SUMMARY
The bacterial pathogen Vibrio cholerae uses toxin-coregulated pili (TCP) to colonize the human intestine, causing the severe diarrheal disease cholera. TCP are long, thin, flexible homopolymers of the TcpA subunit that self-associate to hold cells together in microcolonies and serve as the receptor for the cholera toxin phage. To better understand TCP’s roles in pathogenesis, we characterized its structure using hydrogen/deuterium exchange mass spectrometry and computational modeling. We show that the pilin subunits are held together by tight-packing of the N-terminal α-helices, but loose packing of the C-terminal globular domains leaves substantial gaps on the filament surface. These gaps expose a glycine-rich, amphipathic segment of the N-terminal α-helix, contradicting the consensus view that this region is buried in the filament core. Our results explain extreme filament flexibility, suggest a molecular basis for pilus:pilus interactions, and reveal a previously unrecognized therapeutic target for V. cholerae and other enteric pathogens.
INTRODUCTION
The molecular structures of Type IV pili are of great interest due to their central role in bacterial pathogenesis and their attractiveness as targets for vaccines and therapeutics. Type IV pili are homopolymers of pilin subunits present on many Gram-negative bacteria and at least one Gram-positive organism (Varga et al., 2006). They are typically several microns in length but less than 10 nm in diameter. Type IV pili are essential to virulence for many pathogenic bacteria, where they mediate a wide range of functions, including motility, microcolony and biofilm formation and immune escape (Hegge et al., 2004; Jurcisek et al., 2007; Kirn et al., 2000; Merz et al., 2000). Type IV pilin subunits share a conserved N-terminal amino acid sequence in an extended α-helix that mediates filament assembly (Craig et al., 2004). The amino acid sequences of the C-terminal globular domains vary substantially among Type IV pilins and define the diverse and in some cases unique pilus functions. The molecular structures of the Type IV pilus filaments should provide important clues to the mechanisms by which they accomplish their functions, and may help to identify specific targets for vaccines and therapeutics. These structures have proved challenging to obtain due to the insolubility of the pilin monomers, and the size, flexibility and length heterogeneity of the pilus filaments. Several high resolution structures of Type IV pilin subunits or subunit fragments have been obtained by x-ray crystallography and nuclear magnetic resonance spectroscopy (Audette et al., 2004; Craig et al., 2003; Craig et al., 2006; Hazes et al., 2000; Keizer et al., 2001; Parge et al., 1995; Ramboarina et al., 2005; Xu et al., 2004), yet only one structure of an intact Type IV pilus filament has been determined to date, for the gonococcal (GC) pilus from Neisseria gonorrhoeae (Craig et al., 2006), by combined cryo-electron microscopy (cryo-EM) reconstruction, x-ray crystallography and computational docking.
Vibrio cholerae toxin-coregulated pili (TCP) are Type IV pili that self-aggregate, bringing the bacteria together in microcolonies that protect them from host defenses and concentrate their secreted cholera toxin (Taylor et al., 1987). Pilus:pilus interactions are essential for V. cholerae colonization of the human small intestine. TCP also act as highly specific receptors for the cholera toxin phage (CTXΦ), which can infect non-pathogenic Vibrio species and confer virulence by providing the genes that encode the cholera toxin subunits (Waldor and Mekalanos, 1996). The crystal structure of a soluble, monomeric form of the TCP pilin subunit, TcpA, was solved to 1.3 Å resolution (Craig et al., 2003). This TcpA monomer lacks the hydrophobic N-terminal 28 residues needed for polymerization to filaments. In the full-length Pseudomonas aeruginosa PAK pilin and N. gonorrhoeae GC pilin crystal structures, this segment forms the protruding N-terminal half (α1-N) of an extended α-helix, α1 (Craig et al., 2003; Parge et al., 1995). Residues 1–28 are predicted to be α-helical in TcpA as well, due to their homology with the PAK and GC pilins. In all pilin structures solved to date, the C-terminal half of α1, α1-C, is embedded in a β-sheet within the globular domain. In the GC pilus filament structure, α1 forms a helical array that is buried in the filament core and anchors the globular domains on the pilus surface (Craig et al., 2006). It is widely held that α1 is completely buried in all Type IV pili based on its conserved hydrophobic amino acid sequence and considerable spectroscopic, biochemical and genetic data. Beyond the conserved structural core formed by α1 and the β-sheet, the pilin structures vary substantially in two regions: the αβ-loop that lies between α1-C and the β-sheet, and the D-region, situated between two conserved, disulfide-bonded cysteines. Residues important for pilus:pilus interactions were identified in the D-region of TcpA (Kirn et al., 2000). The αβ-loops and D-regions of numerous other Type IV pilins also play key roles in pilus functions (reviewed in Craig et al., 2004).
