Summary
Background
Cohesin proteins link sister chromatids and provide the basis for tension between bi-oriented sister chomatids in mitosis. Cohesin is concentrated at the centromere region of the chromosome despite the fact that sister centromeres can be separated by 800nm in vivo. The function of cohesin at sites of separated DNA is unknown.
Results
We provide evidence that the kinetochore promotes the organization of pericentric chromatin into a cruciform in mitosis such that centromere-flanking DNA adopts an intramolecular loop, while sister chromatid arms are paired intermolecularly. Visualization of cohesin subunits by fluorescence microscopy revealed a cylindrical structure that encircles the central spindle and spans the distance between sister kinetochores. Kinetochore assembly at the apex of the loop initiates intra-strand loop formation that extends approximately 25kb (12.5kb on either side of the centromere). Two centromere loops (one from each sister chromatid) are stretched between the ends of sister kinetochore microtubules along the spindle axis. At the base of the loop there is a transition to intermolecular sister chromatid pairing.
Conclusions
The C-loop conformation reveals the structural basis for sister kinetochore clustering in budding yeast, kinetochore bi-orientation, and resolves the paradox of maximal inter-strand separation in regions of highest cohesin concentration.
Introduction
During mitosis, the eukaryotic cell constructs a bipolar array of microtubule (MTs) that serves as the machinery to segregate duplicated chromosomes. The centromere on each sister chromatid specifies the assembly of the kinetochore, a DNA-protein complex that interacts with the plus-ends of kinetochore MTs (kMTs). Sister kinetochores can attach to MTs emanating from either pole, leading to configurations where sister kinetochores are attached to opposite poles (amphitelic), same poles (syntelic) or one kinetochore attached to both poles (merotelic). The correct MT arrangement that persists is the one where sister kinetochores are attached to opposite poles. Tension produced by amphitelic attachment is the probable basis for the stability of this configuration. The budding yeast S. cerevisiae has only one MT attachment per kinetochore and is an ideal system to characterize the force producing mechanisms and tension elements that reside at the interface of kinetochore-MT attachments.
The physical linkage of sister chromatids is the mechanism for generation of tension for amphitelic attachment. This linkage is mediated by a multisubunit complex cohesin, composed of two members of the SMC (structural maintenance of chromosomes) family of ATPases, Smc1 and Smc3, and two non-SMC subunits, Mcd1/Scc1 and Scc3 [1, 2]. Cohesin is associated with chromosomes from G1 in the cell cycle until the onset of anaphase. It has been assumed that cohesin promotes association between sister chromatids (intermolecular linkage), and that is the basis for tension when sister chromatids are oriented to opposite spindle pole bodies. The Scc1 subunit disappears from chromosomes when sisters separate at the metaphase/anaphase transition. Scc1 is cleaved by separase upon anaphase onset. The discovery of cohesin dispelled the view that sister chromatids might be held via intercatenation of sister DNAs that was resolved at anaphase due to microtubule pulling forces.
Cohesins can form ring-shaped structures in vitro, leading to several hypotheses that describe how these proteins connect sister chromatids [1, 2]. These include the embrace model, in which the complex forms a ring around sister DNA helices, the snap model in which each cohesin complex binds a single DNA helix and linkage occurs through the association of two complexes, and the bracelet model in which cohesin complexes oligomerize to wrap around sister DNA helices.
Genome wide chromatin immunoprecipitation (ChIP) in budding yeast has revealed the predominant sites of cohesin binding [3, 4]. Most notable is the finding that cohesin is enriched ∼3-fold in a 20−50kb domain flanking the centromere, relative to the concentration of cohesin on chromosome arms. Although the location of cohesin along the length of the yeast chromosome has been established, little is known about how the concentration of cohesin within pericentric chromatin contributes to the fidelity of chromosome segregation.
The ability to visualize budding yeast chromosomes in live cells revealed that sister centromeres/kinetochores are separated before anaphase, as occurs in mammalian cells. Repeated arrays of the lac operator (E. coli lacO) were inserted into the yeast genome. Introduction of lac repressor-GFP allowed visualization of specific chromosomal domains [5]. Placement of the lacO array at varying distance from the centromere revealed that chromosome arms were closely apposed, while pericentric chromatin is stretched poleward in mitosis, prior to anaphase onset (Fig. 1)[6-9]. Sister centromeres on a single chromosome oscillate relative to each other, and often are separated by distances of up to 800nm. The oscillation in separation distance suggests that the peri-centromere regions of the chromosome are elastic, stretching in response to their dynamic kMT attachments. Using GFP-fusion proteins to mark centromeres of all chromosomes (centromeric histone H3 variant Cse4 [9, 10], and the inner kinetochore component Mtw1 [6]), it was found that sister kinetochores are organized into two lobes on either side of the equator of the metaphase spindle. This bipolar alignment is indicative of sister centromere separation prior to anaphase. Subsequent visualization of a number of kinetochore proteins and examining their behavior after photobleaching [11] has substantiated the finding that sister centromeres are pulled apart by sister kinetochore pulling forces in metaphase. The major paradox in the field is the accumulation and function of cohesin at sites of sister chromatid separation.
Figure 1. Organization of a mitotic chromosome.
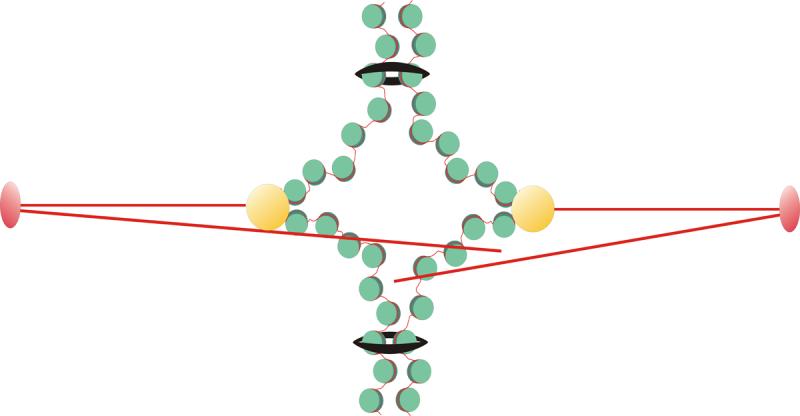
Chromosome arms are closely apposed and held together via cohesin (black rings). Sister kinetochores (yellow) are attached to kinetochore microtubules (red) and the pericentric chromatin is stretched toward the spindle poles. There are 16 chromosomes in yeast, and 16 kinetochore microtubules in each spindle half. Cohesion between sister chromatids provides a mechanism to resist microtubule pulling forces and generate tension at centromeres. The function of cohesin in pericentric chromatin is not well understood.
