a 5′ overhang fill-in). Deletions are denoted by Δ next to the number of nucleotides resected from either end. The number of times (n) that the identical junction sequences were recovered from independent transformants is indicated at right, when applicable.
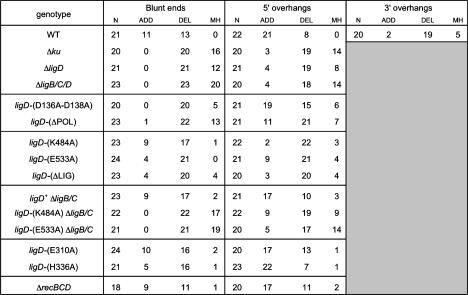
Figure 4.
Summary of unfaithful NHEJ events. M. smegmatis genotypes are indicated in the left column. The total number (N) of unfaithfully repaired junctions that were sequenced for each strain and DSB end configuration is specified, along with the number of repair events that entailed nucleotide additions (ADD) or deletions (DEL) at the repair junctions. The incidence of ligation at microhomologies (defined as two or more positions of nucleotide identity flanking the junction) is reported for each data set.
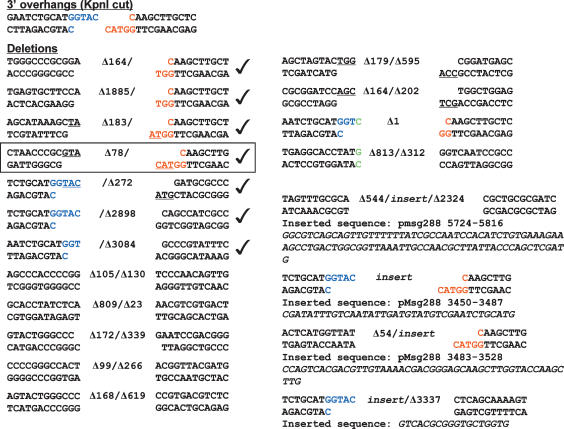
In contrast, the analysis of unfaithful NHEJ at complementary 3′ overhang DSBs (a scenario not studied previously in mycobacteria) implied an entirely different mode of mutagenic repair that was dominated by deletion formation (Fig. 5). Nine of 20 repairs entailed unidirectional deletion (with retention of some or all of the original KpnI site nucleotides at the other end), and nine of 20 events involved bidirectional deletions. Only 10% of the junctions (two of 20) had a nucleotide insertion. In one case, a single nucleotide was added at a partially trimmed KpnI 3′ overhang; in the second case, a single nontemplated nucleotide was added in the context of a bidirectional deletion. Four of the mutagenic 3′ overhang repairs involved insertion of a DNA segment that, in three of four cases, derived from a distant site in the reporter plasmid (Fig. 5).
Figure 5.
Outcomes of unfaithful 3′ overhang DSB repair in wild-type M. smegmatis. The nucleotide sequences of NHEJ junctions from 20 independent lacZ− kanR colonies that had been transformed with KpnI-cut linear plasmid DNA are shown. The two halves of the original KpnI restriction site are shown in blue and red. Deletions are denoted by Δ next to the number of nucleotides resected from either end. Inserted nucleotides are colored green. Microhomologies at the junctions are underlined and depicted as resulting from nucleolytic resection and exposure of complementary 3′ extensions, which can anneal to the 3′ single-stranded tail of one of the original KpnI-cut ends. Four of the junctions had sequences inserted between the sealed plasmid ends; three of these sequences correspond to distant sites in the pMSG288-based plasmid. These inserted sequences (and their numerical coordinates in the plasmid) are indicated below the junctions. The junction highlighted in the box exemplifies a likely instance of cross-break templated nucleotide addition to a 3′ single-stranded tail to fill-in a gap prior to strand sealing. Other likely instances of cross-break fill-in are indicated by check marks.
In addition to the strong bias toward deletions, the mutagenic 3′ overhang NHEJ mechanism differed from the blunt and 5′ overhang repair pathways in its more frequent use of ≥2-nt microhomologies at the deletion junctions (five of 20). An exemplary junction, depicted in the box in Figure 5, shows that the microhomology leaves a 1-nt gap between the recessed 5′ end (5′-CAA) of an intact KpnI 3′ overhang and the complementary 3′ microhomology tail (GTA-3′) of other DSB end that incurred a deletion. Because the NHEJ junction (top strand sequence 5′-GTACCAA-3′) has a C residue inserted between the microhomology tail and the original KpnI end, the implication is that the DSB was repaired by cross-break priming of templated gap filling by a DNA polymerase prior to sealing the top strand.
Reliance of NHEJ on Ku and LigD depends on the structure of the DSB ends
The efficiency of blunt NHEJ was reduced by factors of 500 and 300 in M. smegmatis Δku and ΔligD strains, respectively, and the residual blunt DSB repair events observed in these strains were overwhelmingly mutagenic (0.7 and 2.3% fidelity in Δku and ΔligD, respectively) (Fig. 2). Sequence analysis of 20 junctions recovered from lacZ− kanR Δku transformants revealed a very different spectrum of outcomes compared with wild-type mycobacteria, insofar as every repair event entailed a deletion and not a single nucleotide insertion event was detected (Fig. 6A). Thus, we conclude that faithful blunt end joining is Ku-dependent in vivo, as is the addition of nontemplated nucleotides to blunt ends prior to ligation. The majority (17 of 20; 85%) of the deletional repair events in the Δku strain were bidirectional. The deletions ranged in size from 2 to 2125 nt and were notable for the high incidence of microhomologies at the junctions (16 of 20 events), 12 of which were formed at sites consisting of between four and nine identical nucleotides (Fig. 6A). Thus, Ku specifically promotes microhomology-independent ligation of blunt DSBs. Similar findings pertained to the residual blunt NHEJ junctions in the ΔligD strain (Fig. 4; Supplemental Fig. S1), which were exclusively deletional (21 of 21 cases), predominantly bidirectional (18 of 21), and conspicuously reliant on microhomologies (12 of 21 junctions), most of which consisted of between four and 10 identical nucleotides (Supplemental Fig. S1).
Figure 6.
Outcomes of unfaithful blunt and 5′ overhang DSB repair in Δku cells. The nucleotide sequences of NHEJ junctions from independent lacZ− kanR colonies that had been transformed with EcoRV-cut (A) or Asp7181-cut (B) linear plasmid DNAs are shown. The two halves of the original restriction sites are shown in blue and red. Deletions are denoted by Δ next to the number of nucleotides resected from either end. Inserted nucleotides are colored green. Microhomologies at the junctions are underlined and depicted as resulting from nucleolytic resection and exposure of complementary 5′ extensions. The number of times (n) that the identical junction sequences were recovered from independent transformants is indicated at right, when applicable.
