Abstract
A key effector of the canonical Wnt pathway is β-catenin, which binds to TCF/LEF factors to promote the transcription of Wnt target genes. In the absence of Wnt stimulation, β-catenin is phosphorylated constitutively, and modified with K48-linked ubiquitin for subsequent proteasomal degradation. Here, we identify Trabid as a new positive regulator of Wnt signaling in mammalian and Drosophila cells. Trabid show a remarkable preference for binding to K63-linked ubiquitin chains with its three tandem NZF fingers (Npl4 zinc finger), and it cleaves these chains with its OTU (ovarian tumor) domain. These activities of Trabid are required for efficient TCF-mediated transcription in cells with high Wnt pathway activity, including colorectal cancer cell lines. We further show that Trabid can bind to and deubiquitylate the APC tumor suppressor protein, a negative regulator of Wnt-mediated transcription. Epistasis experiments indicate that Trabid acts below the stabilization of β-catenin, and that it may affect the association or activity of the TCF–β-catenin transcription complex. Our results indicate a role of K63-linked ubiquitin chains during Wnt-induced transcription.
[Keywords: Wnt pathway, β-catenin, APC tumour suppressor, deubiquitylation, K63-linked ubiquitin, NZF zinc finger, transcription]
The Wnt signaling pathway regulates numerous processes during normal animal development. It also controls stem and progenitor cell types in adult tissues, including those in the intestinal epithelium (Reya and Clevers 2005). In humans, the majority of sporadic and hereditary colorectal tumors are due to hyperactivation of the Wnt pathway caused by inactivating mutations in APC (Adenomatous polyposis coli) or activating mutations in β-catenin (Bienz and Clevers 2000; Polakis 2000).
In the absence of Wnt stimulation, an active mechanism operates to down-regulate β-catenin, the key effector of the canonical Wnt pathway (Kimelman and Xu 2006). This involves the constitutive phosphorylation of β-catenin by the Axin complex that also includes APC, glycogen synthase kinase 3β (GSK3β), and casein kinase I. Phosphorylated β-catenin is recognized by β-TrCP, an F-box-containing adaptor, which binds to an SCF ubiquitin ligase (Winston et al. 1999). The latter conjugates a K48-linked ubiquitin chain to phosphorylated β-catenin, which earmarks it for subsequent proteasomal degradation (Aberle et al. 1997). Binding of Wnt ligand to its transmembrane receptor leads to Dishevelled-mediated inhibition of the Axin complex; as a consequence, unphosphorylated β-catenin accumulates and associates with TCF/LEF factors in the nucleus, to stimulate the transcription of Wnt target genes. This involves the recruitment of a wide range of transcriptional cofactors to the C terminus of β-catenin, including the chromatin modifier and remodeling factors CBP, Brg-1, and SET1 (Hecht et al. 2000; Takemaru and Moon 2000; Barker et al. 2001; Sierra et al. 2006); TATA-binding and associated factors (Hecht et al. 1999; Bauer et al. 2000); and the transcriptional elongation factor Parafibromin (Mosimann et al. 2006). Through a more N-terminal segment, β-catenin binds to the BCL9–Pygopus complex, to recruit an unknown transcriptional coactivator (Kramps et al. 2002) and/or to facilitate efficient recruitment of β-catenin to TCF target genes (Townsley et al. 2004; de la Roche and Bienz 2007). Thus, the key output of canonical Wnt signaling is a switch in transcription.
The same SCF ubiquitin ligase complex that regulates β-catenin also controls the NF-κB pathway: Following TNF-α stimulation, this complex conjugates a K48-linked ubiquitin chain to phosphorylated IκB, an inhibitor of NF-κB. The subsequent proteasomal degradation of ubiquitylated IκB allows NF-κB to accumulate in the nucleus and to stimulate the transcription of NF-κB target genes (Chen 2005). Notably, several components of the NF-κB pathway are also regulated by K63-linked polyubiquitylation. The TNF receptor-associated factors TRAF2, TRAF6, RIP, and the NEMO subunit of the IκB kinase are modified by K63-linked ubiquitin chains (Wang et al. 2001; Shi and Kehrl 2003; Ea et al. 2006; Wu et al. 2006), which are thought to mediate the recruitment and activation of their effectors. For example, the binding of TAB2 to K63-polyubiquitylated TRAF6, NEMO, and RIP appears to be required for activation of NF-κB (Kanayama et al. 2004). TAB2 binds to its polyubiquitylated substrates via its NZF motif (Npl4 zinc finger), one of many different ubiquitin-binding domains (Hicke et al. 2005) that bind to the commonly recognized hydrophobic “Ile44 patch” of the ubiquitin surface (Alam et al. 2004).
The deubiquitylating (DUB) enzyme A20 antagonizes the NF-κB pathway, by removing the K63-linked polyubiquitylation of RIP, which appears to be necessary for its ability to down-regulate TNF-α-induced NF-κB activity (Wertz et al. 2004). A20 belongs to the OTU (ovarian tumor) family of DUBs, which includes the largely uncharacterized proteins Cezanne and Trabid. The OTU domain is a conserved cysteine protease that possesses DUB activity (Balakirev et al. 2003). Overexpressed Cezanne can also down-regulate NF-κB signaling, whereas overexpressed Trabid had negligible effects on this pathway (Evans et al. 2001).
Here, we report our discovery of Trabid as a new APC-interacting protein and a positive regulator of Wnt responses in mammalian cells and Drosophila. We show that its variant OTU domain exhibits DUB activity, and that it deubiquitylates APC. Its remarkable preference for binding to, and cleaving, K63-linked ubiquitin chains are critical for Trabid’s function in TCF-mediated transcription. Epistasis analysis suggests that the TCF–β-catenin complex is the functional target of Trabid.
Results
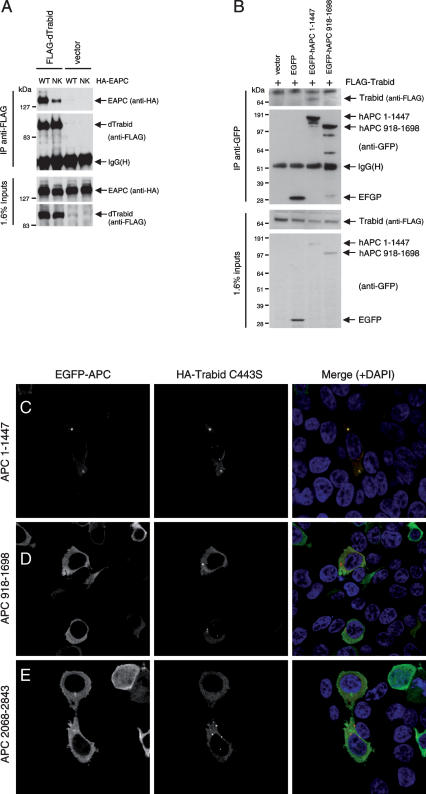
We conducted a yeast two-hybrid (Y2H) screen with the armadillo repeat domain (ARD) of mouse Apc, and counterscreened 13 independent positives with a mutant ARD bearing a point mutation in its putative surface that, in Drosophila, causes inactivation of E-APC (Hamada and Bienz 2002). This revealed two isolates that were sensitive to this mutation (Supplemental Fig. S1A). One of these encodes an internal fragment of mouse Cezanne overlapping its OTU domain (residues 252–403), which shows high sequence similarity to Trabid, and also to dTrbd, the Trabid ortholog in Drosophila and only representative of the A20 OTU family in flies (Supplemental Fig. S1B). The Cezanne Y2H fragment, and a similar OTU-spanning fragment from human Trabid, both coimmunoprecipitate preferentially with wild type rather than mutant ARD in cotransfected 293 cells (Supplemental Fig. S1C). The same selective association is also seen with full-length Flag-dTrbd (Fig. 1A). Furthermore, Flag-Trabid coimmunoprecipitates with coexpressed GFP-APC(1–1447), an ARD-spanning APC fragment similar to APC type 1 truncations typically found in colorectal cancer cells (Rosin-Arbesfeld et al. 2003), but not with coexpressed GFP-APC(918–1698) that lacks the ARD (Fig. 1B). These sequence-specific interactions in yeast and mammalian cells indicate that Trabid, Cezanne, and dTrbd may be bona fide direct ARD ligands.