The pilin subunit packing within the TcpA crystal lattice provided a clue to the TCP filament assembly as the pilin subunits were arranged as helical filaments. In the absence of α1-N, the TcpA subunits were held together by interactions between their αβ-loops and D-regions. A computational model of the TCP filament was constructed by integrating the TcpA crystal structure and its helical packing in the crystal lattice with pilus dimensions and helical symmetry obtained by transmission electron microscopy (TEM) (Craig et al., 2003). The x-ray coordinates for the full-length PAK pilin were used to model positions of the 28 N-terminal TcpA residues. In the TCP filament model, the TcpA subunits are held together by extensive hydrophobic interactions among the N-terminal α-helices. An additional interface was predicted between the αβ-loops and D-regions of the globular domains, and residues known to be involved in pilus:pilus interactions were exposed on the filament surface. Although the TCP model was consistent with existing biological data, the compact packing of the subunits did not explain the extreme flexibility of the filaments and the structure did not adequately explain the mechanisms for TCP functions in cholera infections.
Here we use hydrogen/deuterium exchange mass spectrometry (DXMS) to examine the structure of TCP. By comparing the solvent accessibility of the TCP filament with that of the soluble, N-terminally-truncated TcpA subunit, we could precisely identify regions of the pilin protein that are exposed in the monomeric TcpA subunit but are buried in the intact pilus filament and are hence involved in subunit:subunit interactions. Using the DXMS data, we modified our computational TCP filament model, which we then tested by mutating key residues at predicted subunit:subunit interfaces to see if these would disrupt pilus assembly. The resulting model explains pilus flexibility and suggests a molecular mechanism for pilus bundling in microcolony formation as well as other pilus functions. Surprisingly, rather than being buried in the filament core, a segment of the N-terminal α-helix is exposed in a gap on the filament surface, and may thus play a role in pilus functions in addition to its structural role.
RESULTS
TCP filaments and soluble TcpA monomer used in the DXMS analysis
To investigate the V. cholerae TCP structure, we used DXMS to determine the surface accessibility of amide nitrogens on short overlapping peptides covering the entire length of the TcpA subunit. TCP peptides that demonstrated rapid hydrogen/deuterium exchange lie on the filament surface whereas those that exchanged slowly are buried, either by the protein fold or by subunit:subunit interactions. To distinguish between these two possibilities, DXMS results of the intact TCP filament were compared with those of the monomeric TcpA subunit. Regions of the pilin protein that are buried by protein fold should be similarly inaccessible to deuterium exchange in both the monomeric TcpA and the intact filament, whereas regions that are involved in subunit:subunit interfaces will be accessible in the TcpA monomer but not in the filament.
The soluble TcpA monomer used for DXMS analysis has its N-terminal 28 amino acids (α1-N) replaced with a 21 residue segment comprised of a hexahistidine tag and linker (his-tag/linker, Craig et al, 2003). The TCP filaments using in this study have a mutation in the TcpA subunit, His181→Ala (TCPH181A), which causes them to be shed into the culture supernatant (Kirn et al., 2000) for large-scale purification. TCPH181A filaments from whole cell culture resemble wild type TCP (TCPWT) by negative stain TEM (Fig. 1A and B) and are equally resistant to chymotrypsin proteolysis (Fig. 1C) suggesting that they share the same quaternary structure. Importantly, TCPH181A do not bundle after purification/concentrating with ammonium sulfate (see Experimental Procedures), thus the deuterium labeling is not inhibited by pilus:pilus interactions.