5′ overhang NHEJ efficiency was diminished by factors of 500 and 100 in Δku and ΔligD strains, respectively, compared with wild-type mycobacteria (Fig. 2). Yet, unlike blunt NHEJ, the fidelity of the residual 5′ overhang events (51% in Δku and 62% in ΔligD) was only marginally different from the 43% repair fidelity seen in wild-type cells (Fig. 2). Junction sequencing revealed a shift in unfaithful NHEJ outcomes in the absence of Ku, whereby templated nucleotide additions occurred in only 10% of events (two of 20), compared with 95% in wild-type M. smegmatis (Figs. 4, 6B). The vast majority (19 of 20) of Ku-independent 5′ overhang repairs entailed deletions, most of which were bidirectional (n = 17), and many of which exploited microhomologies at the junctions (n = 14) ranging in size from three to 10 identical nucleotides (Fig. 6B). Residual unfaithful 5′ overhang NHEJ in the ΔligD strain was similarly biased against nucleotide additions (four of 21 events) and in favor of deletions (19 of 21 cases), with most being bidirectional (n = 17) and many relying on microhomologies (n = 8) in the range of two to 10 identical nucleotides (Supplemental Fig. S1). Thus, LigD, like Ku, drives microhomology-independent sealing in vivo.
These drastic effects of ablating Ku and LigD on blunt and 5′ overhang NHEJ contrast with the more modest contributions of Ku and LigD to 3′ overhang NHEJ. The 3′ overhang DSBs were repaired in Δku and ΔligD strains at 24%–27% of wild-type efficiency (Fig. 2). Moreover, all of the kanR Δku and ΔligD transformants were lacZ+ (Fig. 2). These results herald the discovery of a novel Ku/LigD-independent pathway of DSB repair that is specific for 3′ overhangs and operates with 100% fidelity. Because the absolute efficiency of repair of 3′ overhang plasmid DSBs was approximately sevenfold greater than that of blunt plasmid DSBs in wild-type M. smegmatis (7.2% versus 1%) (see Materials and Methods), we can surmise that the Ku-independent NHEJ pathway for 3′ overhang NHEJ is at least as active in vivo as the Ku-dependent mechanism operative on blunt and sticky 5′ overhang ends.
Analysis of NHEJ in a ΔligB/C/D strain implicates LigA in faithful 3′ overhang repair
Previous findings that the M. smegmatis ligB, ligC1/C2, and ligD chromosomal loci could be deleted singly, or in pairs, with no apparent effect on cell growth (Gong et al. 2005) left open the possibility that the mycobacterial ATP-dependent ligases do perform an essential task in vivo, but that any one of them is capable of doing so. To address this issue, we constructed a triple-knockout strain in which the ligB, ligC1/C2, and ligD loci were deleted en masse. The resulting ΔligB/C/D strain was viable and grew as well as the wild-type Mycobacterium under laboratory conditions (data not shown). Thus, LigA is the only essential DNA ligase in the M. smegmatis proteome.
Analysis of NHEJ in ΔligB/C/D cells revealed that the triple-knockout phenocopied Δku and ΔligD with respect to the severe decrements in NHEJ efficiency at blunt and 5′ overhang DSBs, the fidelities of the residual NHEJ events, and the molecular outcomes at the unfaithfully repaired junctions (Figs. 2, 4). Sequence analysis of the mutagenic blunt DSB repair events in ΔligB/C/D cells (Supplemental Fig. S2A) showed that (1) the junctions were devoid of insertions (zero of 23), (2) all of the deletions were bidirectional, and (3) junction formation was microhomology-dependent (in 20 of 23 cases) (Fig. 4). The outcomes of unfaithful repair of 5′ overhang DSBs in ΔligB/C/D cells (Supplemental Fig. S2B) were notable for the relative paucity of nucleotide additions (four of 20) and the consequent abundance of deletions (18 of 20), predominantly bidirectional and most (n = 14) with microhomologies at the junctions (Fig. 4).
The instructive finding was that 3′ overhang NHEJ was no more affected in ΔligB/C/D cells (26% of wild-type NHEJ efficiency, with 100% fidelity) than in the ΔligD single-knockout strain (Fig. 2). This result signifies that neither LigB nor LigC1 nor LigC2 is responsible for the sealing step of the Ku-independent 3′ overhang NHEJ pathway. We impute that function, by default, to mycobacterial LigA, the only known residual ligase in the ΔligB/C/D strain.
Different effects of inactivating versus deleting the LigD POL domain
To gain deeper insights to the role of the LigD POL domain in mycobacterial NHEJ, we compared the effects of ablating polymerase catalysis by mutation of the active site to the effects of deleting the entire POL domain. Alanine substitutions for the metal-binding residues in the POL active site abolish LigD catalysis of templated and nontemplated nucleotide additions without affecting the strand-sealing function of LigD (Gong et al. 2005; Pitcher et al. 2005; Zhu et al. 2006). Previously, we exchanged an allele encoding a polymerase-defective mutant of M. smegmatis LigD—ligD-(D136A–D138A)—for the wild-type ligD gene at the native chromosomal locus (Zhu et al. 2006). Using the improved NHEJ reporter assay, we find that inactivating the LigD polymerase in the ligD-(D136A–D138A) strain elicited only a modest decrement in the efficiency of blunt NHEJ (to 29% of wild-type) but resulted in a significant increase in the fidelity of blunt NHEJ—to 93%, compared with 57% in wild-type cells (Fig. 2). The increased fidelity was attributable to the ablation of nontemplated single nucleotide additions from the pool of sequenced unfaithful NHEJ junctions (Supplemental Fig. S3A). In the absence of polymerase activity, all unfaithful repair events involved deletions (Fig. 4; Supplemental Fig. S3A). These results confirm our previous inferences that LigD itself is the direct catalyst of nontemplated nucleotide additions during blunt NHEJ in vivo (Zhu et al. 2006).
We extended the analysis of NHEJ in the ligD-(D136A–D138A) strain to 5′ overhang and 3′ overhang DSB ends and noted that the efficiency of 5′ overhang and 3′ overhang NHEJ was unaffected by loss of LigD polymerase activity (Fig. 2) The salient findings were that (1) the fidelity of 5′ overhang repair was higher (81%) in the ligD-(D136A–D138A) strain than in wild-type cells (43% fidelity) (Fig. 2), and (2) despite the absence of LigD polymerase activity, the majority of the unfaithful repair events (18 of 21) still involved templated nucleotide additions to one or both 5′ overhang ends (Fig. 4; Supplemental Fig. S3B). These results demonstrate that mycobacteria have another template-dependent polymerase that is capable of contributing to Ku-dependent mutagenic NHEJ at 5′ overhang DSBs when LigD polymerase is impaired. Conceivably, the backup polymerase is less efficient than LigD POL itself in filling in the 5′ overhangs, so that the balance between sealing of the unmodified Asp7181 ends by the LigD LIG component, versus nucleotide addition prior to LigD sealing, is tilted toward the former scenario, thus accounting for the increased overall fidelity of 5′ overhang NHEJ in the ligD-(D136A–D138A) strain.