Results
A cylindrical array of cohesin in mitosis
We have determined the 3-dimensional distribution of cohesin in mitotic cells using two core cohesin components Smc3 and Mcd1/Scc1 and a spindle pole body protein (Spc29) expressed from their endogenous promoters and fused to green or red fluorescent protein, respectively (GFP, RFP). In sagittal section (side view of the mitotic spindle) Smc3-GFP and Scc1-GFP are concentrated in two fluorescent lobes between the poles of the mitotic spindle (Smc3-GFP, Spc29-RFP Fig. 2A; Scc1-GFP, Spc29-CFP Suppl. Fig. 2A). The peak intensity of fluorescence is 2.23±0.43 times the intensity of cohesin in nuclear regions away from the spindle. In transverse section (end-on view of the mitotic spindle) Smc3-GFP forms an apparently hollow ring centered about the spindle axis (Smc3-GFP, Spc29-RFP, Fig. 2B). There are numerous cohesin subunits concentrated in a cylindrical array around the mitotic spindle. To determine the height of the cylindrical array, linescans were drawn through the long axis of fluorescence in Fig. 2A. The distance between the half maximum positions of the fluorescence intensity from one end of the cylinder to the other is 586±105 nm (Fig. 2A, n=42, schematic in Fig. 2G). The width of the cylindrical array was determined by taking 3D stacks of images through sagittal (Fig. 2C) and transverse (Fig. 2D) planes and measuring the distance between maximal fluorescence values of each peak (sagittal, Fig. 2E, G; transverse, Fig. 2F, H). The distance between the bi-lobed peaks of fluorescence is 293±56 nm (sagittal plane, Fig. 2E, G) versus 365±51 nm (transverse plane Fig. 2F, H). The distance between the two peaks is constant through the entire height of the cylinder (3D stacks of transverse sections at 100nm steps, Fig. 2D). The decreased width measurement through the short axis of the bilobed fluorescence in sagittal section (Fig 2A, C, E) reflects variation that arises from off-centered sections. If the aperture of the cylindrical array was less than 350nm in diameter, the objective point spread function (Airy disk) would preclude the appearance of a hole in the cohesin fluorescence (Suppl. Fig. 1B). This pattern of cohesin fluorescence is indicative of a cylindrical array ∼365nm in width and ∼586nm in height.
Figure 2. Distribution of Smc3-GFP in metaphase.
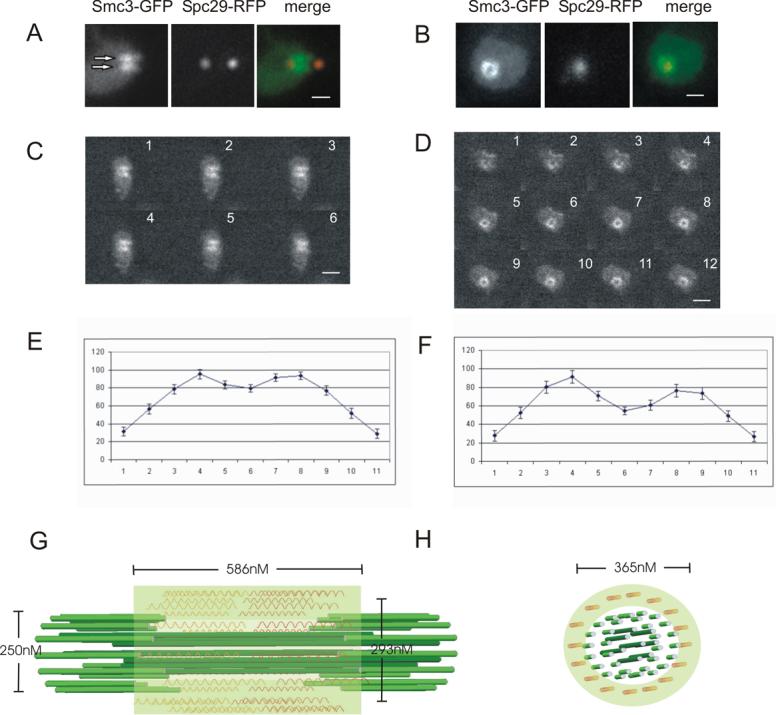
A. Smc3-GFP is concentrated between the spindle pole bodies in metaphase. Two oblongate lobes of fluorescence (arrows in Smc3-GFP image, green in overlay) with a dimmer area between are aligned between the spindle poles in a sagittal view of the spindle (Spc29-RFP, red in overlay). Spindle length=1.2μm. (scale bar = 1 μm). B. A cylindrical array of Smc3-GFP is visible in a transverse view of the spindle. Note the circular distribution Smc3-GFP (green in overlay) relative to the spindle pole body (Spc29-RFP, red in overlay). C. Confocal serial sagittal sections of Smc3-GFP. D. Confocal serial transverse sections of Smc3-GFP. The bi-lobed fluorescence is qualitatively visible above background through ∼600 nm (on average 6.4 ± 0.9 100nm steps, n=8) from the sagittal view and ∼1000 nm (on average 7.8 ± 1.9 100nm steps, n=10) from the transverse view. A cylinder with a diameter of ∼350nm and height of ∼600nm would be seen through greater than 4 100nm steps in the sagittal view, and greater than 7 100nm steps in the transverse view when the decreased resolution due to the PSF of the microscope objective in the z-axis is taken into account. The number of steps in the sagittal and transverse views is consistent with the fluorescence measurements of the cylinder's height and diameter. E. Linescan through the oblongate lobes of Smc3-GFP fluorescence in the sagittal view. Error bars are S.E.M. (average of 53 cells). F. Linescan through Smc3-GFP fluorescence in the transverse view. Error bars are S.E.M. (average of 22 cells). Arbitrary fluorescence units (y-axis) are plotted vs. distance in pixels (x-axis, 65 nm/pixel). G. Model for pericentric cohesin in sagittal view. The mitotic spindle is comprised of 32 kinetochore MTs (16 in each half spindle, lt. green) and 8 interpolar MTs (4 from each pole, dk. green), 250nm in diameter. The distribution of cohesin is depicted as a transparent cylinder 586 nm in height and 293 nm in width. Pericentric DNA associated with cohesin is depicted as springs (orange) that span the distance between kinetochore microtubules. H. Model for pericentric cohesin in transverse view. Spindle microtubules (green) are surrounded by the cylindrical array of cohesin (transparent green). The position of pericentric DNA (orange) is based upon cohesin binding pericentric chromatin. The diameter of the cylindrical array in transverse section is 365nm.