Figure 1.
Association between Trabid and APC proteins. (A,B) Immunoprecipitations between Flag-dTrbd and Drosophila APC (HA-E-APC) (A) or Flag-Trabid and an N-terminal truncation of human APC, GFP-APC(1–1447) (B), in cotransfected 293 cells, as indicated in panels. (C–E) Immunofluorescence of 293 cells cotransfected with HA-Trabid C443S and different GFP-tagged APC fragments, as indicated, fixed and stained with α-HA antibody (red in merge) and DAPI (blue in merge, to reveal the nuclei; see also Supplemental Fig. S5). Recruitment of GFP-APC(1–1447) into C443S puncta was observed in 9% of transfected cells (n = 100); neither APC(918–1698) nor APC(2068–2843) colocalized with these puncta (n = 100).
To gain further support for this, we used a colocalization assay, exploiting a Trabid mutant (C443S) (see below; Fig. 3, below) that forms distinct cytoplasmic puncta in cells: A significant fraction of transfected 293 cells (9%) show recruitment of APC(1–1447) into these puncta (Fig. 1C), but we never see recruitment of APC(918–1698) or of a C-terminal APC fragment (Fig. 1D,E). These results suggest that the association between APC and Trabid may be conditional or labile, which could explain why we were not able to detect reliably the association of endogenous APC and Trabid.
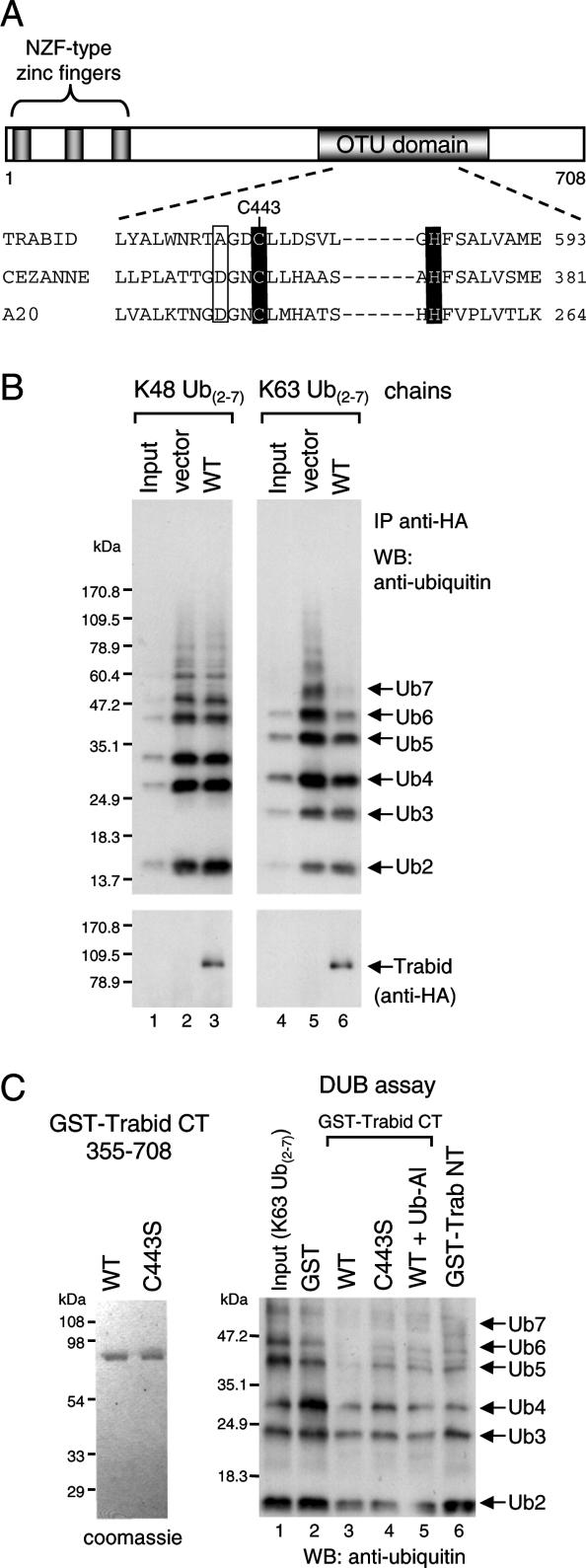
Figure 3.
Binding of Trabid’s NZF motifs to K63-linked ubiquitin. (A) Alignments of various NZF motifs, as indicated, with invariant cysteines shaded in black and other invariant residues in gray; the first cysteines (underlined) were mutated to alanine in the 3xCysNZF mutant. The T10 F/Y11 Φ22 signature for ubiquitin binding (Alam et al. 2004) is indicated by arrowheads (Φ, aliphatic amino acid; residue numbers refer to the position in this alignment); mutated TY residues are boxed. (B, Top) Ubiquitin-binding assays, with wild-type and mutant HA-Trabid immunoprecipitated from transfected 293T cells; asterisk indicates ubiquitylated protein coimmunoprecipitated with wild type and C443S; this signal is eliminated in the double-mutant C443S 3xNZFmut whose ubiquitin binding is abolished (see also D); background band, IgG heavy chain. (Below) The same blot was reprobed with α-HA antibody to monitor expression levels. (C) Ubiquitin-binding assays, with GST alone or GST-tagged Trabid(1–354) (see also Fig. 2C), expressed in bacteria (shown in bottom panel), and incubated in vitro with K48- or K63-linked ubiquitin; note the strong binding preference for K63-linked chains (shown in lanes 5,6). (D, top) Ubiquitin-binding assays as in C, with wild-type or mutant Trabid(1–200) (spanning the three tandem NZF motifs) incubated with K63-linked ubiquitin; TY > LV mutation of individual NZF (lanes 4–6), two NZF motifs (lanes 7–9), or all three NZF motifs (lane 10). (Middle panel) Long exposure of the same blot (bottom part only), to visualize residual binding of the NZF1 + 2 and NZF1 + 3 mutants to Ub4. (Bottom panel) The same blot was reprobed with α-GST antibody to monitor levels (revealing some breakdown; see also C). Note that the minimal module binding to K63-linked ubiquitin chains consists of at least two linked NZF motifs. (E,F) Immunofluorescence of 293 cells cotransfected with C443S and C443S-NZF1 + 2, fixed and stained as in Figure 1C–E.
Intriguingly, we also found that HA-Trabid coimmunoprecipitates with Flag-tagged β-TrCP, an adaptor between SCF E3 ligase and Axin–APC complex (Kitagawa et al. 1999), and even more strongly with a dominant-negative version (called ΔF) that cannot bind to the SCF ligase (Supplemental Fig. S2). We believe that this association between Trabid and β-TrCP is mediated by APC since we cannot detect direct binding between Trabid and β-TrCP in vitro (data not shown).
We conclude that Trabid is a bona fide APC-interacting protein that associates with its ARD in a sequence-specific fashion. We note that this ARD, like the ARD of β-catenin, has multiple ligands, including a regulatory subunit of protein phosphatase 2A (Seeling et al. 1999), itself a positive regulator of Wnt signaling (Ratcliffe et al. 2000); a guanine nucleotide exchange factor called Asef, whose association with APC appears to control cell migration (Kawasaki et al. 2000, 2003); and another sequence-specific ligand called AMER1, a membrane-associated protein that regulates APC’s subcellular distribution (Grohmann et al. 2007).