To gauge the effects of deleting the POL domain, we replaced the native ligD gene with a new allele—ligD-(ΔPOL)—encoding only the segment of the LigD protein that embraces the PE and LIG domains. The absence of the POL domain does not impact on in vitro catalysis of nick sealing by the LigD LIG domain (Pitcher et al. 2005; Zhu and Shuman 2005a). The effect of deleting POL on NHEJ efficiency at blunt DSBs was profound (0.61% of wild type) and virtually phenocopied the drastic decrement elicited by complete ablation of LigD or Ku (Fig. 2). The key finding was that loss of the whole POL domain was 35-fold more deleterious to blunt NHEJ efficiency than was simple loss of the polymerase activity (Fig. 2). The fidelity of blunt NHEJ in ligD-(ΔPOL) cells (5.8%) was 10-fold lower than in wild-type bacteria, again mimicking the effects of a full ΔligD-null mutation, rather than the increased fidelity seen in the polymerase-defective strain. The outcomes of unfaithful blunt NHEJ in the ligD-(ΔPOL) strains were notable for the strong skew toward deletions (22 of 23 events, 13 of which involved microhomologies) and the paucity of single nucleotide insertions between otherwise intact EcoRV ends (one of 23 events) (Fig. 4; Supplemental Fig. S4A). Although rarely encountered when LigD POL is absent, the latter type of junction implies that another mycobacterial polymerase is able to add a nontemplated nucleotide to a blunt DNA end.
The POL domain deletion exerted similar suppressive effects on the efficiency of 5′ overhang NHEJ (to 4.6% of the wild-type level), albeit not to the full extent seen with the ΔligD-null mutant, but in stark contrast to the lack of effect of polymerase-inactivating point mutations on NHEJ efficiency (Fig. 2). The increased fidelity of 5′ overhang NHEJ in ligD-(ΔPOL) cells (85%) mimicked that seen in the ligD-(D136A–D138A) strain. Here too, templated nucleotide additions were well represented at the repair junctions (11 of 21 events, entailing full or partial fill-in at one end plus a deletion at the other end), notwithstanding the lack of the LigD POL domain (Fig. 4; Supplemental Fig. S4B). Compared with the outcomes in the ligD-(D136A–D138A) strain, the NHEJ products in ligD-(ΔPOL) cells were notable for the absence of junctions that were filled-in at both ends (Supplemental Figs. S3B, S4B).
The effect of the POL domain deletion on 3′ overhang NHEJ was to elicit only a threefold decrement in repair efficiency, without affecting the intrinsically high fidelity. In this pathway, ligD-(ΔPOL) phenocopied Δku and ΔligD.
These results prompt the conclusion that the LigD POL domain is an essential structural component of the Ku-dependent mycobacterial NHEJ apparatus, for reasons having naught to do with its polymerase activity.
Effects of inactivating versus deleting the LigD LIG domain
The LIG domain of bacterial LigD resembles other ligases in that it catalyzes strand joining via a series of three nucleotidyl transfer reactions (Akey et al. 2006; Tomkinson et al. 2006). In the first step, attack on the α phosphorus of ATP by ligase results in the formation of a covalent ligase-adenylate intermediate in which AMP is linked via a phosphoamide bond to a lysine side chain (Lys484 in the case of M. smegmatis LigD; Lys481 in M. tuberculosis LigD). In the second step, the AMP is transferred to the 5′ end of the 5′ phosphate-terminated DNA strand to form a DNA-adenylate intermediate. In the third step, the ligase catalyzes an attack by a DNA 3′-OH on the DNA-adenylate to join the two polynucleotides and liberate AMP.
Prior studies have shown that changing Lys481 to alanine in the context of full-length M. tuberculosis LigD abolished the strand sealing reaction in vitro, by precluding formation of the LigD-AMP intermediate (Pitcher et al. 2005; Akey et al. 2006). The lysine-to-alanine change had no effect on the DNA polymerase activity of LigD (Akey et al. 2006). Here we tested the effects of the equivalent ligase-inactivating K484A mutation in M. smegmatis LigD on the efficiency and fidelity of blunt and 5′ overhang NHEJ in vivo, verifying with the improved plasmid-based assay that LIG inactivation had only modest effects on blunt and 5′-overhang NHEJ efficiencies in vivo (37% and 27% of the wild type, respectively), compared with the severe reductions in the ΔligD strain (Fig. 2).
The salient finding was that LIG inactivation drastically affected the fidelity of NHEJ at blunt DSBs, reducing it to 0.5% from the value of 57% seen in wild-type mycobacteria (Fig. 2). The spectrum of outcomes of mutagenic blunt NHEJ in the ligD-(K484A) strain (Supplemental Fig. S5A), comprising single nucleotide additions at otherwise intact blunt EcoRV sites to yield +1 frameshifts (six of 23), single nucleotide additions at one EcoRV end and unidirectional deletion of the other end (three of 23), and various unidirectional or bidirectional deletions (including three with uptake of distant plasmid sequences between the junction ends), was fairly similar to the mutagenic outcomes observed in wild-type cells. These results demonstrate that (1) LigD is not the exclusive source of ligase activity for Ku-dependent NHEJ in mycobacteria, and (2) reliance on a “backup” ligase virtually abolishes direct joining of two unprocessed blunt DSB ends; hence the drop in fidelity. It is conceivable that the backup ligase is inherently less proficient than LigD in blunt ligation or that access of the backup ligase (acting in trans) to the sealable ends is slowed compared with LigD (acting in cis), thereby allowing more time for nucleotide additions or deletions to occur before the joining step is executed.
Similar themes emerged from the analysis of 5′ overhang NHEJ outcomes in the ligD-(K484A) strain, where fidelity was reduced to 5.1%, compared with 43% in wild-type cells (Fig. 2). The unfaithfully repaired junctions were strongly skewed toward bidirectional deletions (in 20 of 22 events) and away from templated nucleotide insertions at the 5′ overhangs (only two of 20 events) (Fig. 4; Supplemental Fig. S5B), which is counter to the abundance of templated fill-ins seen in wild-type mycobacteria. We favor the idea that the slow action of the backup ligase provides more opportunity for nuclease action prior to sealing.
We showed previously that the lysine-to-alanine mutant of Mycobacterium LigD, though inert in sealing 3′-OH/5′-PO4 DNA nicks, retains the capacity to seal a preformed nicked DNA-adenylate intermediate (Akey et al. 2006). Therefore, it is conceivable that the LigD-K484A protein catalyzes the step of phosphodiester formation during NHEJ in vivo but relies on a different ligase to form a DNA-adenylate at the 5′ end of the processed repair junction. To probe this scenario, we introduced a different mutation at the ligase active site of M. smegmatis LigD—an alanine in lieu of an essential glutamate (Glu533) in nucleotidyltransferase motif III. The equivalent Glu-to-Ala change in M. tuberculosis LigD abolishes the ligase-adenylylation reaction (step 1) and the sealing of a preformed DNA-adenylate intermediate (step 3) (Akey et al. 2006). We observed that blunt and 5′ overhang NHEJ efficiencies in the M. smegmatis ligD-(E533A) strain were 13% and 11% of wild type, respectively (Fig. 2). These values are 42-fold and 14-fold higher than in the ΔligD-null strain, yet they are several-fold lower than the efficiencies seen in ligD-(K484A) (Fig. 2). Our results are consistent with a backup ligase providing DNA-adenylate intermediates for sealing by LigD-K484A, but they also underscore that a backup ligase can also function in NHEJ when all the reactions of LigD LIG are blocked. The ligD-(E533A) strain displayed very low fidelity of blunt NHEJ (0.42%). The outcomes of mutagenic repair included nontemplated single nucleotide additions to a blunt EcoRV end (four of 24 events), unidirectional deletions (11 of 24) and bidirectional deletions (10 of 24), some of which involved uptake of distant plasmid sequences at the junctions (Fig. 4; Supplemental Fig. S6A). The ligD-(E533A) strain also displayed reduced fidelity of 5′ overhang NHEJ (8.6%). Analysis of the unfaithful 5′ overhang repair junctions revealed deletions from at least one end in each case (n = 21) (Fig. 4; Supplemental Fig. S6B). Templated nucleotide additions at one of the 5′ overhang ends occurred in eight of 21 repair events (Supplemental Fig. S6B), a higher incidence than in the ligD-(K484A) strain.