To determine the relationship of the cylindrical array of cohesin with respect to kinetochore microtubules and the metaphase spindle, we examined strains containing Smc3-GFP and Ndc80-Cherry (Ndc80, an outer kinetochore complex member)(Fig. 3A,B) or Tub1-CFP (Fig. 3C, D). In the sagittal view, clusters of Ndc80-Cherry cap the fluorescent cohesin lobes (Fig. 3A) while the valley of SMC3-GFP fluorescence coincides with the position of interpolar microtubules (Fig. 3C). In the transverse view, Ndc80-Cherry is surrounded by a ring of Smc3-GFP (Fig. 3B) and the spindle is centrally aligned (Fig. 3D). Thus Smc3-GFP is distributed cylindrically around central spindle interpolar microtubules and spans ∼75% the distance between separated clusters of kinetochores in metaphase (586 nm cohesin vs. 800 nm kinetochore clusters).
Figure 3. Position and functional requirements for the cylindrical array of cohesin.
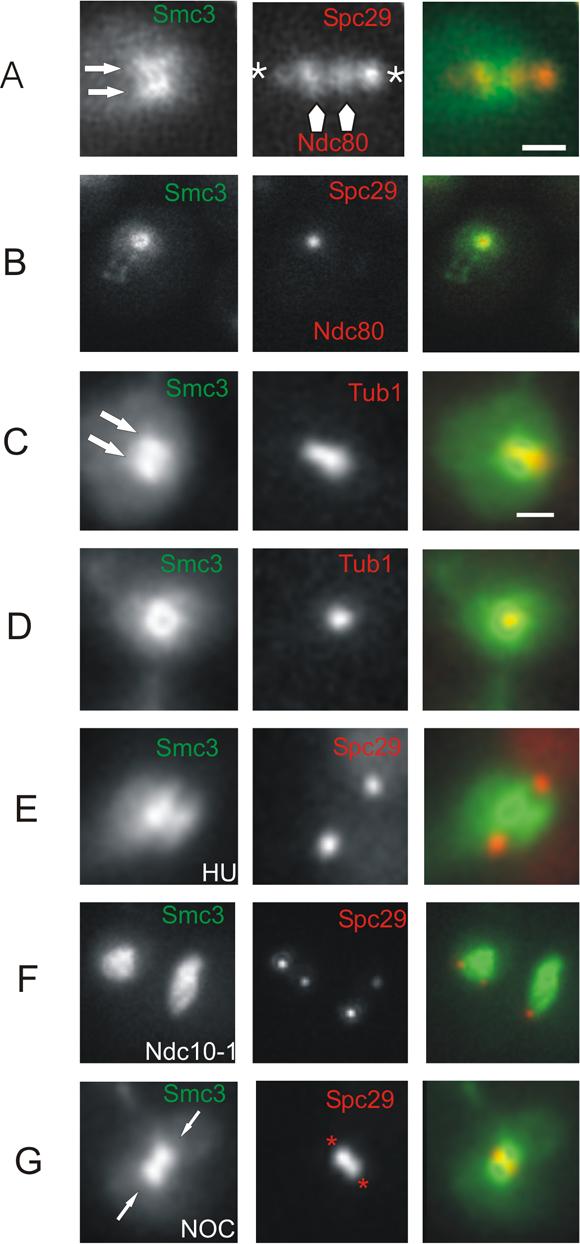
A. Co-localization of Smc3-GFP (left), Ndc80-Cherry (arrows) and Spc29-RFP (asterisks)(middle) in sagittal section. The overlay (right) is a merge of both channels, Smc3-GFP green; Spc29-RFP and Ndc80-Cherry red. B. Co-localization of Smc3-GFP, Ndc80-Cherry and Spc29-RFP in transverse section. Smc3-GFP appears as a cylinder. Ndc80-Cherry resides within the cohesin cylindrical array. C. Co-localization of Smc3-GFP and Tub1-CFP in sagittal section. The bi-lobed fluorescence of Smc3-GFP encircles Tub1-CFP (red in merge). D. Co-localization of Smc3-GFP and Tub1-CFP in transverse section. The cylindrical array of Smc3-GFP encircles Tub1-CFP (red in merge). E. Smc3-GFP and Spc29-RFP localization following incubation with hydroxyurea (HU) for 3 hours. A bipolar spindle is formed (middle) and Smc3-GFP is concentrated in a bi-lobed structure (left) that lies between the two spindle poles. Smc3-GFP green, Spc29-RFP red in merge. F. Smc3-GFP distribution in the absence of functional kinetochores. Cells containing a temperature sensitive allele of ndc10−1 were grown at the restrictive temperature. There is no structural organization of Smc3-GFP. Smc3-GFP green, Spc29-RFP red in merge. G. Smc3-GFP distribution following 2 hour incubation in 20 μg/ml nocodazole (NOC). The spindle has collapsed as evidenced by the appearance of adjacent spindle pole bodies that mark the former spindle axis (middle, asterisks in red). Smc3-GFP remains concentrated proximal to the spindle poles in nocodazole-arrested cells (arrows in GFP panel and overlay). Two lobes of Smc3-GFP flank a dimmer area in which the collapsed spindle poles lie. Linescan through Smc3-GFP following spindle collapse is similar in shape to linescans of Smc3-GFP in untreated cells (see Suppl. Fig. 4). The fluorescence intensity of Smc3-GFP lobes in cells with collapsed spindles is 2−3× brighter than untreated (see Suppl. Fig. 4). Smc3-GFP green, Spc29-RFP red in merge.
Cohesin is associated with chromosomes from G1, promoting cohesion upon replication and persisting until the onset of anaphase. In live cells, Smc3-GFP is seen to accumulate near the SPB in S-phase (Suppl. Fig. 1D-F). However the cylindrical array is only apparent following DNA replication and bipolar spindle formation. To address the extent of DNA replication required for the cylindrical array we examined Smc3-GFP in cells treated with hydroxyurea (HU) (Fig. 3E). In the presence of HU only early origins of replication fire, and the bulk of chromosome replication is severely delayed or arrested [12]. Centromere DNA replication is under the control of early firing origins. Upon HU treatment cells arrest with bipolar spindles and bi-oriented chromosomes, visualized by two centromere proximal lacO spots, reflecting the replication of centromere DNA (Suppl. Fig. 2A) [6]. The dimensions and intensity of Smc3-GFP are indistinguishable in HU treated vs. non-treated cells (Fig. 3E). Replication of centromeric regions and subsequent biorientation is therefore sufficient for establishment of cohesin into a cylindrical array surrounding the central spindle.