Trabid cleaves preferentially K63-linked ubiquitin chains
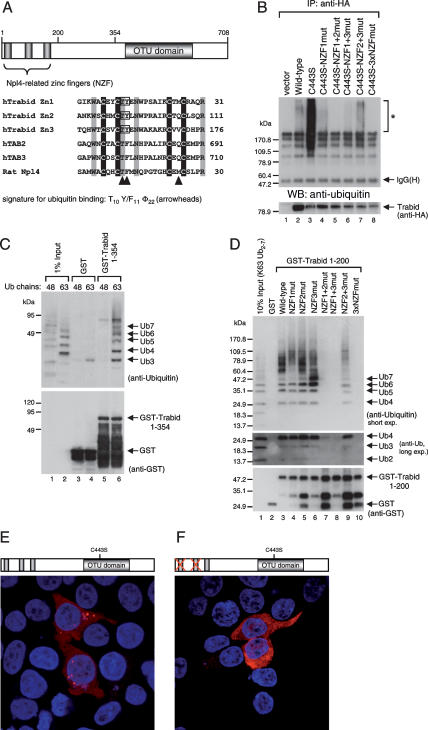
One defining feature of Trabid orthologs is a conserved D > A substitution three residues upstream of the putative catalytic C443 residue (Fig. 2A, boxed). Other cysteine proteases exhibit an aspartate in this position, which is part of the catalytic Asp Cys His triad (Makarova et al. 2000; Balakirev et al. 2003), though in the A20 OTU domain, a more N-terminal aspartate occupies the active site (Komander and Barford 2008). We asked whether this variant OTU domain of Trabid has DUB activity.
Figure 2.
Cleavage of K63-linked ubiquitin by Trabid. (A) Domains of human Trabid and alignment of the invariant cysteines and histidines (shaded in black) of three human OTU family members, with putative catalytic cysteine (C443 in Trabid) and histidine residues shaded, and the D > A substitution characteristic of Trabid orthologs boxed. (B) DUB assays, with wild-type and mutant HA-tagged Trabid immunoprecipitated from transfected 293T cells, as indicated, incubated with K48- or K63-linked ubiquitin (Ub2–7). (C) In vitro DUB assays, with wild-type and mutant GST-tagged C- or N-terminal fragments of Trabid expressed in bacteria (left; see also Fig. 3C), incubated with K63-linked ubiquitin (Ub2–7; right). (Ub-Al) Ubiquitin aldehyde.
To test this, we incubated immunoprecipitated HA-Trabid from 293T cells in vitro with synthetic ubiquitin chains (consisting of two to seven monomers; Ub2–Ub7) linked via ubiquitin K48 or K63. HA-Trabid cleaved K63-linked but not K48-linked ubiquitin; the activity was predominantly directed toward the longer chains in the mixture (Ub6, Ub7) (Fig. 2B). As expected, mutation of the catalytic cysteine (C443S) (Fig. 2A) blocked the DUB activity, as did mutation of the first invariant cysteine in the third NZF motif (C155A) (see below; Fig. 3); both mutants were expressed at levels comparable with the wild type (Supplemental Fig. S3; data not shown).
To rule out that the observed DUB activity was due to coprecipitating proteins, we conducted in vitro DUB assays with bacterially expressed GST-tagged OTU domain (GST-Trabid CT) (Fig. 2C). Wild-type GST-Trabid CT, but not C443S, cleaved K63-linked ubiquitin chains (Fig. 2C, cf. lanes 3and 1,4–6), while an N-terminal fragment spanning the NZF motifs (GST-Trabid NT, amino acids 1–354) was inactive (Fig. 2B, lane 6). Thus, the C terminus of Trabid exhibits DUB activity without its N-terminal NZF motifs (possibly because this in vitro assay involves relatively high protein concentrations). Importantly, preincubation of GST-Trabid CT with ubiquitin aldehyde, a specific DUB inhibitor, significantly reduced its DUB activity (Fig. 2B, lane 5). We conclude that the OTU domain of Trabid possesses DUB activity with a preference for K63-linked ubiquitin.
Trabid exhibits a striking binding preference for K63-linked ubiquitin chains
A second defining feature of Trabid orthologs are their three tandem NZF motifs, each of which bears the T10 F/Y11 Φ22 signature of ubiquitin-binding NZF motifs (Fig. 3A); based on previous structural analysis of the Np14–ubiquitin complex (Alam et al. 2004), these are predicted to be NZF surface residues that participate directly in ubiquitin binding. We generated mutations in these signature motifs (TY > LV) of HA-Trabid, and also in the first invariant cysteines of its NZF motifs, individually or combined, and tested the binding of these mutants to synthetic ubiquitin chains in vitro after immunoprecipitation from transfected 293T cells. HA-Trabid bound to K63-linked but not K48-linked Ub4–Ub6 (Supplemental Fig. S4A, note that Ub4–6 were enriched in the precipitate compared with the input), but less binding was observed with the NZF mutants, whereas C443S showed somewhat increased binding (Supplemental Fig. S4A, lanes 7–12). Strikingly, C443S, but not the NZF mutants, coprecipitated an abundance of endogenous proteins from 293T cell lysates that are also detectable, albeit to a lesser extent, in the wild-type precipitates (Fig. 3B; Supplemental Fig. S4A, asterisks). These are likely to represent ubiquitylated substrates bound to Trabid’s NZF fingers, trapped by the catalytically dead OTU domain of this C443S mutant (as observed previously with catalytically dead Cezanne) (Evans et al. 2003). We confirmed this by combining C443S with NZF mutations, which eliminated the signal from endogenous ubiquitylated proteins (Fig. 3B, see also D).
To demonstrate direct ubiquitin binding by the NZF fingers, we used in vitro pull-down assays with bacterially expressed GST-Trabid NT, which revealed a remarkable preference for K63- versus K48-linked ubiquitin (Fig. 3C; Supplemental Fig. S4B). This was somewhat surprising, given the lack of specificity in ubiquitin binding seen with other (individual) NZF fingers (Alam et al. 2004; Kanayama et al. 2004). We thus wondered whether Trabid’s preference for K63-linked chains might be due to the tandem linkage of its three NZF motifs (Fig. 3A).
This is the case: Individual NZF mutations did not affect ubiquitin binding (Fig. 3D, though note the binding bias of NZF3mut toward shorter chains). However, simultaneous mutation of two NZF motifs strongly reduced binding (Fig. 3D, lanes 7–9), while no binding was detectable after mutation of all three NZF motifs (Fig. 3D, lane 10). None of these mutants bound to K48-linked ubiquitin (data not shown). Thus, the minimal module conferring selective binding to K63-linked ubiquitin comprises at least two linked NZF motifs.
Another striking property of C443S is its ability to produce distinct puncta in the cytoplasm of transfected 293 cells (Fig. 3E), in addition to diffuse staining throughout the cytoplasm and nucleus (as seen with wild-type Trabid) (see below; Supplemental Fig. S5). No puncta are seen with C443S double mutants, which also bear NZF mutations that eliminate their ubiquitin binding (Fig. 3F), suggesting that the C443S puncta represent sites of concentrated ubiquitylated protein. So far, we have not been able to identify independent markers for these sites.
Trabid is required for TCF-dependent transcription
We used RNAi in different human cell lines to examine the roles of Trabid and Cezanne in Wnt signaling. Affinity-purified antibody against the Trabid N terminus revealed that Trabid is expressed in 293 cells, both in the cytoplasm and nucleus (Supplemental Fig. S5A,B). This subcellular distribution does not appear to change after Wnt stimulation, although Trabid is more nuclear in the colorectal cancer cell line SW480 (Supplemental Fig. S5C), which expresses truncated APC (Morin et al. 1997) that accumulates to some extent in the nucleus (Rosin-Arbesfeld et al. 2000, 2003). Similar patterns of subcellular distribution are seen with overexpressed HA-Trabid (Supplemental Fig. S5D–F).
Next, we tested the specificity and efficiency of depletion of different siRNAs after transfection of 293T cells with HA-Trabid or HA-Cezanne (Supplemental Fig. S6). We also established that Trabid- but not Cezanne-specific siRNAs reduced the level of endogenous Trabid transcripts (Supplemental Fig. S6A) and protein in these cells (Fig. 4A).
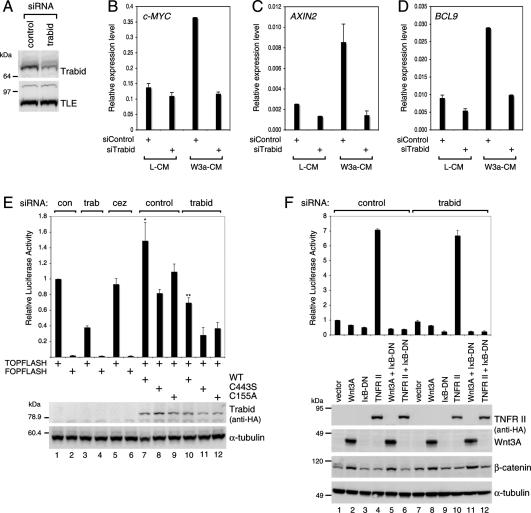
Figure 4.