To discriminate the contributions of the LigD ligase catalytic activity versus potential structural contributions of the LIG domain, we constructed a M. smegmatis mutant in which a ligD-(ΔLIG) allele (truncated to delete the C-terminal LIG domain) replaced the native ligD gene. Deletion of the LIG domain has no impact in vitro on the polymerase activity of the rump POL-PE version of the LigD protein (Gong et al. 2005). The ligD-(ΔLIG) strain resembled the ligase-defective strain with respect to its 12% efficiency of blunt NHEJ, which was overwhelmingly mutagenic (0.58% fidelity) (Fig. 2). The unfaithfully repaired blunt DSBs included a minority of single nucleotide insertions to the EcoRV ends to yield +1 frameshifts (three of 23 events) and a majority of deletions (20 of 23), most of which were bidirectional (Fig. 4; Supplemental Fig. S7A). The ΔLIG version of LigD apparently favored bidirectional deletions, which were relatively rare (one of 22) during blunt DSB repair in wild-type cells (Fig. 3). These results provide unambiguous evidence for a backup NHEJ ligase in a mutagenic pathway of blunt NHEJ.
The ligD-(ΔLIG) strain performed 5′ overhang NHEJ at one-sixth the efficiency of wild-type M. smegmatis. The 22% fidelity of 5′ overhang repair in the absence of a LIG domain was higher than the 5.1% and 8.6% fidelities noted for the ligase catalytic-defective LigD point mutants (Fig. 2), suggesting that the presence of an inactive LIG structural domain in the NHEJ complex might be an impediment to direct sealing of the sticky 5′ overhang ends by the backup ligase. The outcomes of mutagenic 5′ overhang NHEJ in the ligD-(ΔLIG) strain resembled those seen in ligD-(K484A) (Fig. 4) with respect to the prevalence of bidirectional deletions (18 of 20 junctions) and the low incidence of fill-in nucleotide additions (two of 20) accompanying a deletion at the other DSB end (Supplemental Fig. S7B).
The effects of inactivating LigD ligase or deleting the LIG domain on 3′ overhang NHEJ were modest (Fig. 2). Efficiencies of 42%–52% were higher than the 27% value in the ΔligD mutant, reflecting a partial decrement in the Ku-dependent route of 3′ overhang NHEJ (which accounts for about three-fourths of the 3′ overhang repairs in wild-type cells) when LigD sealing activity was lost. NHEJ at 3′ overhang DSBs was still extremely faithful (97%–99%) in the absence of LigD ligase (Fig. 2).
An ATP-dependent ligase provides the backup for LigD LIG
Which of the other mycobacterial ligases catalyzes the substantial residual levels NHEJ at blunt and 5′ overhang DSBs in the absence of LigD LIG? To address this question, we constructed M. smegmatis strains in which the ligB and ligC loci were deleted, and the ligD locus was either wild type (ligD+) or substituted by the ligase-defective ligD-(K484A) or ligD-(E533A) alleles. NHEJ efficiencies in the ligD+ ΔligB/C strain were as high or slightly higher than in wild-type cells, and the fidelities of NHEJ were similar to wild-type fidelities for the repair of blunt, 5′ overhang, and 3′ overhang DSB ends (Fig. 2). The spectrum of unfaithful repair outcomes at blunt and 5′ overhang DSB ends in the ligD+ ΔligB/C strain was fairly similar to that of wild-type cells (Supplemental Fig. S8). Thus, LigB and LigC appear not to be major contributors to NHEJ of linear plasmid DSBs when wild-type LigD is present.
The instructive findings were that the substantial residual blunt and 5′ overhang NHEJ activities observed in the ligD-(K484A) and ligD-(E533A) single-mutant strains were abolished in a ΔligB/C background (Fig. 2). Yet, 3′ overhang NHEJ persisted at one-fourth the wild-type efficiency in the ligD-(K484A) ΔligB/C strain, phenocopying ΔligB/C/D (Fig. 2). We conclude that (1) LigA cannot provide backup sealing activity for blunt or 5′ overhang NHEJ in M. smegmatis, and (2) the backup sealing function is provided by one of the ATP-dependent mycobacterial ligases, most probably LigC (see Discussion).
The residual blunt DSB repair events in the ligD-(K484A) ΔligB/C and ligD-(E533A) ΔligB/C strains were all unfaithful, always involved deletions, and relied heavily on microhomologies at the junctions (Supplemental Figs. S9A, S10A). These outcomes resemble those seen in other mutant strains (Δku, ΔligD, or ΔligB/C/D) in which blunt NHEJ was suppressed (Fig. 4). Similarly, the 0.12% efficiency of 5′ overhang NHEJ in ligD-(K484A) ΔligB/C and ligD-(E533A) ΔligB/C cells and the 52%–59% fidelities matched the other grossly NHEJ-defective mutants (Fig. 2). The junctions of the unfaithfully repaired 5′ overhang DSBs in ligD-(K484A) ΔligB/C and ligD-(E533A) ΔligB/C cells (Supplemental Figs. S9B, S10B) included many templated fill-ins but were most remarkable for the high incidence of bidirectional deletions that were joined with the aid of microhomologies. These outcomes were similar to those in Δku, ΔligD, or ΔligB/C/D cells (Fig. 4).
Effects of PE inactivation indicate that LigD is not the catalyst of deletion formation
There is ambiguity in the literature concerning the nature of the 3′ end-modifying functions of bacterial LigD and their possible roles in NHEJ. Doherty and colleagues reported that full-length M. tuberculosis LigD has a DNA 3′ exonuclease activity that they attributed to the PE domain based on the finding that a mutation within the PE domain suppressed the DNA exonuclease activity in vitro (Della et al. 2004). These investigators subsequently found that the isolated PE domain of M. tuberculosis LigD had no detectable DNA 3′ exonuclease activity (Pitcher et al. 2005). The high prevalence of deletions at repaired plasmid DSB ends in E. coli cells expressing mycobacterial Ku and LigD was cited in support of a model whereby a LigD nuclease is responsible for deletion formation in vivo (Malyarchuk et al. 2007).