Stability of pericentric cohesin
Upon anaphase onset, a subunit of cohesin (Scc1) is cleaved by separase [13], the spindle elongates (anaphase B) and sister kinetochores migrate to opposite poles (anaphase A). Anaphase onset is defined as spindle elongation (13−14 min, Fig. 4A). The fluorescence intensity of pericentric and arm cohesin was plotted as a function of time from metaphase to anaphase (Fig. 4A graph). Cohesin fluorescence decreased on average 66% between two and six minutes upon the onset of anaphase (n=6). Pericentric cohesin is lost at or slightly before pole separation (Fig. 4A, top). No overall change in Smc3 arm fluorescence is detected from metaphase to the end of anaphase.
Figure 4. Cohesin stability.
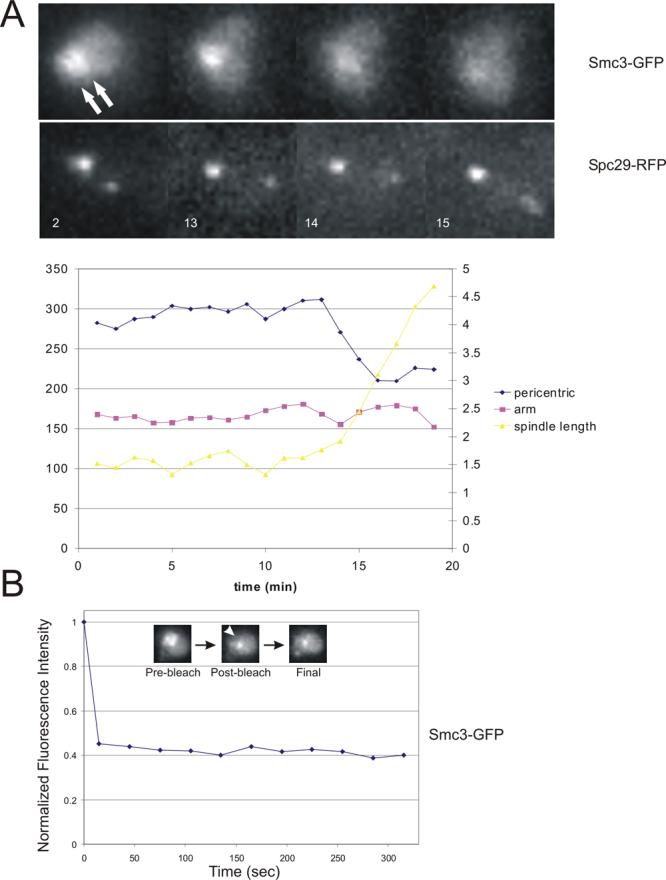
A. Loss of the cylindrical array in early anaphase. A timelapse series of Smc3-GFP is shown as cells progress from metaphase to anaphase. The concentration of Smc3-GFP fluorescence is visible in metaphase between the spindle poles (2min, top left Smc3-GFP; bottom, left Spc29-RFP)(minutes indicated in bottom left corner). The fluorescence intensity between the two poles decreases from 13−14 min as the spindle elongates in anaphase (increase in distance between Spc29-RFP spindle poles, bottom panel). The difference in fluorescence intensity of spindle poles reflects the maturation time for RFP fluorescence in the new pole (to the right, bottom panel). Fluorescence intensity of Smc3-GFP was determined as described in Materials and Methods and plotted as a function of time in the bottom graph (left axis in arbritrary units, fluorescence intensity; right axis in microns, spindle length). Slightly before or concomitant with spindle elongation the fluorescence intensity between the spindle poles decreases. B. Stability of cohesin in the cylindrical array. Fluorescence Recovery after Photobleaching (FRAP) was measured to determine cohesin stability in the pericentric region in cells expressing Smc3p-GFP. An argon laser attached to an inverted fluorescence microscope was directed to a portion of Smc3-GFP between the spindle poles (prebleach). GFP within a diffraction limited spot (∼0.25micron) was bleached (post-bleach). Images were acquired every 30 seconds for 5min following photobleaching. As shown in the graph no recovery of Smc3-GFP fluorescence was detected. Cohesin fluorescence recovery above the background was apparent in only 2 of 17 cells
The number of cohesin complexes holding the pericentric DNA structure together is crucial for understanding the dynamic extension-relaxation behavior of pericentric chromatin observed during metaphase centromere movements. Using a comparative measurement of Smc3p-GFP fluorescence signal (to a known number of 2 Cse4 molecules per kinetochore [14], we estimate that there are 108 +/− 40 (n=7) molecules in the pericentric region of cells containing Smc3-GFP (Suppl. Table 1). The concentration of cohesin increases in the absence of tension to 222 +/− 103 molecules/pericentric region (n=11). These measurements translate to a minimum of 3.5 cohesin complexes per C-loop (108 Smc3 molecules/32 centromeres). Based on centromere dynamics to deduce the amount of DNA in each C-loop under tension in metaphase [9], we estimate that there is 1 cohesin complex every 4 kb, or 1 complex every 20 nucleosomes. The measurements are consistent with distribution of cohesin from chromatin immunoprecipitation experiments [4, 15] and the ∼2.5 fold increase in cohesin along pericentric chromatin in the absence of tension [16].
Considering that sister kinetochore separation is dynamic, we have addressed whether cohesin within the pericentric chromatin is stably bound. Fluorescence Recovery after Photobleaching (FRAP) was used to quantitatively monitor cohesin stability in metaphase cells expressing Smc3-GFP (Fig. 4B). In metaphase, one lobe of the GFP-cohesin cylinder was selectively targeted with a 200 ms laser exposure, while the other lobe retained fluorescence. Fluorescence of the bleached area was measured at 30 second intervals for five minutes. Comparison of integrated intensity measurements from the two sides of the cylinder revealed that cohesin fluorescence recovered above the background in only 2 of 20 cells analyzed. No significant fluorescence loss in the unbleached lobe was detected (data not shown). These results suggest that once assembled cohesin is stably bound to pericentric chromatin. In contrast, FRAP of histone H2B-GFP (an exchangeable component of the nucleosome [17]), revealed that 37 % +/− 12 % of H2B was dynamic in 10 out of 10 cells analyzed (exchanging with t1/2 = 67 seconds +/− 16 seconds) (data not shown).