Requirement of Trabid for efficient TCF-mediated transcription. (A) Western blot showing siRNA depletion of endogenous Trabid in 293 cells (internal control, α-TLE). (B–D) Real-time quantitative RT–PCR assays, after transfection of 293 cells with siRNAs as in A, monitoring transcript levels of the Wnt target genes c-MYC, AXIN2, and BCL9, as indicated, after treatment of cells with control (L-CM) or Wnt3A-conditioned (W3a-CM) medium. (E) TOPFLASH luciferase assays, after two subsequent transfections of SW480 cells with siRNAs, with or without re-expression of wild-type and mutant HA-tagged ΔsiRNA Trabid rescue constructs, as indicated; control FOPFLASH values are also shown. Relative luciferase values are shown (statistical significance, [*] P < 0.01; [**] P < 0.05). (Below) Western blots showing HA-Trabid expression from one representative experiment (α-tubulin, loading controls). (F) NF-κΒ-dependent luciferase reporter assays in 293T cells transfected with siRNAs as in A and cotransfected with the expression vectors as indicated. (Below) Western blots showing expression levels. Error bars in this and subsequent figures indicate standard deviations from the mean, from two or three independent experiments (performed in duplicate).
We then measured the expression levels of two well-established transcriptional targets of Wnt signaling, c-MYC (He et al. 1998; van de Wetering et al. 2002; Sansom et al. 2007) and AXIN2 (Jho et al. 2002; Lustig et al. 2002), by using quantitative RT–PCR on RNA isolated from 293 cells (which we found to be more Wnt-responsive than 293T cells). We confirmed that the transcript levels of c-MYC and AXIN2 were induced more than twice in cells transfected with control siRNA after exposure to Wnt3A (Fig. 4B,C). We also confirmed that the transcript levels of BCL9 are induced more than twice by Wnt3A (Fig. 4D), as described recently (M. de la Roche and M. Bienz, in prep.). Importantly, the stimulation of all three Wnt target genes was blocked in Trabid-depleted cells (Fig. 4B–D) while the values of the control HPRT transcripts did not change significantly after Wnt stimulation (data not shown). Trabid depletion also reduced the expression of AXIN2 and BCL9 somewhat in unstimulated 293 cells (Fig. 4C,D), reflecting a low level of constitutive Wnt pathway activity in these cells. Thus, Trabid is required for the efficient transcription of endogenous TCF target genes.
We also monitored the effects of Trabid depletion on SW480 colorectal cancer cells, using the TOPFLASH assay (based on a luciferase reporter linked to multiple TCF-binding sites), a highly specific and quantitative readout of Wnt pathway activity (Korinek et al. 1997). Trabid depletion reduced the TOPFLASH values to ∼40% of control cells, whereas Cezanne depletion had no effect (Fig. 4E). The activity of a luciferase reporter containing mutated TCF-binding sites (FOPFLASH) was unaffected (Fig. 4E). Conversely, overexpression of HA-Trabid in SW480 cells, but neither of C155A nor C443S, increased the TOPFLASH values slightly relative to the controls (Fig. 4E, lanes 1,7–9). Thus, Trabid but not Cezanne is required for efficient TCF-mediated transcription in these APC-mutant colorectal cancer cells.
Given the strong functional link of Trabid relatives to NF-κB signaling (see above), we also tested whether NF-κB-dependent transcription is sensitive to Trabid depletion. We transfected 293T cells with a luciferase reporter containing multiple NF-κB-binding sites and stimulated them by cotransfection of the TNF receptor II (TNFR-II), which resulted in an approximately seven times increase of NF-κB-dependent transcription (compared with the vector control), which was unaffected in Trabid-depleted cells (Fig. 4F, lanes 4,10). Further controls in this experiment included cotransfection with a dominant-negative IκB (IκB-DN) protein that effectively blocked the TNFR-II induction and with Wnt3A, which did not affect this NF-κB reporter assay (Fig. 4F). Thus, Trabid is not required for NF-κB-mediated transcription, consistent with the previous conclusions based on Trabid overexpression (Evans et al. 2001).
DUB activity and ubiquitin binding are important for Trabid’s function in TCF-dependent transcription
To rule out off-target effects of Trabid siRNAs, we conducted rescue experiments in Trabid-depleted SW480 cells with re-expressed Trabid constructs bearing silent mutations that rendered them refractory to the siRNAs (ΔsiRNA constructs) (Supplemental Fig. S6C). Wild-type HA-TrabidΔsiRNA restored TOPFLASH activity to ∼70% of matched controls (Fig. 4E, lanes 1,3,10), but neither C443SΔsiRNA nor C155AΔsiRNA were able to restore any activity (Fig. 4E, lanes 3,11,12), despite being expressed at similar levels to the wild type (Fig. 4E, bottom panel). Similar results were observed in Wnt3A-stimulated 293 cells (Supplemental Fig. S7). Thus, the ubiquitin-binding and DUB activities of Trabid are required for its function in TCF-mediated transcription in SW480 colorectal cancer cells.
Epistasis analysis indicates that Trabid acts downstream from stabilized β-catenin
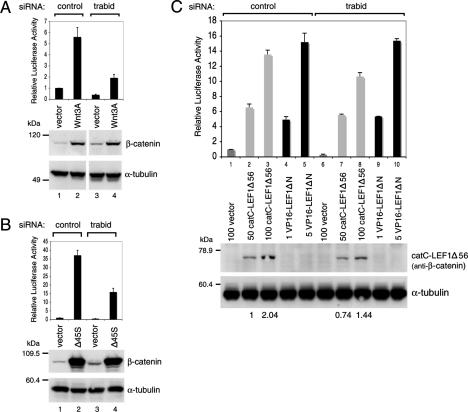
We conducted epistasis experiments, based on TOPFLASH assays in Trabid-depleted 293T cells, to identify the level within the Wnt signaling cascade at which Trabid acts. First, we examined cells transfected with a plasmid encoding Wnt3A, which stimulated TOPFLASH approximately six times, and induced the accumulation of β-catenin (Fig. 5A). In Trabid-depleted cells, the TOPFLASH values were reduced to ∼30% of the control, but the Wnt3A-induced accumulation of β-catenin was unaffected (Fig. 5A, lanes 2,4), suggesting that Trabid acts below the stabilization of β-catenin.
Figure 5.
Epistasis analysis of Trabid’s function in the Wnt pathway. TOPFLASH assays in 293T cells transfected with siRNAs as in Figure 4F and cotransfected with empty vector, HA-Wnt3A (A), stabilized Flag-β-catenin (Δ45S) (B), or 50–100 ng of catC-LEF1Δ56 or 1–5 ng VP16-LEF1ΔN chimeras (C), as indicated (the expression levels of the two chimeras were chosen to result in comparable transactivation). (Below) Western blots from a representative experiment to monitor expression levels of endogenous or overexpressed proteins (α-β-catenin antibody was used to detect catC-LEF1Δ56; VP16-LEF1ΔN was undetectable at these low expression levels); numbers below the blot in C indicate relative levels of catC-LEF1Δ56 expression (normalized to lane 2).
Given its role in ubiquitin conjugation, and its association with Trabid (Supplemental Fig. S2), we also asked whether β-TrCP might be affected by Trabid depletion. Dominant-negative β-TrCP (ΔF) induced the TOPFLASH activity in a dose-dependent manner, similarly to previous results (Kitagawa et al. 1999), but this was reduced more than two times in Trabid-depleted cells (Supplemental Fig. S8A). Similarly, the high TOPFLASH activities due to Dishevelled overexpression or to inhibition of GSK3β by LiCl were reduced to <50% after Trabid depletion, whereas the stabilization of β-catenin was unaffected under these conditions (Supplemental Fig. S8B,C).