Studies in our laboratory of LigD PE domains from Pseudomonas, Agrobacterium, and Mycobacterium indicate that LigD is a bifunctional 3′ ribonuclease/3′ phosphatase that resects a short tract of 3′ ribonucleotides on a primer-template substrate to the point at which the primer strand has a single 3′ ribonucleotide remaining (Zhu and Shuman 2005b, 2006, 2007; Zhu et al. 2005). The failure to digest beyond this point reflects a requirement for a 2′-OH group on the penultimate nucleoside of the primer strand, which explains why no DNA 3′ exonuclease was detected in our assays. The ribonucleotide resection activity resides within the PE domain and is the result of at least two component steps: (1) The 3′-terminal nucleoside is first removed to yield a primer strand with a ribonucleoside 3′-PO4 terminus, and (2) the 3′-PO4 is hydrolyzed to a 3′-OH. The PE domain also catalyzes hydrolysis of the 3′-PO4 of an all-DNA primer-template substrate. The phosphodiesterase and phosphomonoesterase activities are both dependent on the presence and length of the 5′ single-strand tail of the primer-template substrate. These properties distinguish LigD PE from other 3′ end-processing enzymes and suggested to us that PE activity might come into play during gap repair or remodeling of DSBs with recessed 3′ ends, though not as the likely culprit in forming the large deletion tracts prevalent during unfaithful repair of blunt and 5′ overhang DSBs in vivo in wild-type M. smegmatis.
To address the role of the PE domain in vivo, we constructed M. smegmatis strains in which the native ligD locus was substituted by the mutant alleles ligD-(E310A) or ligD-(H336A). Glu310 is conserved in LigD PE domains from diverse bacteria and is specifically essential for the polynucleotide 3′ phosphomonoesterase activity, though not for the phosphodiester cleavage step of ribonucleotide resection (Zhu et al. 2005; Zhu and Shuman 2006). The efficiencies and fidelities of blunt, 5′ overhangs, and 3′ overhang DSB repair in the ligD-(E310A) strain were virtually identical to wild-type M. smegmatis (Fig. 2). Moreover, the prevalence of deletions at one or both ends during unfaithful blunt or 5′ overhang NHEJ was no less in ligD-(E310A) than in wild-type bacteria (Fig. 4; Supplemental Fig. S11). His336 is also strictly conserved in LigD PE domains and is essential for both the 3′ phosphomonoesterase and 3′ phosphodiesterase activities (Zhu et al. 2005; Zhu and Shuman 2006). The efficiencies of blunt, 5′ overhangs, and 3′ overhang DSB repair in the ligD-(E310A) strain were minimally affected (60%–87% of wild type), and the fidelities were virtually identical to wild-type M. smegmatis (Fig. 2), The incidence (and lengths) of deletions during unfaithful blunt or 5′ overhang NHEJ was no less in ligD-(H336A) than in wild-type M. smegmatis (Fig. 4; Supplemental Fig. S12). These results exclude LigD PE as the source of the nuclease for deletion formation during mutagenic NHEJ of linear plasmid DSBs in vivo.
RecBCD is not the principal agent of deletion formation
Bacterial RecBCD is a heterotrimeric nuclease/helicase that rapidly and processively resects DSB ends in vivo and in vitro (Singleton et al. 2004). An M. smegmatis ΔrecBCD mutant deleted for all three subunits is viable (Stephanou et al. 2007). Here we found that ablation of RecBCD had virtually no effect on the efficiency or fidelity of blunt, 5′ overhang, or 3′ overhang NHEJ (Fig. 2). The spectrum of unfaithful NHEJ outcomes at blunt and 5′ overhang DSBs in the ΔrecBCD mutant was similar to wild-type mycobacteria (Fig. 4). In particular, there was no decline in the prevalence or size of the deletion tracts (Supplemental Fig. S13), indicating that RecBCD is not the principal catalyst of deletion formation during mycobacterial NHEJ in vivo.
Discussion
The studies presented above support a new and more complex view of mycobacterial NHEJ instead of the simple two-component model (Della et al. 2004), which held that Ku and LigD are necessary and sufficient to perform all the physiological reactions of NHEJ in vivo. Whereas it is the case that the Ku and LigD proteins are stringently required for the repair of blunt and 5′ overhang DSBs in Mycobacterium, the Ku/LigD pathway is but one of two options available to rectify cohesive 3′ overhang DSBs. We estimate that three-fourths of the 3′ overhang NHEJ events in wild-type cells flux through the Ku/LigD system, while one-fourth of the repairs are catalyzed by a novel mechanism. Our experiments show that only the POL domain of LigD is strictly required for the Ku-driven NHEJ pathway, in which POL plays a structural role that is genetically separable from its ability to catalyze nucleotide additions at DSB ends.
The working model of bacterial NHEJ is that Ku binds to the broken DNA ends and recruits LigD. Physical interaction of Ku and LigD has been demonstrated by detection of a DNA–Ku–LigD ternary complex by native gel electrophoresis and identification of Ku in a genome-wide yeast two-hybrid screen for LigD interaction partners (Weller et al. 2002; Gong et al. 2005). Both assays implicate the POL domain as the major point of contact between LigD and Ku (Pitcher et al. 2005; N.C. Stephanou and M.S. Glickman, unpubl.). This engenders the prediction that, absent the POL domain, the LigD protein composed of PE and LIG domains would not be effectively recruited to Ku-bound DSB ends. The failure of the “backup” ATP-dependent ligase to work in this case suggests that POL might have a function beyond recruitment of the sealing module. Indeed, the recently reported crystal structure of the mycobacterial POL domain bound to DNA (Brissett et al. 2007) provides a key insight to the NHEJ mechanism, by implicating LigD POL in synapsis of the broken DNA ends. Accordingly, the core of the NHEJ complex could plausibly consist of the toroidal Ku homodimer engaged near the DSB end with the LigD POL domain tethered to Ku on the side facing toward the break (Fig. 7A).
Figure 7.