Chromosome conformation at the centromere
If the cylindrical distribution of cohesin reflects the geometric array of bi-oriented sister chromatids in mitosis, then separated sister kinetochores and flanking pericentric chromatin may be paired via intramolecular rather than intermolecular contacts [18]. To examine the conformation of pericentric DNA in vivo we utilized an inverse PCR strategy to map chromosome conformation (3C) [19]. Inverse primer pairs to map the conformation of chromatin are shown in Fig. 5A (Pericentric chromatin P1, P2 and P3; Arm chromatin A, 75 kb from the centromere). Upstream and downstream primer pairs P1u, P1d and Au, Ad are separated by the same physical distance (15kb) in the genome. Chromatin was fixed by treating cells with formaldehyde, the chromatin digested with XbaI and ligated under dilute conditions to minimize intermolecular reactions. Each primer is ∼200bp downstream from an XbaI site resulting in PCR products of 400bp when fragments containing sites complementary to the respective oligonucleotides (P1u + P1d and Au + Ad) ligate. To quantitate random association due to thermal motion, ligation was also performed in the absence of cross-linking. Primer pairs from the chromosome arm (Au, Ad) provide control template to account for template yield in each experimental preparation.
Figure 5. 3C assay for intramolecular looping.
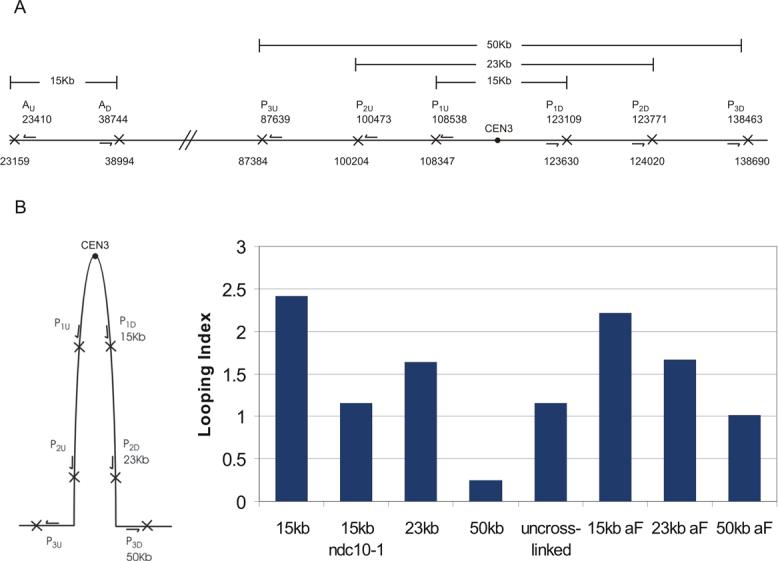
A. The schematic shows the position of oligonucleotide primers on chromosome III (arrows) relative to the centromere (filled circle). Each pair of oligonucleotides (P1u,P1d and Au,Ad; pericentric vs. arm chromatin) extend away from each other on the linear chromosome. XbaI sites are indicated by (X) downstream each oligonucleotide primer. In the linear chromosomal configuration, these oligonucleotides will not prime DNA synthesis following the 3C assay. If there is intramolecular looping (as diagrammed in B, left), the P1u,P1d oligonucleotides will prime DNA synthesis. The products from PCR reactions following cross-linking, restriction digestion and ligation were quantified as described in Material and Methods. B. (left) Schematic representation of the extent of the intramolecular loop. (right) Looping index for each experimental sample. The looping index accounts for differential efficiency of PCR reactions with primer set P versus A at an equivalent ratio of input template (see complete description in Suppl. Fig. 3). A looping index of 1 indicates equal concentration of input template for pericentric and arm products respectively. Experimental samples (WT-wild-type; alpha factor, ndc10−1, and uncrosslinked) were prepared as described in Materials and Methods.
In the absence of cross-linking the ratio of PCR products from the pericentric vs. arm chromatin (primer pairs P versus A) was 1.25 ±0.15 (Suppl. Fig. 3B). This ratio could reflect greater ligation efficiency between fragments spanning the XbaI junction flanked by P1u, P1d versus Au, Ad, or more efficient PCR reaction with primer pairs P1u, P1d versus Au, Ad. To address the source of the difference and to ensure that the PCR reactions were linear over the range of input DNA, we constructed the template for inverse primer pairs P1u, P1d and Au, Ad. Templates were constructed by amplifying DNA from each primer to its respective XbaI site (P1u → XbaI, P1d → XbaI; and Au → XbaI, Ad → XbaI, as shown in Fig. 5A). The fragments were digested with XbaI and appropriate fragment pairs ligated (P1u + P1d; Au + Ad), and amplified with primer pairs P1u + P1d or Au + Ad. The yield of each product was quantitated by gel electrophoresis and absorbance at A260. PCR was performed over a range of template DNA concentration (Suppl. Fig. 3A). At an input ratio of 1:1 P:A template, the mean ratio of PCR products was 1.21. This value was constant over a 10-fold range of input DNA concentration (Suppl. Fig. 3A). The ratio of the PCR products using primer pairs P versus A in the uncrosslinked sample (above) reflect equal concentration of the respective pericentric (P) or arm (A) templates following the sample preparation.
The experimental Pericentric:Arm (P:A) product ratios plotted against reconstructed P:A input ratios over the range of 0.3 − 5.0 are shown in Suppl. Fig. 3A. There is a linear relationship over a 10-fold range (0.3−2.0 picograms) of template DNA. All subsequent analysis was performed within the linear range of these 3C PCR reactions. The plot accounts for the different PCR efficiency of the primer sets and provides a standard for calibrating the degree of intramolecular looping from the experimental sample. We thus define a “looping index” as the ratio of P:A normalized for differences in PCR efficiency. A looping index of ∼1.0 reflects an equal concentration of input pericentric and arm products, indicative of an equal propensity for arm or pericentric chromatin to loop (uncrosslinked sample, Fig. 5B; P/A product ratio of 1.25, Suppl. Fig. 3B). For example, in the cross-linked samples amplified with the 15kb primer pair (7.5 kb on either side of CEN3), there is a P/A product ratio of 1.96 (Suppl. Fig. 3B) and a looping index of 2.41 (Fig. 5B). Thus pericentric chromatin DNA is 2.4× more prone to adopt an intramolecular loop (C-loop) relative to arm DNA (Fig 5B). This increase in looping index (2.41) is comparable to the increased cross-linking efficiency reported by Dekker et al. [19] for the chromosome III centromere.