Figure 1. Comparison of morphology and protease susceptibility for TCPWT and TCPH181A.
Transmission electron micrographs of (A) TCPWT and (B) TCPH181A showing similar morphologies and bundling characteristics, though TCPWT bundle to a greater extent. (C) Coomassie-stained SDS-PAGE of chymotrypsin-digested TCPH181A (lanes 1, 4 and 7), TCPWT (lanes 2, 5 and 8) and the TcpA monomer (lanes 3, 6 and 9) in the absence (0) and presence (+) of chymotrypsin at a chymostrypsin:protein mass ratio of 3:1. Both pilus forms are equally resistant to chymotrypsin proteolysis, remaining intact after a 60 min digestion. In contrast, monomeric TcpA was rapidly degraded at an early time point.
Solvent accessibility of overlapping peptides in TCP filaments
Non-deuterated TCP filaments were used to determine the denaturation and reducing conditions that would produce a protease “fragmentation map” of overlapping peptides covering the entire TcpA sequence. These conditions also serve to quench deuterium exchange in the DXMS experiments. Reduced and denatured TCP filaments were digested on a pepsin column and peptide fragments were separated and their masses were determined using liquid chromatography-mass spectrometry (LC-MS). High-quality overlapping peptides covering the entire TcpA sequence were identified, with the exception of the first three residues (Fig. 2A). For the DXMS experiments, TCP filaments were incubated in deuterated buffer for 10 to 3000 seconds, then treated as above to determine the degree of deuterium incorporation for each peptide fragment. Figure 3A shows the percent deuteration averaged over six time points for select non-overlapping peptides covering the entire TcpA amino acid sequence. Solvent-exposed regions of the TcpA protein are clearly indicated by their high levels of deuteration (colored orange or yellow in Fig. 3), whereas regions that are buried, either by the protein fold or by subunit:subunit interactions, were poorly-labeled over the 3000 s time course (black or blue). Much of α1 is buried in the filament, as is the C-terminal half of α2, the extended loop following α2, the β-sheet and α3. Surprisingly, a segment in the middle of α1-N is solvent-exposed, as is the N-terminal half of α2 and much of the D-region, including α4.
Figure 2. Overlapping peptides identified by pepsin digest and LC-MS.
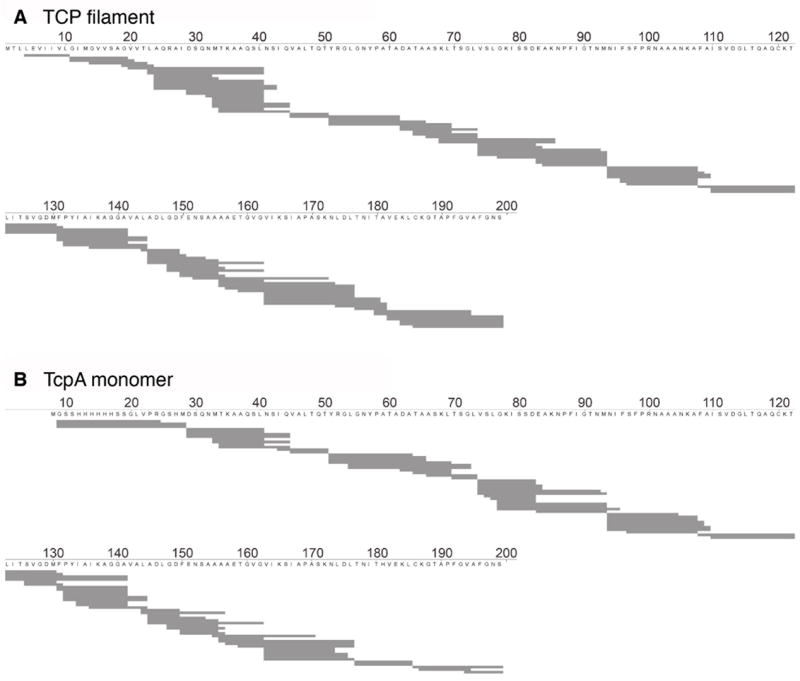
Peptide maps of (A) the TCP filament and (B) the TcpA monomer. Amino acids are numbered according to the mature TcpA sequence.