Finally, we transfected 293T cells with β-cateninΔ45S (Δ45S), an activated nonphosphorylatable form of β-catenin (Morin et al. 1997), which resulted in >35 times stimulation of TOPFLASH (Fig. 5B). However, in Trabid-depleted cells, the TOPFLASH values were reduced to ∼40% (Fig. 5B, lanes 2,4). Consistent with this, Trabid depletion in HCT-116 colorectal cancer cells (that harbor the same Δ45S mutation) (Morin et al. 1997) resulted in an ∼50% reduction of their TOPFLASH activity compared with control cells (Supplemental Fig. S9). This corroborates our conclusion that Trabid is required for the transcriptional activity rather than the stabilization of β-catenin.
LEF1 chimeras bearing heterologous transactivation domains (TADs) function independently of Trabid
Chimeras between LEF1 and heterologous TADs potently stimulate TCF-dependent transcription (Hsu et al. 1998; Ishitani et al. 2005). We asked whether their activities would depend on Trabid. We first tested a chimera between LEF1 and the C terminus of β-catenin (the main recruitment domain for transcriptional coactivators) (see above). This chimera (called catC-LEF1Δ56 since it lacks its β-catenin-binding domain) (Hsu et al. 1998) mediates strong dose-dependent transactivation of TOPFLASH in transfected 293 cells (Fig. 5C, lanes 1–3), as shown previously (Hsu et al. 1998). This activity was barely affected by Trabid depletion (Fig. 5C, lanes 7,8). Indeed, if the slightly reduced expression levels of the chimera in Trabid-depleted cells are taken into account (Fig. 5C, values below panels), then the activity of catC-LEF1Δ56 is judged to be independent of Trabid.
We also tested another chimera—namely, LEF1—without its N terminus fused to the TAD from the viral protein VP16 (VP16-LEF1ΔN), which stimulates TCF-dependent transcription (Fig. 5C, lanes 4,5) as shown previously (Ishitani et al. 2005). Again, VP16-LEF1ΔN was independent of Trabid, showing essentially the same dose-dependent transactivation of TOPFLASH irrespective of Trabid depletion (Fig. 5C, lanes 9,10). These results argue that Trabid is neither required for the activity of LEF1 itself (or of its regulators, such as TAK1/NEMO-like kinase; Ishitani et al. 2003) nor for its natural coactivators or those recruited by the VP16 TAD. Instead, Trabid appears to affect the association of LEF1 with β-catenin, which is bypassed by direct linkage in these chimeras.
Trabid depletion results in hyperubiquitylation of APC
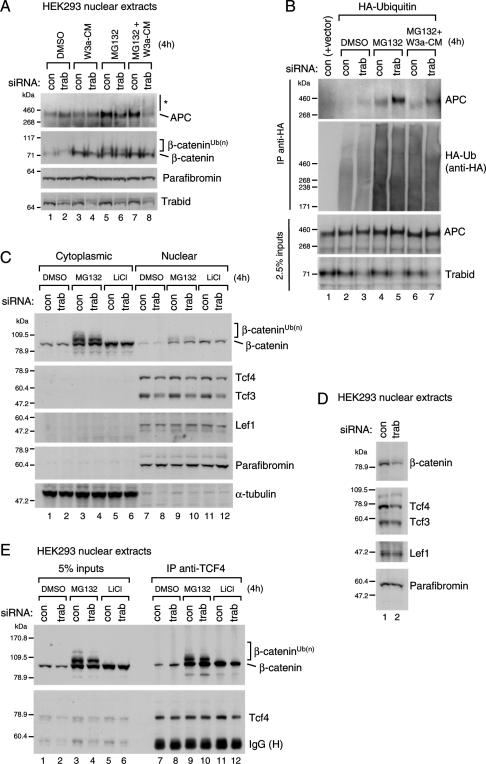
To identify a substrate for Trabid’s DUB activity, we asked whether Trabid depletion would affect the ubiquitylation of Wnt signaling components. We first focused on APC to which Trabid binds (Fig. 1) and that has been shown to be ubiquitylated and subject to proteasome-mediated degradation (Choi et al. 2004). Note also that APC can be nuclear (Rosin-Arbesfeld et al. 2000, 2003) and is recruited to TCF target genes after Wnt stimulation to attenuate their transcription (Sierra et al. 2006), in addition to its main function in the cytoplasm promoting the phosphorylation and degradation of β-catenin (see above), which appears to be unaffected by Trabid (Fig. 5A,B). We thus depleted Trabid in 293 cells stimulated with Wnt3A and prepared nuclear fractions to examine specifically nuclear APC.
This revealed a slight increase of the nuclear APC levels in Trabid-depleted cells, both before and after Wnt stimulation (Fig. 6A, lanes 1–4). Furthermore, the band corresponding to full-length APC decreased after Trabid depletion, and a smear of higher-molecular-weight (HMW) APC bands appeared, particularly noticeable in Wnt-stimulated cells that had been pretreated with the proteasome inhibitor MG132 (Fig. 6A, lanes 6,8, asterisk). This HMW smear may thus reflect ubiquitylated APC. Under the same conditions, HMW bands corresponding to ubiquitylated β-catenin were also apparent (Aberle et al. 1997); however there was no obvious change of these (Fig. 6A, lanes 5–8), consistent with our conclusion that Trabid acts below the stabilization of β-catenin.
Figure 6.
Hyperubiquitylation of APC in Trabid-depleted cells. Western blots of nuclear or cytoplasmic fractions 24 h after transfection of 293 (A,B,D) or 293T (C) cells transfected with siRNAs (A–D) (as in Fig. 4F) and HA-ubiquitin (B); treated for 4 h with DMSO (control), 10 mM MG132, and/or 20 mM LiCl or Wnt3A-conditioned medium as indicated; and probed sequentially with the antibodies indicated on the right. (E) Coimmunoprecipitations from nuclear fractions from 293T cells (200 μg), prepared as in C, as indicated.
To demonstrate directly that Trabid affects the ubiquitylation of APC, we conducted the same analysis after overexpressing HA-ubiquitin, followed by immunoprecipitation with anti-HA antibody, to selectively examine the ubiquitylated protein pool. This revealed an increase, and upshift, of the APC antibody-reactive bands in Trabid-depleted cells, especially noticeable after MG132 treatment (Fig. 6B, top panel, note also the subtle upshifts of the APC bands in the input lanes, middle panel). We conclude that APC becomes hyperubiquitylated after Trabid depletion, indicating that APC is a direct enzymatic target of Trabid.
We also examined other nuclear Wnt signaling components in the same way, including β-catenin and TCF factors. However, we only found minor changes in Trabid-depleted cells: a slight increase of the cytoplasmic levels of β-catenin, but a slight decrease of its nuclear levels (Fig. 6C,E [lanes 11,12], D; see also Fig. 5) and of the predominant TCF factors in these cell lines, TCF3 and TCF4 (Fig. 6C [lanes 7,8], D). In contrast, LEF1 (expressed at low levels in these cells) and the β-catenin-binding protein Parafibromin remained unchanged (Fig. 6C,D). None of the proteins showed HMW bands as observed with APC, with the exception of β-catenin (Fig. 6C,E, brackets) whose ubiquitylation, as already mentioned, did not appear to change in Trabid-depleted cells (Fig. 6C,E).
The observed reductions of nuclear β-catenin and TCF levels in Trabid-depleted cells might explain why the TCF-mediated transcription is reduced under these conditions. However, this is not the case: Overexpression of activated β-catenin or TCF factors does not restore normal levels of TOPFLASH activity in Trabid-depleted cells (Supplemental Fig. S6; data not shown), nor does overexpression of BCL9 (whose Wnt-induced expression is blocked after Trabid depletion) (Fig. 4D).
Finally, we examined whether Trabid depletion affects the interaction between TCF and β-catenin by using coimmunoprecipitation of endogenous nuclear proteins. There was no detectable change in the levels of TCF4-associated β-catenin (Fig. 6E, lanes 7–12), suggesting that Trabid is neither essential for recruitment of β-catenin to TCF4 nor does it affect the steady-state level of TCF-associated β-catenin. However, our analysis would not allow us to detect changes in the dynamics of complex formation or dissociation (see Discussion).