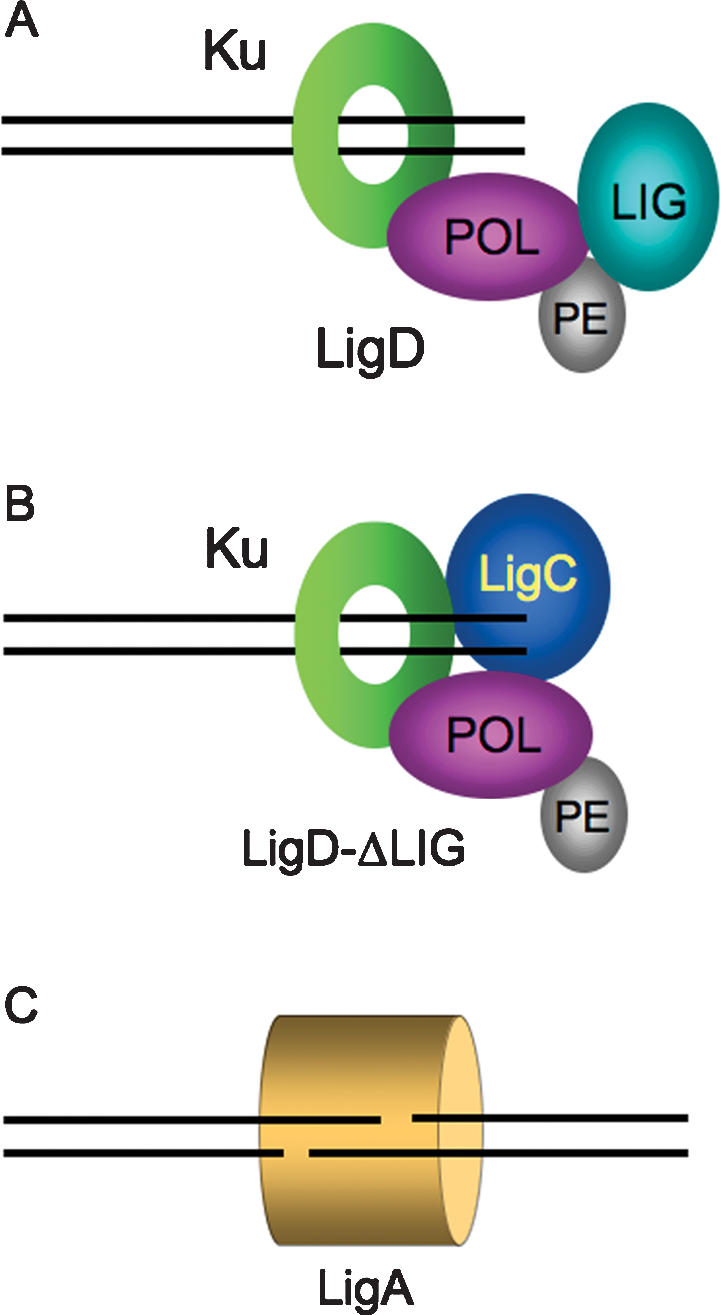
Multiple NHEJ pathways in mycobacteria. Two alternative pathways of Ku-dependent blunt DSB repair are depicted in A and B. (A) In wild-type cells, the NHEJ apparatus is assembled at the DSB via contacts between the Ku homodimer (which encircles the DNA as a toroidal clamp) and the POL domain of LigD. The choice between faithful and unfaithful DSB repair reflects a kinetic balance between immediate sealing by the LIG domain (in the context of a synaptic complex with the other DSB end), nontemplated single nucleotide addition by the LigD POL domain, and end resection by an unknown mycobacterial nuclease. (B) Absent the LigD LIG domain (or when LigD ligase activity is suppressed), LigC provides backup sealing activity for blunt and 5′ overhang NHEJ. Because all blunt/5′overhang NHEJ requires the POL domain, we surmise that LigC interacts with the Ku–POL complex. Virtually all NHEJ events that proceed via this pathway are mutagenic. (C) M. smegmatis has a Ku-independent NHEJ pathway specific to sticky 3′ overhang DSBs that is nonmutagenic and, being active in ΔligB/C/D cells, apparently relies on LigA. It is not known if other accessory factors participate in this novel pathway.
When full-length wild-type LigD is assembled in the complex, the outcomes of NHEJ are strongly dependent on the structure of the initial DSB ends. Blunt and complementary 5′ overhang DSBs are repaired with a high incidence of mutation (43%–57%), while 3′ complementary overhang repair is remarkably faithful (4% error rate if one considers only the fraction of 3′ overhang repairs that are Ku-dependent). The present study shows how the decision between faithful and unfaithful NHEJ at a particular DSB end is determined by a balance between LigD ligase and LigD polymerase activities and the action of mycobacterial nuclease(s) that resect the DSB ends. A genetic manipulation that abolishes LigD polymerase activity decreases the mutation frequency of blunt NHEJ (from 43% to 7%) by eliminating the nontemplated single nucleotide additions that are a major source of infidelity in wild-type mycobacteria. Loss of LigD polymerase activity also decreases the mutation frequency of 5′ overhang NHEJ (from 67% to 19%), yet templated fill-ins of the 5′ overhang ends are still well-represented among the residual unfaithful repair events; they are apparently catalyzed by a different mycobacterial polymerase. It is satisfying to note that the NHEJ reactions impacted by the polymerase-inactivating LigD mutations in vivo (nontemplated blunt addition and short templated fill-in) are the ones at which LigD POL is most adept in vitro (Gong et al. 2005; Pitcher et al. 2005; Zhu and Shuman 2005a).
LigD POL is also capable of adding nucleotides in vitro to ssDNA primers and to DNA duplexes with 3′ single-stranded overhangs (Della et al. 2004; Zhu and Shuman 2005a; Brissett et al. 2007), which are analogous to the KpnI-cut plasmid ends used here to study NHEJ in vivo. The enhancing effect of a nearby 5′-PO4 terminus on POL binding to a gapped DNA duplex (Pitcher et al. 2007c) is especially pertinent to explain many of the mutagenic 3′ overhang repair junctions formed in wild-type M. smegmatis. In these cases (indicated by check marks in Fig. 5), there is potential for POL-assisted cross-junction synapsis to form a gap that could then be filled-in by templated nucleotide addition (Brissett et al. 2007). Although our studies of 3′ overhang NHEJ in vivo provide evidence for such a mechanism, they underscore that nucleotide additions to a 3′ tailed duplex represent only a tiny fraction of the total flux of 3′ overhang plasmid DSB repair. In the plasmid-based assay, we did not observe any unfaithful junctions containing an extra nucleotide at otherwise unresected KpnI ends, indicating that nontemplated addition to a 3′ single-stranded tail is either unlikely to occur or does not eventuate in repair (in contrast to nontemplated blunt-end additions, which are quite common in mycobacterial NHEJ). NHEJ events entailing incorporation of nontemplated nucleotides at 3′ overhang chromosomal DSBs can be detected when relentless selection pressure is applied so that survival of M. smegmatis depends on accumulating mutations at the repair junction (Stephanou et al. 2007).
Interdiction of the LigD ligase activity drastically increases the mutation frequency of blunt and 5′ overhang NHEJ and strongly skews the outcome toward deletions, indicating that delayed or inefficient strand sealing by the backup ligase affords more opportunity for DSB end resection by nuclease action. Our findings suggest that the backup ligase can either provide DNA-adenylate that can be sealed by the LigD K484A mutant or provide all sealing activities when LigD LIG is deleted (albeit with diminished efficiency). LigC emerges as the best candidate for the backup ligase (Fig. 7B) because of its close structural and functional similarities to the LigD LIG domain, including (1) relatively feeble activity in sealing nicked DNA duplexes; (2) propensity to accumulate the DNA-adenylate intermediate during DNA nick-sealing in the presence of ATP; and (3) the ability of LigC to catalyze plasmid DSB repair in vitro in the presence of Ku (Gong et al. 2004; Zhu and Shuman 2007). In contrast, LigB is an efficient nick-sealing enzyme, by virtue of an N-terminal DNA-binding domain that is lacking in LigD and LigC (Fig. 1A), and it does not accumulate DNA-adenylate during strand joining in vitro (Gong et al. 2004).