Figure 3. Deuterium accessibility for peptides in the TCP filament and TcpA monomer. (A).

Average deuteration levels for non-overlapping peptides in the TCP filament. Bars span the length of each peptide and are colored according to their average deuteration over six time points (10, 30, 100, 300, 1000, 3000 s), reported as a percentage of a fully deuterated peptide, as shown in the color key. The first two residues of each peptide are not shown as they are fully deuterated at all time points. The TcpA secondary structure is indicated above the graph: blue coils represent α-helices and red arrows represent β-strands. (B) TcpA crystal structure colored as in (A) according to its deuterium accessibility in TCP filament. Segments colored grey represent regions not covered by non-overlapping peptides. Residues Cys120 and Cys186, which delineate the D-region, are shown as a ball-and-stick representation. The N-terminal 28 residues were modeled onto the TcpA structure using the coordinates for the PAK pilin N-terminus (Craig et al., 2003). Discrete structural regions and residues used for filament modeling and/or mutational analysis are labeled. (C) Comparison of deuteration levels for identical peptides in the TcpA monomer (upper lines) and the TCP filament (lower lines), mapped onto the amino acid sequence of TcpA. The amino acid sequences are identical for the monomer and the filament for residues 29–199, with the exception of residue 181, which is an alanine in the TCP filament (TCPH181A). Secondary and other structural features are indicated above the sequence. Peptides are shown for six time points: 10, 30, 100, 300, 1000, 3000 s, from top to bottom. Each peptide is labeled by the residues it spans, with its average percent labeling over the six time points shown in parentheses.
α1 and the αβ-loop form the only subunit:subunit interfaces in the TCP filament
The solvent accessibility of the TCP filament was compared to that of the TcpA monomer to identify regions that were buried only in the filament form: these are the regions that form subunit:subunit interactions. Because the TcpA monomer lacks the N-terminal 28 residues, only residues 29–199 can be directly compared between the two samples. A complete overlapping fragmentation pattern was obtained for the TcpA monomer using the same denaturation and reducing conditions as for the TCP filaments (Fig. 2B). DXMS experiments were carried out under these conditions. Fourteen peptides were selected from both the soluble TcpA monomer and the TCP filament samples based on having identical amino acid sequences. In total, these peptides cover 80% of the common amino acid sequence. Surprisingly, both samples showed very similar patterns of deuteration over much of the protein length (Fig. 3C), indicating that large regions of the protein are equally solvent-accessible in the monomer and filament forms, and thus are not involved in subunit:subunit interactions. Peptides that show similar low levels of deuteration in both the TcpA monomer and the filament indicate regions of TcpA that are buried by protein folding rather than by subunit:subunit interactions. Two discrete regions of the pilin subunit showed substantially reduced levels of deuteration in the TCP filament compared to the TcpA monomer, indicating subunit:subunit interfaces: residues 31–50, which covers most of α1-C; and residues 68–83 of the αβ-loop.
α1-C
Peptides 31–40 and 47–50 are moderately well-labeled in the TcpA monomer (average percent deuteration: 32.8 and 26.2, respectively), whereas corresponding peptides in the TCP filament are protected from hydrogen/deuterium exchange (16.7% and 10.2%, respectively) (Fig. 3C). Although residues 41–46, cannot be directly compared due to the lack of a common peptide, peptides in this region of the TcpA monomer are also moderately exposed (data not shown), whereas peptides in this region of the TCP filament are only minimally deuterated for all time points measured (Fig. 3A). Thus, α1-C is buried by interactions with neighboring subunits in the TCP filament.