Drosophila Trabid is a positive regulator of the Wingless response
To test the function of the single Trabid ortholog of Drosophila in its Wingless response, we deleted the trbd locus (CG9448) by homologous recombination. To our surprise, trbd-null mutants were viable and fertile, suggesting that trbd may be redundant with another gene.
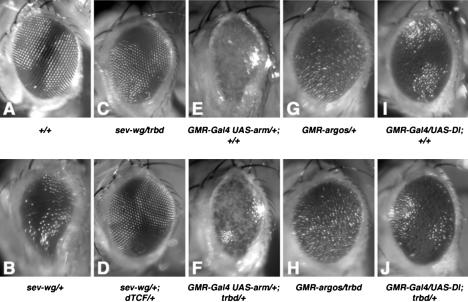
We thus tested trbd function in a more sensitive assay by asking whether lowering the dose of Trabid by half (in trbd heterozygotes) would affect the rough eye phenotype due to ectopic Wingless in the eye imaginal disc (Fig. 7A,B; Brunner et al. 1997). Indeed, trbd heterozygosity suppressed this phenotype (Fig. 7C, cf. B), similarly to dTCF heterozygosity (Fig. 7D). trbd heterozygosity also suppressed the rough eye phenotype due to overexpressed Armadillo (the β-catenin of Drosophila) to some extent (Fig. 7F, cf. E). In contrast, we observed no genetic interactions between trbd and components of EGF receptor signaling, a pathway that controls differentiation in the developing fly eye (Freeman 1997): The rough eye phenotype due to activation or inhibition of this pathway remained unchanged in trbd heterozygotes (Fig. 7G,H; data not shown). Similarly, there was no interaction between trbd and the Notch pathway, whose stimulation also causes rough eyes (Fig. 7I,J). These results suggest that dTrbd is required for an efficient Wingless response, but not for other signaling responses in the eye, consistent with our results in human cell lines.
Figure 7.
Function of dTrbd in the Wingless response of Drosophila. Eyes from control y w flies (A), or from flies after expression, in the larval eye disc of Wingless (B–D), Armadillo (E,F), Argos inhibitor of EGF receptor (G,H), or Delta agonist of Notch receptor (I,J). (A,B,E,G,I) +/+. (C,F,H,J) trbd/+. (D) dTCF3/+. trbd heterozygosity suppresses the rough eye phenotype due to ectopic Wingless or Armadillo (C,F), but not that due to ectopic Argos (H), Rhomboid (not shown), or Delta (J).
Discussion
To date, the only known function of ubiquitylation in the Wnt pathway relates to protein turnover—especially that of its key effector β-catenin—typically conferred by K48-linked ubiquitin chains. Our discovery of Trabid, a DUB enzyme that exhibits a remarkable preference for K63-linked ubiquitin, and its functional link to APC and the Wnt response of mammalian and Drosophila cells implies a distinct role of ubiquitylation in this pathway, namely, one mediated by K63-linked ubiquitin chains. These chains are likely to affect the activity rather than the stability of proteins, and we discuss below how Trabid may affect the transcriptional response to Wnt signaling by antagonizing the ubiquitylation of APC.
The preference of Trabid for K63-linked ubiquitin
We showed that Trabid is a bona fide DUB and that it cleaves preferentially K63-linked ubiquitin. This activity resides in its OTU domain, which possesses intrinsic DUB activity, as has been shown previously for other members of the OTU family (Evans et al. 2001, 2003; Balakirev et al. 2003; Wang et al. 2004; Wertz et al. 2004).
Trabid exhibits an unprecedented selectivity for binding to K63- versus K48-linked ubiquitin chains (Fig. 3), which is conferred by a minimal module of at least two linked NZF motifs. In contrast, an individual NZF motif—e.g., that of TAB3—binds indiscriminately to K48- and K63-linked ubiquitin chains (Kanayama et al. 2004), consistent with structural analysis that revealed the commonly used surface of ubiquitin for NZF binding (Alam et al. 2004). Notably, the three specificity-conferring NZF motifs of Trabid are not only a defining feature of Trabid orthologs, but also a fairly unique one: Although NZF domains with ubiquitin-binding signatures are found in a number of proteins, arrays of such NZF fingers are rare. It is conceivable that tandem NZF arrays confer specificity for K63-linked ubiquitin by topological restriction: K48-linked chains are known to form relatively compact structures (Eddins et al. 2007), whereas K63-linked chains are likely to be extended.
Our evidence suggests that the NZF motifs of Trabid contribute to its function in TCF-mediated transcription (Fig. 4E), although they are not required for its intrinsic DUB activity in vitro (Fig. 2C). These motifs may have a targeting function in vivo, similarly to the ubiquitin binding by the TAB adaptors, which is thought to facilitate the recruitment of ubiquitylated substrates to the TAB-associated TAK1 kinase (Kanayama et al. 2004).
APC as an enzymatic target of Trabid
Our epistasis analysis indicated that Trabid acts below the stabilization of β-catenin. Consistent with this, we detected only minor effects of Trabid depletion on the ubiquitylation of β-catenin (by K48-linked ubiquitin) and on its steady-state levels (Figs. 5B, 6). These results argue against a direct function of Trabid in proteasomal degradation.
In contrast, we discovered that APC becomes hyperubiquitylated in Trabid-depleted cells (Fig. 6A,B), indicating that APC is not only a binding partner of Trabid, but also a substrate of its DUB activity. Thus, at least one function of Trabid is to antagonize the ubiquitylation of APC. This is likely to involve K63-linked ubiquitin chains, given Trabid’s preference for cleaving these chains (Fig. 2B). Also, the hyperubiquitylation of APC in Trabid-depleted cells, despite being more pronounced after MG132 treatment, neither seems to promote the degradation of APC itself (Fig. 6A,B), nor that of its key target, β-catenin (Figs. 5B, 6). It thus appears that the deubiquitylation of APC by Trabid has a regulatory output (rather than antagonizing the proteasomal turnover of APC mediated by Axin in the absence of Wnt stimulation; Choi et al. 2004). For example, it might regulate the activity of APC in transcription (see below). Further work will be required to clarify the molecular nature of these APC ubiquitylations, and their effects on APC’s stability and/or activity.
The function of Trabid in TCF-mediated transcription
Our results from the epistasis analysis and LEF1 chimeras suggest a role of Trabid in TCF-mediated transcription. Consistent with this, preliminary results from chromatin immunoprecipitation experiments indicate that Trabid is associated with the c-myc promoter and upstream sequences (H. Tran, unpubl.). Trabid’s prime physiologically relevant substrate could be APC itself, which is recruited (together with β-TrCP) to TCF target genes to repress their transcription during sustained Wnt signaling (Sierra et al. 2006). If so, this would imply that Trabid inactivates APC; hyperubiquitylation of APC in Trabid-depleted cells would result in its hyperactivation, possibly leading to hyperrepression (or precocious repression) of Wnt target genes, which would explain why the Wnt response is attenuated in these cells. Alternatively, the prime functional target of Trabid might be an unknown nuclear protein, a negative regulator like APC whose deubiquitylation results in its inactivation or a positive regulator of Wnt-induced transcription that becomes activated as a result of Trabid’s DUB action; in the latter case, hyperubiquitylation would be expected to result in inactivation.
How does Trabid affect the TCF-mediated transcription? We were unable to detect a change in the steady-state association between β-catenin and TCF in Trabid-depleted cells (Fig. 6E). Nevertheless, our results from the LEF chimeras suggest that Trabid may affect the dynamic association or dissociation of the TCF–β-catenin complex: These chimeras might bypass the requirement for Trabid because of their direct linkage to TADs, which allows them to recruit (or retain) coactivators efficiently—possibly more efficiently than normal TCF factors whose transcriptional activity during Wnt signaling relies on transient, and perhaps labile, association with limited amounts of nuclear β-catenin. It is therefore conceivable that Trabid increases, for example, the rate of TCF–β-catenin complex formation or the lifetime of its activity.