Our results indicate that the PE activity of LigD does not catalyze deletion formation and does not obviously participate in plasmid-based NHEJ with either blunt or 5′ overhang DSB substrates. What then is the purpose of the PE activity? The DNA 3′ phosphatase function could be relevant to repair scenarios in which the inciting damage produces 3′ modified ends that cannot be either extended by a polymerase or sealed by a ligase. For example, DNA breaks with 3′-PO4 termini can be generated directly by nucleolytic cleavage or ionizing radiation, or as intermediates formed during base excision repair. Because 3′-PO4 ends would probably not arise during NHEJ of transfected linear plasmids, it is sensible that PE-inactivating mutations of mycobacterial LigD would not affect NHEJ in this in vivo assay format.
The identity of the mycobacterial nuclease(s) responsible for deletion formation during blunt and 5′ overhang NHEJ remains to be determined. Here we provided genetic evidence that RecBCD is neither the sole catalyst nor the predominant agent of DSB end resection. Either RecBCD is unable to degrade the DSBs destined for NHEJ—e.g., because they might be protected by Ku—or there are other mycobacterial nucleases available to generate deletions at DSB ends when RecBCD is absent. With respect to the latter model invoking functional redundancy, it is intriguing that the ΔrecBCD-null strain is no more sensitive than wild-type M. smegmatis to killing by UV irradiation (Stephanou et al. 2007), whereas E. coli is acutely reliant on RecBCD to withstand UV damage.
It is not clear what purpose is served by mycobacterial NHEJ being so highly error-prone in the repair of complementary 5′ overhang and blunt DSB ends, while simultaneously maintaining high fidelity for NHEJ at complementary 3′ overhangs. Although mutagenic DSB repair might seem counterproductive, it could have advantages in certain stress situations such as stationary phase or starvation (Ponder et al. 2005). Specifically, given the choice between certain death from a chromosomal break and survival via NHEJ with a single gene mutation, the quiescent mycobacterial cell seems better off opting for survival. NHEJ might even play a role in mycobacterial resistance to isoniazid, a first-line drug for treatment of human tuberculosis, which is conferred by loss-of-function mutations in the katG gene (Zhang et al. 1992).
The advantages to the Mycobacterium of highly faithful 3′ overhang NHEJ are uncertain, but this unique feature is apparently exploited by the mycobacteriophages Omega and Corndog, which depend on host LigD for plaque formation (Pitcher et al. 2006). Neither virus requires the host Ku, but each virus encodes its own Ku homolog. Phage infection requires the ligase activity of LigD, but not the polymerase activity. LigD is needed to circularize the mycobacteriophage genome, which entails sealing of complementary 4-base 3′ single-strand overhangs at the ends of the linear phage DNA (Pitcher et al. 2006). It is sensible that sealing of the phage 3′ overhang ends ought to be error-free (as we demonstrate here for plasmid 3′ overhangs) in order to allow for proper site-specific cleavage of the replicated progeny phage DNAs into unit-genome-length linear molecules. It is unclear why the LigD-independent pathway of faithful 3′ overhang NHEJ described here (Fig. 7C) is not available to the infecting mycobacteriophage. We speculate that the mycobacteriophage-encoded Ku might be deposited at the ends of the linear phage genome during DNA encapsidation, where it is poised to direct sealing of the injected phage DNA down the LigD-dependent pathway during the next round of infection. Mycobacteriophage Ku, in common with the Ku-like Gam protein of bacteriophage Mu (d’Adda di Fagagna et al. 2003), is a homodimer that binds to linear DNA ends and protects them from exonucleolytic digestion (Pitcher et al. 2006).
Materials and methods
LigD mutants
Plasmid pET-MsmLigD encodes the M. smegmatis LigD polypeptide fused to an N-terminal His10 tag (Gong et al. 2005). Missense mutations that selectively ablate LigD’s polymerase (D136A–D138A), phosphoesterase (E310A or H336A), or ligase (K484A or E533A) activities were introduced into the ligD gene by the two-stage PCR overlap extension method using pET-MsmLigD as the template. An N-terminal truncation mutant, ligD-(ΔPOL), was generated by PCR amplification with a sense-strand primer that introduced an NdeI site overlapping a new translation start codon in lieu of the codon for Glu289. A C-terminal truncation mutant, ligD-(ΔLIG), was generated by PCR amplification with an antisense-strand primer that introduced a stop codon in lieu of amino acid 449. The mutated PCR products were inserted into pET16b, and the inserts were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning.
Plasmid pMSG346 (marked with selectable and counterselectable hyg and sacB genes) was used to perform allelic exchange at the ligD locus of a M. smegmatis ΔligD strain (Gong et al. 2005). pMSG346 contains 503 base pairs (bp) of genomic DNA 5′ of the ligD ORF and 490-bp of genomic DNA 3′ of the ORF, with an NdeI site introduced at the start codon and a BamHI site introduced 37 bp 3′ of the stop codon. The mutated ligD genes described above were excised from the pET vector and inserted between the NdeI and BamHI sites of pMSG346. The resulting plasmids were transformed into the ΔligD strain; allelic exchange was executed by the two-step selection/counterselection strategy (Braunstein et al. 2001). A control allelic exchange was performed with pMSG346 containing the wild-type ligD gene. Restoration of the ligD locus was confirmed in each case by Southern hybridization.
ΔligB/C/D triple knockout
The ligD gene was deleted in a M. smegmatis ΔligB/C strain (Gong et al. 2005) by two-step allelic exchange for the ligD knockout cassette employed previously to make the ΔligD strain (Gong et al. 2005). The resulting ΔligB/C/D mutant was then used to reintroduce the following alleles at the ligD locus: wild type (ligD+), ligD-(K484A), and ligD-(E533A). The initial ligD deletion and subsequent allelic exchanges of the ligD locus in the ΔligB/C background were confirmed by Southern hybridization with diagnostic probes.
Improved NHEJ reporter plasmids
The original NHEJ reporter plasmid, pMSG288 (Gong et al. 2005), was modified by insertion of a 3.8-kbp EcoRV fragment from a different plasmid into the unique EcoRV site within the lacZ gene (Fig. 1B) or by insertion of a 3-Kbp Asp7181 fragment from another plasmid into the unique Asp7181 site in the lacZ gene (Fig. 1C). For the linear NHEJ substrates, plasmid was sequentially digested with KpnI + PsiI (3′ overhangs), Asp7181 + PsiI (5′ overhangs), or EcoRV + XbaI (blunt ends). The digestion products were gel-purified using Qiagen gel purification kit.