The αβ-loop
The second region that differs markedly in deuterium labeling between the TcpA monomer and the TCP filament is a segment on the αβ-loop between residues 68–83, covering the C-terminal half of α2 followed by a short α-helical turn. Two peptides are directly comparable for this region: peptides 68–73 and 76–83 are moderately well-labeled in the monomer (27.2% and 31.2% average deuteration, respectively) yet they are relatively inaccessible to deuterium exchange in the TCP filament (12.0% and 18.9%, respectively, Fig. 3C). Furthermore, a peptide preceding and partially overlapping this segment, residues 66–69, is well-labeled in both samples (41.0% for the monomer, not shown, and 50.3% for the TCP filament, Fig. 3A), which suggests that the buried region of the αβ-loop starts at residue 70. This protruding region of the αβ-loop was predicted to interact with the D-region on neighboring subunits in our previously-published TCP model based on the interactions observed between TcpA subunits in the crystal lattice (Craig et al., 2003). However, our DXMS data show that the D-region is well-labeled in both the subunit and filament and thus does not interact with the αβ-loop (see below), so residues ~70–83 must interact elsewhere with another region of a neighboring TcpA subunit.
α1-N
Although the deuterium accessibility of the N-terminal half of α1, α1-N, in the TCP filament cannot be directly compared with that of the TcpA monomer, which lacks this segment, the results are nonetheless extremely informative. Like α1-C, much of α1-N does not readily undergo hydrogen/deuterium exchange. Peptide 6–10 is almost completely inaccessible, as are peptides spanning residues 26–50 (Fig. 3A, B). This is not unexpected as the N-terminal α-helix is predicted to be buried in the filament core in our TCP model, and is buried in the GC pilus cryo-EM structure (Craig et al., 2003; Craig et al., 2006). However, peptides spanning residues 13–23 are surprisingly well-labeled (~50–64%), indicating that this segment of α1-N is solvent exposed. Thus, any model for TCP filament assembly must bury most of the N-terminal α-helix, but expose residues 13–23.
The D-region is exposed in the TCP filament
The D-region comprises a large portion of the TcpA globular domain and is delineated by disulfide-bonded cysteines at residues 120 and 186 (Fig. 3B). We had previously predicted that α4 (residues 179–184), which lies within the D-region on a protruding edge of the globular domain, interacted with the αβ-loop of a neighboring subunit (Craig et al., 2003). But this is clearly not the case, as a peptide overlapping with α4 (residues 176–181) is the most highly-labeled peptide in the TCP filament (Fig. 3A, B). The first third of the D-region (residues 120–138) forms part of the β-sheet and α3, which are buried in both the filament and the monomer. However the second two-thirds of the D-region (residues ~145–182) represent the most solvent-accessible region of the protein in both samples (Fig. 3) and is therefore not involved in subunit:subunit interactions. In fact, peptide 158–162 is even more readily-labeled in the TCP filament (51.2%) than in the monomer (39.3%), as are peptides 147–155 and 165–176 flanking this region. This unexpected result is likely an artifact of the his-tag/linker at the N-terminus of the TcpA monomer interacting with peptide 158–162 and its surrounding region and protecting them from deuterium exchange. In support of this, peptides spanning the his-tag/linker of the monomer are only moderately accessible to deuterium exchange (26.3%, data not shown), suggesting that this segment interacts with the globular domain of TcpA. This interaction is not likely to be tight or well-ordered since the his-tag/linker segment was not resolved in the TcpA crystal structure (Craig et al., 2003).