To determine the physical length of the intramolecular loop, we designed additional primer pairs spanning 23kb (P2u, P2d) and 50kb (P3u, P3d) of pericentric DNA (∼11.5 and 25 kb on either side of CEN3, respectively) (Fig. 5A, B). The Pericentric:Arm product ratio of uncrosslinked DNA varies with each primer pair (1.02 ± and 2.01 ± for the 23kb and 50kb primer pairs respectively, see Suppl. Fig. 3B). Following crosslinking, the Cen:Arm product ratio increased by 24% increase (1.64 looping index) for the 23kb primer pair, and decreased 43% (0.25 looping index) for the 50kb primer pair. The looping index for the 50kb primer pair in the absence of DNA replication is 1.01 (alpha factor 50kb, Fig. 5B). The 0.25 looping index obtained in logarithmic phase growth indicates that the conformation of chromatin 25kb on either side of the centromere is anti-correlated with intramolecular looping and may be constrained in its ability to adopt certain conformations. These results extend conclusions from examination of sister chromatids with lacO operators at various positions from the centromere [9] and indicate that intramolecular looping extends beyond 11.5 kb but not 25kb on either side of the centromere.
The intramolecular pericentric loop is completely dependent upon kinetochore function. Ndc10 is one of the centromere DNA binding factors (CBF3) and is essential for kinetochore formation [20]. The looping index for pericentric chromatin in ndc10−1 mutant cells at the restrictive growth temperature is 1, equivalent to the uncross-linked sample (Fig. 5B and Suppl. Fig 3). As Ndc10 has been shown to bind additional sites in the genome [21], we utilized an alternative strategy disrupt kinetochore function. Induction of a transcriptional promoter adjacent to the centromere results in loss of segregation function [22]. Cells containing a conditionally functional centromere (GALCEN3) were transferred to media containing galactose to activate the GAL1 promoter. The looping index of pericentric chromatin flanking an inactive centromere was 1.04 (Table 1 and Suppl. Fig. 3A). Ndc10 is depleted at the GALCEN locus [23], and therefore intramolecular looping of pericentric chromatin is dependent upon a functional kinetochore.
Table 1.
| Condition/Fragment Size | Looping Index |
|---|---|
| WT 15kb | 2.41 |
| WT 23kb | 1.64 |
| WT 50kb | 0.25 |
| mcd1−1 15kb | 1.54 |
| ndc10−1 15kb | 1.15 |
| ndc10−1, aF 15kb | 1.15 |
| aF 15kb | 2.31 |
| aF 23kb | 1.67 |
| aF 50kb | 1.01 |
| galcen 15kb | 1.18 |
| uncrosslinked | 1.15 |
Looping Index for wild-type and mutant strains. The looping index is a measure of the propensity for two regions of chromosome III to interact following crosslinking, restriction digestion and ligation. The looping index is detailed in the text and Suppl. Fig. 3.
Upon loss of kinetochore function, there is concomitant loss of the cylindrical cohesin array. ndc10−1 mutants at restrictive temperature no longer organize cohesin into a cylinder around the mitotic spindle (Fig. 3F). Likewise, upon disruption of microtubule attachment in the Ndc80 outer kinetochore complex (nuf2−45 mutants), Smc3-GFP is randomly distributed in the nucleus and visible as puncta of various size and position relative to the spindle poles (Suppl. Fig. 1C).
To determine whether sister chromatids are required for C-loop formation, we examined the structure of pericentric chromatin in cells arrested prior to DNA replication (via alpha-factor treatment). The looping index for the 15kb primer pair (7.5kb on either side of the centromere) was 2.31 in cells treated with alpha-factor (Table 1, Fig. 5B and Suppl. Fig. 3A), and the loop extends a comparable physical distance to that observed in logarithmic growing cells (Fig. 5B, WT 23kb 1.64, aF 23kb 1.67). In contrast, the looping index for 50kb of pericentric DNA (0.25) increased to 1.01 in αF-treated cells (Fig. 5B and Suppl. 3B). Thus DNA sequences 25kb on either side of CEN3 exhibit random associations prior to DNA replication, comparable to those measured in uncrosslinked controls at 50kb (Suppl. Fig. 3B). Upon replication, chromatin at the base of the C-loop is held via cohesin-mediated sister chromatid linkages and the efficiency of ligation 25kb on either side of CEN3 drops precipitously (Fig. 5B).
An alternative strategy to address the role of sister chromatids is through the use of mutations in cohesin subunit Mcd1/Scc1. Mcd1 is expressed late in G1 and is largely absent from chromosomes prior to Start [24, 25]. mcd1−1 mutants arrest with a metaphase like spindle and prematurely separated sister chromatids. The looping index is 1.54 in mcd1−1 mutants (Table 1 and Suppl. Fig. 3A). The reduction in looping index reflects a reduction in the number or length of intramolecular loops in the population, or increased distance between the two strands. While cohesin is not required for loop formation, cohesin does contribute to the stability, extent or proximity of the intramolecular loops.
Cohesin contributes to spindle length control
The physical arrangement of pericentric chromatin in intramolecular loops predicts that chromatin may be a mechanical component of the spindle [8, 26-28]. Bouck and Bloom have shown that reduction of histone expression leads to increased spindle length in metaphase [26]. If cohesin contributes to the physical properties of pericentric chromatin, reduction of cohesin is likewise expected to influence metaphase spindle length. Spindle length was measured in mcd1−1 mutants grown at permissive and restrictive conditions. mcd1−1 mutants arrest primarily in mitosis, but the mutation does not prevent Anaphase A (chromosome to pole movement) or exit from mitosis [29]. To examine spindle length in cells prior to anaphase onset we introduced lacO arrays into the LYS2 gene on chromosome II in mcd1−1 mutants [30]. Only cells with separated sister chromatids in metaphase (separated lacO spots, but spots not at spindle poles) were examined. Metaphase spindle length was 1.44 ± 0.32 (n=61) at 25°C. Spindle length increased to 2.35 ± 0.77 (n=27) upon shift to 37°C. This is comparable to the increased in spindle length upon reduction of histone H3 (from 1.47 ± .28 n=71 to 2.33 ± 0.40, n=77 [26]). A corollary to the hypothesis for pericentric chromatin as an extensible element of the spindle is that upon spindle collapse the chromatin should relax and the cylindrical distribution should be compacted. To examine the distribution of Smc3-GFP in the absence of tension, cells were treated with nocodazole to depolymerize microtubules and collapse the spindle (Fig. 3G). The concentration of cohesin increases in the vicinity of the spindle poles following spindle collapse (Fig. 3G and Suppl. Fig. 4). These data are indicative of the inward recoil of the pericentric chromatin relative to spindle poles upon spindle collapse, and consistent with the finding that cohesin accumulates within pericentric chromatin in the absence of tension [16].