Ubiquitylation is beginning to emerge as a versatile controlling factor in transcriptional switches, including regulatory ubiquitylation events such as monoubiquitylation of histones (Muratani and Tansey 2003). To our knowledge, there are as yet no known regulatory transcription events that rely on K63-linked ubiquitin, although DNA repair can be regulated by K63-linked ubiquitylation (of PCNA) (Hoege et al. 2002). The turnover of regulatory ubiquitylations via DUBs may be as important as the ubiquitylations themselves, one example being the negative regulation of H2B monoubiquitylation by Ubp8, a DUB associated with the yeast histone acetylation SAGA complex (Henry et al. 2003). Interestingly, in vitro transcription from a β-catenin-dependent chromatin template was attenuated by a DUB inhibitor (Sierra et al. 2006).
A role of Trabid in cancer and stem cell compartments?
Given its requirement for efficient TCF-mediated transcription in colorectal cancer cell lines, Trabid has potential as a molecular target for inhibitory drugs in colorectal cancer. Obviously, more work is required to validate this target, but Trabid appears attractive because of (1) its apparent specificity for Wnt responses, (2) its enzymatic activity as a protease, and (3) its catalytic pocket that is likely to be unique among OTU domain proteases (Fig. 2A; Nanao et al. 2004; Komander and Barford 2008).
Wnt signaling has a key role in maintaining intestinal stem cell compartment (Reya and Clevers 2005). Furthermore, a recent compilation of gene expression data sets has identified relatively few genes (<40) commonly up-regulated in a variety of stem cell types (including skin bulge, hematopoietic, embryonic, and neural stem cells), which include TCF3 and TCF4 as well as Trabid (Blanpain et al. 2004). Notably, TCF3 plays a role in maintaining skin stem cells in an undifferentiated state (Nguyen et al. 2006), and TCF4 is required for maintaining crypts in the intestinal epithelium (Korinek et al. 1998). Given the high Trabid levels in these stem cell compartments and Trabid’s role in TCF-mediated transcription, it is conceivable that Trabid may have a function in stem cell self-renewal or differentiation.
Materials and methods
Plasmids and antibodies
pHM6-HA-Trabid and pHM6-HA-Cezanne have been described (Evans et al. 2001, 2003); point mutants were created using QuickChange (Stratagene) and confirmed by DNA sequencing. For GST-Trabid constructs, PCR-amplified Trabid cDNA fragments were cloned between EcoRI and XhoI of the bacterial expression vector pGEX-6P-1. APC and E-APC plasmids were generated previously (Hamada and Bienz 2002; Rosin-Arbesfeld et al. 2003), except for APC(1–1447) (kindly provided by J. Worm). LEF1 chimeras have been described (Hsu et al. 1998; Ishitani et al. 2005).
The following antibodies were used: α-TCF4, α-TCF3/4, α-ubiquitin (Upstate Biotechnology); α-β-catenin, α-Flag M2 (Sigma); α-Parafibromin (Bethyl Laboratories); α-HA 3F10 (Roche); α-LEF1, α-TLE, α-APC Ali-1228 (Santa Cruz Biotechnology); α-α-tubulin (Abcam). Polyclonal rat antibodies were generated (by Eurogentec) to an N-terminal fragment of Trabid (amino acids 1–354) fused to GST and purified from bacteria. The crude rat α-Trabid serum was used at 1:100 dilution (Fig. 4A; Supplemental Fig. S5A) or after affinity purification against Trabid antigen (1:500 for Western blotting; 1:100 for immunofluorescence) (Fig. 6A,B; Supplemental Fig. S5B,C).
Cells, transfections, and treatments
HEK293, HEK293T, SW480, and HCT-116 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. siRNA (Dharmacon) duplexes used to target Trabid and Cezanne mRNAs are as follows (only sense strand is shown): Trabid #1, AGAGGCUUCUUCAAUAA UAdTdT; Trabid #2, AGAGGUGUCUCAACAAGCAdTdT; Cezanne #1, GAAUCUAUCUGCCUUUGGAdTdT; Cezanne #2, AGACUUCCGCAGCUUCAUAdTdT. The negative control siRNA was from Ambion (no. AM4611). Transfection of siRNA duplexes and plasmid DNA was performed using Lipofectamine 2000 (Invitrogen).
TOPFLASH and NF-κB reporter assays
Cells were seeded in 12-well plates at a density of 0.8 × 105 cells per milliliter and allowed to attach to the dish surface for 12–16 h before transfection. For RNAi experiments, cells were first transfected with siRNAs (100 nM final concentration) and, after 24 h, retransfected with siRNAs and 250 ng of pTOPFLASH or pFOPFLASH, 20 ng of pRL-CMV (Renilla luciferase internal control), and 1.0 μg of the effector plasmid DNA, unless specified otherwise. For NF-κB assays, cells were transfected with a luciferase reporter containing 4× NF-κB-binding sites (Clontech) and, where indicated, pEAK12-HA-TNFRII and pEAK12-IκBαΔN. Total amounts of DNA per transfection were equalized with empty vector. After a further 24 h, cells were harvested and lysed in passive lysis buffer (Dual Luciferase Reporter Assay System; Promega). Protein concentration was determined with a Coomassie-based reagent (Pierce); 10 μg of total protein were used per sample (for TOPFLASH assays or Western blotting). For transcription assays, relative luciferase activities were determined (TOPFLASH divided by Renilla control values), and the resulting value from control-transfected cells was set arbitrarily to 1.
Immunoprecipitations and GST pull-down assays
Adherent cells were washed twice with PBS and incubated in NP40 lysis buffer (50 mM Tris-HCl at pH 7.4, 120 mM NaCl, 1% NP-40, 1 mM EDTA, 5 mM Na3VO4, 5 mM NaF, 0.5 μg/mL aprotinin, 1 μg/mL leupeptin) for 10 min on ice. Cells were collected by scraping and were centrifuged at 14,000 rpm for 10 min (4°C). Immunoprecipitations were performed using 500–700 μg of cell extracts precleared with a 50% solution of protein G-Sepharose beads (100 μL; Zymed), before incubation with 4 μg of the indicated antibody for 2 h at 4°C. Antibody–antigen complexes were captured with 100 μL of 50% protein G beads for 1 h at 4°C.
GST pull-down assays were performed with 1 μg of bacterially expressed GST or GST-tagged fragments of Trabid incubated in a 50% solution of glutathione-Sepharose 4B beads (50 μL; Amersham) with or without K48- or K63-linked ubiquitin chains (Ub2–7; Affiniti) for 1 h at 4°C in NP40 lysis buffer (see above). Alternatively, 20 μL of protein G-Sepharose beads bound with immunoprecipitated HA-Trabid protein was used (Fig. 3B; Supplemental Fig. S4A). Precipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting. Nuclear and cytoplasmic fractions were prepared as described (Caruccio and Banerjee 1999).
DUB assays
Assays were performed as described (Wang et al. 2004), with modifications. GST-tagged fragments of Trabid were expressed in Escherichia coli BL21(DE3) and purified with glutathione-Sepharose beads (Amersham) according to the supplier’s instructions. Purified protein (0.5 μg) was incubated with 0.5 μg of synthetic ubiquitin chains, as above, in 20 μL of buffer (150 mM KCl, 50 mM HEPES at pH 7.4, 10 mM DTT, 5% glycerol, 0.01% Triton X-100) for 60 min at 37°C. Alternatively, HA-Trabid was used after immunoprecipitation from transfected 293T cells (20 μL of protein G-Sepharose beads; see above). Reactions were terminated with 2× SDS-PAGE sample buffer, resolved by SDS-PAGE, and analyzed by Western blotting. Ubiquitin aldehyde was purchased from Calbiochem.
Real-time quantitative RT–PCR
293 cells were grown in 12-well plates, transfected with siRNAs and, after 24 h, incubated with Wnt3A-conditioned (from mouse L cells stably expressing Wnt3A) or control medium for a further 6 h. Cells were harvested in ice-cold PBS and used directly for cDNA synthesis with the SuperScript III kit as instructed (Invitrogen). Duplicate cDNA samples were amplified with the TaqMan Universal PCR Master Mix and gene-specific TaqMan TAMRA probes in an ABI Prism 7900HT real-time PCR machine (Applied Biosystems). Standard curves were based on the housekeeping gene HPRT whose expression was not affected by Wnt3A treatment. Results are expressed as fractions of HPRT values.