Modification of the TCP model based on DXMS results
Based on the DXMS data, we modified our earlier TCP model, which was derived from: (i) the monomeric TcpA crystal structure; (ii) the filament dimensions and helical symmetry; and (iii) subunit:subunit interactions observed in the TcpA crystal lattice (PDB code 1OR9 (Craig et al., 2003)). In this earlier model, the subunits are arranged in a left-handed 1-start helix with a rise of 7.5 Å and a rotation of 140°, and the filament has a maximum diameter of 88 Å measured from the outermost atom centers. The TcpA subunits are oriented such that their N-terminal α-helices form a hydrophobic core, leaving a small channel (<9 Å) running through the center of the filament axis. Subunits are held together by interactions among the N-terminal α-helices and by an interface between the αβ-loop on one subunit and the D-region of a neighboring subunit that runs along the strands of the left-handed 3-start helix, creating a relatively smooth filament surface. The orientation of the subunits and the overall architecture of the TCP filament model are similar to that of the N. gonorrhoeae Type IV pilus cryo-EM reconstruction (Craig et al., 2006). DXMS analysis of V. cholerae TCP allowed us to refine our computational model and position the TcpA subunits more precisely within the filament (see Experimental Procedures).
Our new TCP model shown in Figure 4 and Movie S1 differs from our previous model in the following ways: (i) the N-terminal α-helices now completely fill the filament core, leaving no space for a central channel or for water diffusion (Fig. 4A, B); (ii) the filament is held together primarily by interactions between the N-terminal α-helices (Fig. 4A–E), with much of the globular domain being solvent-exposed; (iii) no interface is formed between the αβ-loop of one subunit and the D-region of a neighboring subunit – instead, the αβ-loop interacts with α1-N and α3 of a neighboring subunit (Fig. 4C, F); and (iv) the surface of the filament is no longer smooth, but has a pronounced gap between subunits, exposing a segment of α1-N, and the D-region of each subunit forms a bump (Fig. 4B–D, G).
Figure 4. Refined TCP filament model. (A).
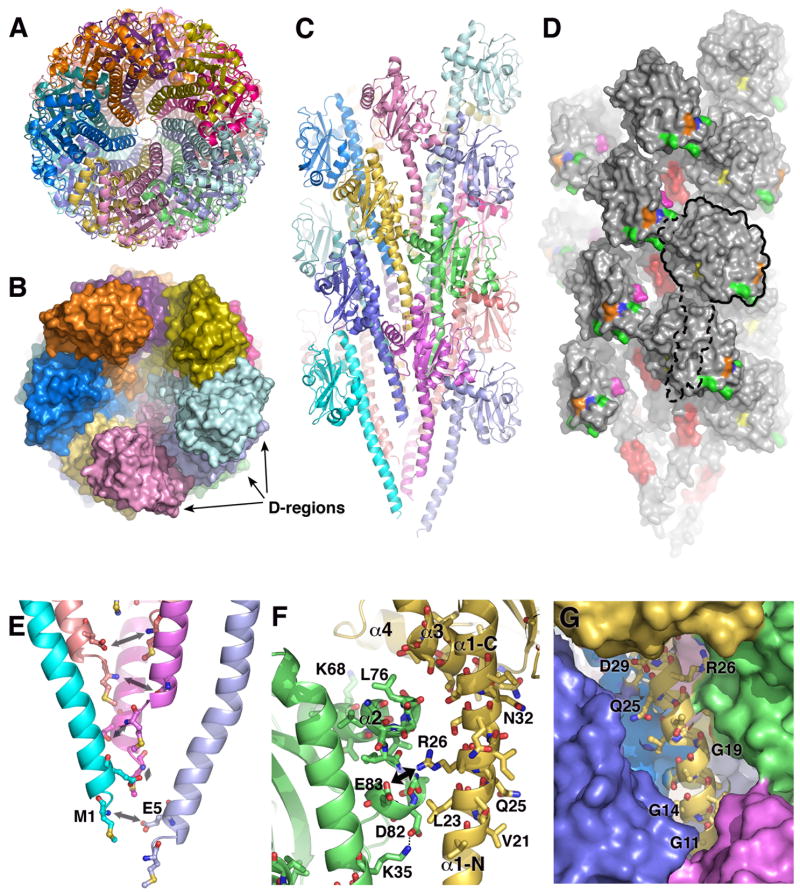
TCP model viewed as a ribbon diagram from the distal end of the filament shows the packing of the N-terminal α-helices. (B) Surface representation of the TCP model shows the N-terminal α-helices completely filling the filament core, leaving no solvent-filled channel for deuterium exchange. The D-regions protrude from the filament surface giving it a bumpy appearance. (C) Side view of the TCP model as a ribbon and (D) surface representation with a single subunit outlined. Residues 13–23 of α1-N, which were shown to be readily-labeled by deuterium, are colored red. His181 is shown in blue, Asp113 is yellow, Glu158 is orange, Asp129 is magenta and additional residues in the D-region shown by Kirn et al. (2000) to be involved in pilus:pilus interactions are green (Asp175, Glu183 and Lys187). (E) Close-up of the N-termini to show the putative electrostatic interaction between