Discussion
The simple budding yeast spindle with its complete genomic sequence including the centromeres is an excellent model from which to deduce the contribution of microtubules and chromosome organization to spindle function. There are 16 kinetochore microtubules emanating from each SPB, on average 0.35μm in length in the half spindle and 4 interpolar microtubules ∼1μm in length emanating from each spindle pole. Kinetochores from each of the 16 chromosomes cluster into a diffraction limited spot, and upon bi-orientation appear as two clusters in mitosis. We provide evidence for the structural basis for kinetochore bi-orientation. Pericentric cohesins are organized in a cylindrical array around the metaphase spindle. Each sister chromatid adopts an intramolecular loop (C loop) with centromere DNA at the apex, attached to the MT, that extends toward sites of intermolecular cohesion between sister chromatids 25kb from the 125pb centromere core (Fig. 6A). Pericentric chromatin spans the distance between clusters of 16 bi-oriented kinetochores, resulting in a cylindrical array of the 32 pericentric regions within the mitotic spindle (Fig. 6B). This results in a cruciform configuration between sites of microtubule attachment and sister chromatid pairing (Fig. 6A). These loops of pericentric DNA together with cohesin may provide the mechanical linkage between separated sister kinetochores.
Fig. 6. Model of the organization of cohesin and pericentric chromatin in metaphase.
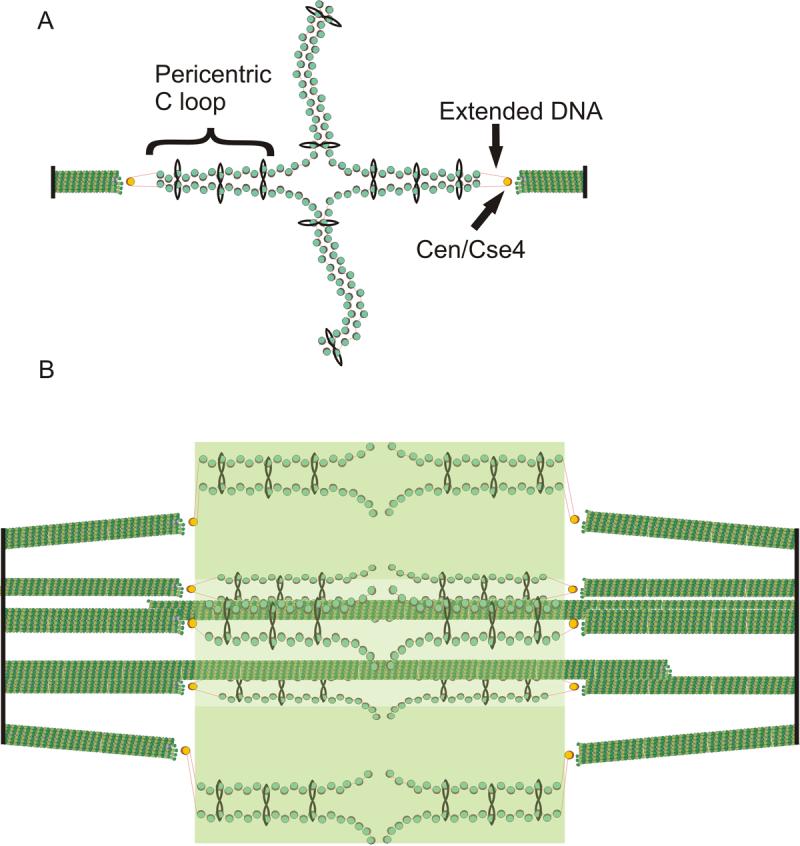
(A) DNA of each sister chromatid is held together via intramolecular bridges that extend approximately 11.5kb on either side of the centromere. There is a transition from intra- to intermolecular linkages resulting in a cruciform structure. (B) 5 (of 16) bioriented sister chromatids are shown with two (of 8) interpolar microtubules. We have proposed that the transition between intramolecular looping and intermolecular cohesion is mobile and on average 7kb from the centromere core [18]. DNA adjacent to the centromere may extend to its B-form length in vivo (as described in text, depicted as red lines) linking the centromere at kinetochore microtubule plus-ends to strands of intramolecularly paired pericentric chromatin and cohesin that are displaced radially from spindle microtubules. Microtubules (green rods), spindle pole bodies (black rods), 125 bp centromere (wrapped around the Cse4-containing histone in yellow), nucleosomal chromatin (green histone cores wrapped around DNA in red), cohesin (black circles). The fluorescence distribution of cohesin is depicted in transparent green. Pericentric chromatin from each of the 16 chromosomes is displaced 70−90nm radially from the central spindle microtubules. The entire spindle is composed of 32 kinetochore microtubules and 8 pole-pole microtubules.
The discovery of intramolecular looping at the centromere provides a solution to the major paradox in understanding the accumulation of cohesin at sites of separated sister DNA strands. Cohesin is organized into a supramolecular cylindrical array encompassing the mitotic spindle. From the fluorescence distribution and the spread of light through the objective (point spread function) we estimate the dimensions of the cylindrical array to be approximately 340nm in diameter by 586nm in length. The diameter of the cohesin cylinder is approximately 70−80nm larger than measured for the diameter of the spindle microtubules in yeast by electron microscopy [31], and 220 nm shorter than the distance between clusters of microtubule plus ends (∼800nm) [32]. This difference in diameter is the basis for depicting cohesin and the pericentric C-loops radially displaced from the spindle microtubules (Fig. 6B). A question raised by these findings is how the pericentric chromatin is physically linked to the microtubule plus-end. The centromeric nucleosome cluster, as visualized by Cse4-fluorescence, shows no such cylindrical array, and the cluster is very close to the microtubule plus-end [33]. We can reconcile these findings by proposing that the plus-ends of the kinetochore microtubules are very close to the Cse4 nucleosome, and the adjacent 70−90bp of DNA flanking the Cse4 nucleosome spans the distance to the flanking chromatin (Fig. 6B). There are several features of the DNA flanking the 125bp centromere that are consistent with the idea that this DNA is in a B-form configuration and devoid of protein. Firstly, the concentration of cohesin is reduced at the centromere core and does not increase until 50−100bp away from the centromere [15]. Secondly, nuclease hypersensitive sites (70−90bp) have been mapped to the region immediately flanking the 125bp CEN [34]. Third, the region of pericentric chromatin is hyper-stretched relative to chromosome arms in mitosis [9]. These data suggest that the 70−90bp of DNA adjacent to the centromere may extend to its B-form length in vivo (24−30nm long), linking the centromere at kinetochore microtubule plus-ends to strands of intramolecularly paired pericentric chromatin that are displaced radially from spindle microtubules (Fig 6B).