Drosophila genetics
A deletion of the trbd locus (CG9448) was generated by homologous recombination. Briefly, the BAC clone RP98-30F1 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute, Oakland, CA) spanning the trbd locus and its flanking regions was used as template for PCR reactions. Primers 5′-ACTGGTACCGTCGAAGTTGGTTGAAGATACG-3′ and 5′-AATGCGGCCGCACAGGGCAATATTTCGTTGC-3′ were used to add Acc65I and NotI restriction sites to a 3.5-kb DNA fragment immediately upstream of the target locus. This fragment was then amplified by PCR and subcloned into the same restriction sites of an ends-out homologous recombination vector pW25 (Gong and Golic 2004). Similarly, a 3.5-kb DNA fragment immediately downstream from the target locus was subcloned into the same vector, after adding AscI and BsiWI restriction sites with the primers 5′-ATTGGCGCGCCGACG TATGTATATCATATCCG-3′ and 5′-ACTCGTACGTGCTGT GCTACCTGACGCAC-3′, respectively. Transgenic flies of the resulting targeting construct were generated by standard germline transformation, and crosses for targeting were carried out as described (Rong and Golic 2001; Gong and Golic 2003); translocation of white+ to the target chromosome was confirmed. Targeted deletion of trbd was confirmed by PCR.
trbd/TM3 or y w control flies were crossed to sev-wg (Brunner et al. 1997), GMR.Argos (Casci et al. 1999), GMR.GAL4 > UAS. Armadillo, or GMR.GAL4 > UAS.Delta (Freeman and Bienz 2001) and grown at 25°C. Eyes of the resulting heterozygous progeny were scored as described (Freeman and Bienz 2001).
Acknowledgments
We thank P. Evans for Cezanne and Trabid expression vectors; J. Worm, F. Randow, M. Matsumoto, and M. Hülsken for other plasmids; M. de la Roche for help with the real-time PCR analysis; and F. Randow for discussions. This study was supported by AICR (grants nos. 03-275 and 07-0040 to M.B.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.463208
References
- Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.L., Sun J., Payne M., Welch B.D., Blake B.K., Davis D.R., Meyer H.H., Emr S.D., Sundquist W.I. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev M.Y., Tcherniuk S.O., Jaquinod M., Chroboczek J. Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A., Chauvet S., Huber O., Usseglio F., Rothbacher U., Aragnol D., Kemler R., Pradel J. Pontin52 and reptin52 function as antagonistic regulators of β-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Lowry W.E., Geoghegan A., Polak L., Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Brunner E., Peter O., Schweizer L., Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Caruccio L., Banerjee R. An efficient method for simultaneous isolation of biologically active transcription factors and DNA. J. Immunol. Methods. 1999;230:1–10. doi: 10.1016/s0022-1759(99)00100-3. [DOI] [PubMed] [Google Scholar]
- Casci T., Vinos J., Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Park S.Y., Costantini F., Jho E.H., Joo C.K. Adenomatous polyposis coli is down-regulated by the ubiquitin–proteasome pathway in a process facilitated by Axin. J. Biol. Chem. 2004;279:49188–49198. doi: 10.1074/jbc.M404655200. [DOI] [PubMed] [Google Scholar]
- de la Roche M., Bienz M. Wingless-independent association of Pygopus with dTCF target genes. Curr. Biol. 2007;17:556–561. doi: 10.1016/j.cub.2007.01.063. [DOI] [PubMed] [Google Scholar]
- Ea C.K., Deng L., Xia Z.P., Pineda G., Chen Z.J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Eddins M.J., Varadan R., Fushman D., Pickart C.M., Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- Evans P.C., Taylor E.R., Coadwell J., Heyninck K., Beyaert R., Kilshaw P.J. Isolation and characterization of two novel A20-like proteins. Biochem. J. 2001;357:617–623. doi: 10.1042/0264-6021:3570617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.C., Smith T.S., Lai M.J., Williams M.G., Burke D.F., Heyninck K., Kreike M.M., Beyaert R., Blundell T.L., Kilshaw P.J. A novel type of deubiquitinating enzyme. J. Biol. Chem. 2003;278:23180–23186. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Freeman M., Bienz M. EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep. 2001;2:157–162. doi: 10.1093/embo-reports/kve019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W.J., Golic K.G. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W.J., Golic K.G. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann A., Tanneberger K., Alzner A., Schneikert J., Behrens J. AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J. Cell Sci. 2007;120:3738–3747. doi: 10.1242/jcs.011320. [DOI] [PubMed] [Google Scholar]
- Hamada F., Bienz M. A Drosophila APC tumour suppressor homologue functions in cellular adhesion. Nat. Cell Biol. 2002;4:208–213. doi: 10.1038/ncb755. [DOI] [PubMed] [Google Scholar]
- He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Hecht A., Litterst C.M., Huber O., Kemler R. Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- Hecht A., Vleminckx K., Stemmler M.P., van Roy F., Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., Hill C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Galceran J., Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol. Cell. Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T., Ninomiya-Tsuji J., Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/β-catenin signaling. Mol. Cell. Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T., Matsumoto K., Chitnis A.B., Itoh M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nat. Cell Biol. 2005;7:1106–1112. doi: 10.1038/ncb1311. [DOI] [PubMed] [Google Scholar]
- Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A., Seth R.B., Sun L., Ea C.K., Hong M., Shaito A., Chiu Y.H., Deng L., Chen Z.J. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Senda T., Ishidate T., Koyama R., Morishita T., Iwayama Y., Higuchi O., Akiyama T. Asef, a link between the tumor suppressor APC and G-protein signaling. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Sato R., Akiyama T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat. Cell Biol. 2003;5:211–215. doi: 10.1038/ncb937. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Xu W. β-catenin destruction complex: Insights and questions from a structural perspective. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem. J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a β-catenin–Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kramps T., Peter O., Brunner E., Nellen D., Froesch B., Chatterjee S., Murone M., Zullig S., Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin–TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., de van Wetering M., Clevers H., Schlag P.M., Birchmeier W., et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Muratani M., Tansey W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nanao M.H., Tcherniuk S.O., Chroboczek J., Dideberg O., Dessen A., Balakirev M.Y. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–788. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes & Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Ratcliffe M.J., Itoh K., Sokol S.Y. A positive role for the PP2A catalytic subunit in Wnt signal transduction. J. Biol. Chem. 2000;275:35680–35683. doi: 10.1074/jbc.C000639200. [DOI] [PubMed] [Google Scholar]
- Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rong Y.S., Golic K.G. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Townsley F., Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Cliffe A., Brabletz T., Bienz M. Nuclear export of the APC tumour suppressor controls β-catenin function in transcription. EMBO J. 2003;22:1101–1113. doi: 10.1093/emboj/cdg105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom O.J., Meniel V.S., Muncan V., Phesse T.J., Wilkins J.A., Reed K.R., Vass J.K., Athineos D., Clevers H., Clarke A.R. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Seeling J.M., Miller J.R., Gil R., Moon R.T., White R., Virshup D.M. Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- Shi C.S., Kehrl J.H. Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2) J. Biol. Chem. 2003;278:15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- Sierra J., Yoshida T., Joazeiro C.A., Jones K.A. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K.I., Moon R.T. The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J. Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F.M., Thompson B., Bienz M. Pygopus residues required for its binding to Legless are critical for transcription and development. J. Biol. Chem. 2004;279:5177–5183. doi: 10.1074/jbc.M309722200. [DOI] [PubMed] [Google Scholar]
- de van Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P., et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Wang C., Deng L., Hong M., Akkaraju G.R., Inoue J., Chen Z.J. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G., Meyer H.H. VCIP135 acts as a deubiquitinating enzyme during p97–p47-mediated reassembly of mitotic Golgi fragments. J. Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack P., Beer-Romero P., Chu C.Y., Elledge S.J., Harper J.W. The SCF β–TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Conze D.B., Li T., Srinivasula S.M., Ashwell J.D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]