the positively-charged N-terminal amine of Met1 and the negatively-charged Glu5 side chain. (F) Close-up of the α2: α1-N/α3 interface, formed between the αβ-loop of one subunit (green) and α1-N and α3 of a neighboring subunit (yellow). The interface is shown as if looking out from the interior of the filament. (G) Close-up of the gap in the TCP filament surface, which exposes the polar face of an amphipathic segment of α1-N, residues 13–23. Key residues, including the three glycines, are labeled.
The N-terminal α-helices pack remarkably well into the filament core, each contacting six neighboring α1s without any steric clashes (Fig. 4A–C). No central channel exists, consistent with most of α1 being completely inaccessible to deuterium. We expect that the precise arrangement of the flexible N-terminal α-helices in the pilus interior will differ somewhat from what we have modeled using rigid body subunits, whose α1-N segments have been fit using the PAK pilin coordinates. Nonetheless, the general arrangement is likely to be maintained. We adjusted the backbone φ and ψ angles of the two most N-terminal residues, Met1 and Thr2, to position the positively-charged N-terminal amide group of Met1 close to the negatively-charged Glu5-Oε1 of the neighboring subunit in the 1-start helix (Fig. 4E). Thus, this model supports our proposal that Glu5 is required in Type IV pilins to neutralize the N-terminus in the otherwise hydrophobic interior of the filament (Craig et al., 2006; Parge et al., 1995).
Apart from the interactions among the N-terminal α-helices, the only other interface holding the subunits together in the TCP filament is formed between segment 70–83 of the αβ-loop, and two segments of a neighboring subunit: residues 23–34 of α1-N; and residues 122–126 of α3 (see green and yellow subunits in Fig. 4C and viewed from the inside of the filament in Fig. 4F). This interface is referred to as the α2: α1-N/α3 interface. Segment 70–83 of the αβ-loop protrudes on one side of the globular domain of TcpA which fits neatly into a nook in a neighboring subunit, interacting with α1-N along residues 23–34. α2 also interacts with α3, which lies at the bottom of the globular domain. α3, comprising of residues 117–128, was not initially predicted from the DXMS data to form an interaction interface as it was poorly-labeled in both the TcpA monomer and in the assembled filament (see peptides 112–122 and 125–131 in Fig. 3C). However, assuming the N-terminal his-tag/linker of monomeric TcpA interacts with segment 158–162 on the outer face of the globular domain, it very likely crosses over α3 in order to reach that segment. Thus it is reasonable to conclude that α3 is partially buried by the his-tag/linker in the TcpA monomer and by the αβ-loop of a neighboring subunit in the TCP filament. Importantly, the α2: α1-N/α3 interface buries the solvent-inaccessible segment of the αβ-loop but leaves α4 solvent-exposed, consistent with the DXMS data. Apart from α1-C and discrete sections of the αβ-loop and α3, the globular domain does not contact neighboring subunits and is freely accessible to solvent (Movie S1).