Cohesin deposition is dependent upon kinetochore function [3]. However cohesin is not essential for intramolecular looping (mcd1−1, Table 1), nor is tension (αF, Fig. 5B and Table 1). In contrast, the loss of the inner centromere binding complex ndc10−1 results in loss of the intramolecular loop. Ndc10 along with the other proteins that constitute the core centromere DNA binding factor (CBF3) bends centromere DNA approximately 60° [35]. This deflection in DNA curvature may favor intramolecular loop formation. Cohesin contributes to the stability and/or extent of pericentric loop formation as evidenced by the fractional decrease in looping index (2.41 to 1.54 in mcd1−1). Cohesin remains concentrated in the vicinity of the spindle poles following spindle collapse with nocodazole (Fig 3G, Suppl Fig 4). The fluorescence intensity per pixel is 2 − 3 times brighter following spindle collapse (Suppl. Fig. 4), indicating that rather than pericentric chromatin unraveling into the nucleus, the chromatin is further condensed or compressed. If pericentric chromatin were inelastic, DNA strands would not be expected to remain aligned tightly with the two collapsed spindle poles. Alternatively, if pericentric chromatin were elastic, then upon loss of tension generated by microtubules the chromatin would condense and retract adjacent to the collapsed spindle poles. The latter is observed experimentally and supports the view that this region behaves as spring-like element that generates an inward force [26-28].
While individual strands of DNA may be quite weak springs, the spring constant of parallel arrays of springs is the sum of the individual spring constants. In addition, the intramolecular pairing of pericentric chromatin confers unique structural properties that may be important during mitosis. A two-fold increase in the radius of a filament increases its resistance to bending 16-fold. The additional loading of cohesin is very likely to reinforce the tensile strength of these loops, as demonstrated for the role of condensin in organizing rigid elastic chromosomes axes [36].
The depiction of the 16 microtubule attachment sites clustered around the mitotic spindle (Fig. 6B) may be relevant to understanding the organization of complex, or regional kinetochores. Centromeres in budding yeast are small (125bp) compared to other fungi (30−40kb S. pombe) and mammalian cells (∼5Mb). In contrast, the number of microtubules/chromosome is 1 in budding yeast, 2−3 in fission yeast and 25−30 in mammalian cells. Why such a large disparity in centromere DNA content, and not in microtubule number? This range of DNA sequences specifying kinetochore formation has lead to the classification of point vs. regional centromeres [37]. If one considers that the centromere is comprised of the site for kinetochore protein binding as well as pericentric flanking DNA, the ratio of pericentric DNA/microtubule may indeed scale throughout phylogeny (20kb of pericentric chromatin / microtubule attachment site in yeast vs. 1Mb of centromeric chromatin/30 microtubule attachment sites in mammalian cells = ∼30kb/attachment site). Furthermore, sister centromere pairs are separated by similar distances when under tension (∼2μm, newt lung cell [38], vs. ∼0.8μm, budding yeast [9]) despite extreme disparity in spindle size. The view of a cylindrical arrangement of pericentric chromatin and clustered kinetochores in yeast may reflect the structural basis for kinetochore function that is conserved throughout phylogeny. The basic subunit organization of the eukaryotic kinetochore is the single attachment site. However multiple attachment sites can be clustered whether they are on separate chromosomes (as in yeast) or within a single chromosome (as in mammals). There is evolutionary and experimental precedence for the idea that kinetochores are clusters of individual attachment sites. Indian muntjac kinetochores (2n=6) are thought to represent a centromere fusion evolved from the smaller Chinese muntjac progenitor (2n=46) [39]. On the experimental side, Zinkowski and Brinkely [40] were able to fragment kinetochores by inducing mitosis with unreplicated genomes. This led to the idea that the mammalian kinetochore is based on a repeat subunit structure. The single microtubule binding site in budding yeast may be the conserved repeat subunit and the cluster of 16 yeast kinetochores may be comparable to one mammalian kinetochore consisting of multiple attachment sites.
In summary, the cylindrical array of two cohesin subunits, Smc3 and Scc1, observed in vivo and the state of pericentric DNA as mapped by chromatin conformation indicate that pericentric chromatin is organized into an intramolecular loop that forms the basis of bi-oriented sister chromatids. The C-loop structure reveals the geometrical basis for kinetochore bi-orientation and resolves the paradox of maximal inter-strand separation in regions of highest cohesin concentration. The data indicate that the chromosome segregation apparatus is a composite structure of two biopolymers, centromere DNA loops and microtubules. C-loops provide the compliant linkage between stiffer kinetochore microtubules. Cohesin contributes to the stability of the C-loops, while the kinetochore provides the mechanical linkage between C-loop DNA and microtubules. This intramolecular loop provides a physical mechanism for biorientation of sister kinetochores. Chromatin buffers mitotic forces on the chromosome throughout cycles of microtubule growth and shortening. The chromatin loops described herein are reminiscent of DNA loops in mammalian kinetochores and may define the fundamental unit for microtubule attachment [40].
Acknowledgments
This work was supported by a grant from the National Institutes of Health GM-32238 to KSB. We thank members of the laboratory for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- 1.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 3.Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 5.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 6.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 7.He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 9.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 12.McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 13.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 14.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamai A, Imoberdorf RM, Strubin M. Continuous Histone H2B and Transcription-Dependent Histone H3 Exchange in Yeast Cells outside of Replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Bloom K, Sharma S, Dokholyan NV. The path of DNA in the kinetochore. Curr Biol. 2006;16:R276–278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 20.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 25.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouck DC, Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol. 2007;17:741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicklas RB. A quantitative study of chromosomal elasticity and its influence on chromosome movement. Chromosoma. 1963;14:276–295. doi: 10.1007/BF00326816. [DOI] [PubMed] [Google Scholar]
- 28.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 29.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 31.Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr., McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson CG, Gardner MK, Paliulis LV, Salmon ED, Odde DJ, Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol Biol Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 35.Pietrasanta LI, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, Hansma H. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci U S A. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almagro S, Riveline D, Hirano T, Houchmandzadeh B, Dimitrov S. The mitotic chromosome is an assembly of rigid elastic axes organized by structural maintenance of chromosomes (SMC) proteins and surrounded by a soft chromatin envelope. J Biol Chem. 2004;279:5118–5126. doi: 10.1074/jbc.M307221200. [DOI] [PubMed] [Google Scholar]
- 37.Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 38.Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci. 1996;109(Pt 12):2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- 39.He D, Brinkley BR. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J Cell Sci. 1996;109(Pt 11):2693–2704. doi: 10.1242/jcs.109.11.2693. [DOI] [PubMed] [Google Scholar]
- 40.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