NHEJ assay
The NHEJ assay was performed and the results were quantified as described previously (Gong et al. 2005; Akey et al. 2006; Zhu et al. 2006). Briefly, between 8 and 108 ng of linearized plasmid DNA (or 3–66 ng of uncut circular plasmid) were electroporated into 400 μL of competent M. smegmatis prepared according to Snapper et al. (1990). The overhang substrates were incubated for 10 min at 65°C to dissociate the sticky ends prior to electroporation. After electroporation, the cells were transferred immediately to 1 mL of Luria-Bertani medium and incubated for 2–4 h at 37°C with constant shaking (150 rpm). Three dilutions of the transformation mixtures were then plated on Middlebrook 7H10 agar medium supplemented with 0.5% glycerol, 0.5% dextrose, 20 μg/mL kanamycin, and 50 μg/mL X-gal. Colony counts and colony color (blue/white) were assessed after incubation for 3 d at 37°C. Efficiency was calculated as the ratio of colonies per nanogram of transformed linear DNA versus colonies per nanogram of circular DNA. Absolute NHEJ efficiency values in wild-type M. smegmatis for repair of blunt, 5′ overhang, and 3′ overhang DSB ends were 1%, 0.6%, and 7.2%, respectively (the values are the averages of four separate experiments). The NHEJ efficiency values for mutant strains were normalized to the efficiency value of wild-type M. smegmatis (defined as 100%) and are reported in Figure 2. Fidelity was calculated as percentage lacZ+ (blue) transformants.
NHEJ plasmid junction sequencing
Pairs of convergently oriented primers flanking the DSB ends of the reporter plasmids were used to PCR-amplify DNA from individual kanR lacZ– transformants and were described previously (Gong et al. 2005). Amplifications were performed with primers at sites located 795 bp and 1252 bp from the original Asp7181/KpnI sites or 792 bp and 693 bp from the original EcoRV sites. In case these primers failed to amplify the repair junction (because NHEJ entailed deletion beyond one or both of the primer sites), a second set of primers was located 1759 bp and 2251 bp from the original Asp7181/KpnI sites or 1911 bp and 3148 bp from the original EcoRV sites. The PCR products were sequence using the primers used for amplification and various nested sets of internal primers located 140 bp and 380 bp on either side of the original restriction sites. In the few cases in which the second set of primers failed to amplify DNA from the M. smegmatis colony, we recovered the plasmid by transformation into E. coli, purified the plasmid DNA, and then sequenced the junction.
Acknowledgments
This research was supported by NIH grant AI064693. J.A. is trainee of the Tri-Institutional MD-PhD program at Weill Cornell Medical College, supported by NIH grant GM07739. S.S. is an American Cancer Society Research Professor.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1631908
References
- Akey D., Martins A., Aniukwu J., Glickman M.S., Shuman S., Berger J.M. Crystal structure and nonhomologous end joining function of the ligase domain of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Brown A.M., Kurtz S., Jacobs W.R. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 2001;183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissett N.C., Pitcher R.S., Juarez R., Picher A.J., Green A.J., Dafforn T.R., Fox G.C., Blanco L., Doherty A.J. Structure of a NHEJ polymerase-mediated synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- di d’Adda Fagagna F., Weller G.R., Doherty A.J., Jackson S.P. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003;4:47–52. doi: 10.1038/sj.embor.embor709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della M., Palmbos P.L., Tseng H.M., Tonkin L.M., Daley J.M., Topper L.M., Pitcher R.S., Tomkinson A.E., Wilson T.E., Doherty A.J. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- Ferreira M.G., Cooper J.P. Two modes of DNA double-strand break repair are reciprocally regulated through fission yeast cell cycle. Genes & Dev. 2004;18:2249–2254. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Martins A., Bongiorno P., Glickman M., Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- Gong C., Bongiorno P., Martins A., Stephanou N.C., Zhu H., Shuman S., Glickman M.S. Mechanism of nonhomologous end-joining in mycobacteria: A low fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- Korycka-Machala M., Rychta E., Brzostek A., Sayer H.R., Rumijowska-Galewicz A., Bowater R.P., Dziadek J. Evaluation of NAD+-dependent DNA ligase of mycobacteria as a potential target for anitbiotics. Anitmicrob. Agents Chemother. 2007;51:2888–2897. doi: 10.1128/AAC.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk S., Wright D., Castore R., Klepper E., Weiss B., Doherty A.J., Harrison L. Expression of Mycobacterium tuberculosis Ku and Ligase D in Escherichia coli results in RecA and RecB-independent DNA end-joining at regions of microhomology. DNA Repair (Amst.) 2007;6:1413–1424. doi: 10.1016/j.dnarep.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller R., Stackebrandt E., Reitz G., Berger T., Rettberg P., Doherty A.J., Horneck G., Nicholson W.L. Role of DNA repair by nonhomologous end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 2007;189:3306–3311. doi: 10.1128/JB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher R.S., Tonkin L.M., Green A.J., Doherty A.J. Domain structure of a NHEJ repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 2005;351:531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Pitcher R.S., Tonkin L.M., Daley J.M., Palmbos P.L., Green A.J., Velting T.L., Brzostek A., Korycka-Machala M., Cresawn S., Dziadek J., et al. Mycobacteriophage exploit NHEJ to facilitate genome circularization. Mol. Cell. 2006;23:743–748. doi: 10.1016/j.molcel.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Pitcher R.S., Brissett N.C., Doherty A.J. Nonhomologous end-joining in bacteria: A microbial perspective. Annu. Rev. Microbiol. 2007a;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- Pitcher R.S., Green A.J., Brzostek A., Korycka-Machala M., Dziadek J., Doherty A.J. NHEJ protects mycobacteria in stationary phase against the harmful effects of dessication. DNA Repair (Amst.) 2007b;6:1271–1276. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Pitcher R.S., Brissett N.C., Picher A.J., Andrade P., Juarez R., Thompson D., Fox G.C., Blanco L., Doherty A.J. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007c;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Ponder R.G., Fonville N.C., Rosenberg S.M. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Shuman S., Glickman M.S. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Snapper S.B., Melton R.E., Mustafa S., Kieser T., Jacobs W.R. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Stephanou N.C., Gao F., Bongiorno P., Ehrt S., Schnappinger D., Shuman S., Glickman M.S. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Tamaguchhi-Iwai Y., Shinohara A., Takeda S. Homologous recombination and nonhomologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson A.E., Vijayakumar S., Pascal J.M., Ellenberger T. DNA ligases: Structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- Wang S.T., Setlow B., Conlon E.M., Lyon J.L., Imamura D., Sata T., Setlow P., Losick R., Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Weller G.R., Kysela B., Roy R., Tonkin L.M., Scanlan E., Della M., Devine S.K., Day J.P., Wilkinson A., di d’Adda Fagagna F., et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- Yakovleva L., Shuman S. Nucleotide misincorporation, 3′-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J. Biol. Chem. 2006;281:25026–25040. doi: 10.1074/jbc.M603302200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Heym B., Allen B., Young D., Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- Zhu H., Shuman S. A primer-dependent polymerase function of Pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J. Biol. Chem. 2005a;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005b;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Shuman S. Substrate specificity and structure-function analysis of the 3′-phosphoesterase component of the bacterial NHEJ protein, DNA Ligase D. J. Biol. Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Shuman S. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 2007;35:3631–3645. doi: 10.1093/nar/gkm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Wang L.K., Shuman S. Essential constituents of the 3′-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Nandakumar J., Aniukwu J., Wang L.K., Glickman M.S., Lima C.D., Shuman S. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl. Acad. Sci. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]