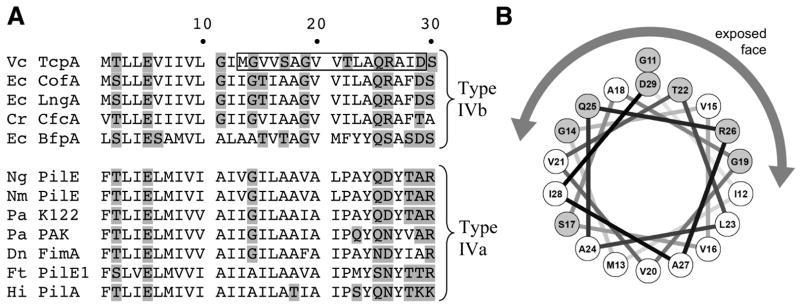
The DXMS data imply that instead of the subunits snugly fitting together to form a smooth, continuous filament surface, the TcpA globular domains are actually packed quite loosely, leaving gaps on the TCP surface (Fig. 4C, D and G). Each gap is framed by four globular domains, forming a cavity that exposes segment ~11–29 of α1-N of the uppermost (i.e. most distal) of these subunits. The most exposed segment of α1-N is residues 13–23, as indicated by the DXMS data (Fig. 3A, B). Although residues 24–29 are also exposed in our model they were not well-labeled, most likely because they are involved in interactions with α-helices and αβ-loops of neighboring subunits. Only one face of this N-terminal segment is solvent exposed in our model, with the back face interacting with α-helices in the interior of the filament, so it is somewhat surprising that this segment is so well-labeled (~50–64% for residues 13–23, see Fig. 3A). This may be in part due to the preponderance of residues with small side chains (Gly14, Ser17, Ala18, Gly19, Ala24), which would allow better access of solvent to the amide nitrogens (Fig. 4G, Fig. 5A). As well, the unusual presence of three glycines within this extended α-helix, at positions 11, 14 and 19, may impart flexibility in this region of α1 and disrupt the α-helical backbone bonding, thereby increasing its solvent accessibility. Although there are many hydrophobic residues in α1-N, segment 13–29 is actually within an amphipathic stretch of the α-helix, with the polar face being solvent-exposed (Fig. 5B). A comparison of the α1-N segments of Type IV pilins reveals that this amphipathic segment, including the preponderance of glycines, is unique to a subset of the Type IV pilins, the Type IVb pilins, which are found on enteric pathogens (Fig. 5A). Thus, the cavity we observe on the TCP filament surface may be a common structural feature of this class of pili, which include bundle-forming pili (BFP) from enteropathogenic Escherichia coli and CFA/III pili from enterotoxigenic E. coli (Giron et al., 1997).
Figure 5. Amino acid sequence alignments of α1-N and helical wheel representation of the amphipathic segment. (A).
Residues 1–30 are shown by their one-letter codes. Polar residues are shaded. Boxed residues indicate the highly deuterated segment of TcpA α1-N. Vc, V. cholerae; Ec, enterotoxigenic E. coli (CofA and LngA) and enteropathogenic E. coli (BfpA); Cr, Citrobacter rodentium; Ng, N. gonorrhoeae; Nm, N. meningitidis, Pa, P. aeruginosa, Dn, Dichelobacter nodosus, Hi, Haemophilus influenzae. (B) Helical wheel representation of residues 11–29 of TcpA. Polar residues are shaded. The polar face of the α-helix corresponds to the exposed face in the TCP model, as indicated.
In the new TCP model, the subunits have been rotated about the filament axis, such that the αβ-loops are turned slightly into the filament core and the D-regions are turned outward, creating a bumpy surface (see Fig. 4B, Movie S1). Several residues in this protruding region were shown by mutational analysis to be involved in pilus bundling and/or microcolony formation (Kirn et al., 2000). The side chains of these residues are among the most protruding side chains of the TcpA subunit (Asp175, Glu183 and Lys187, colored green in Fig. 4D), or lie within the cavity formed between the globular domains (Asp129, colored magenta in Fig. 4D). Pilus:pilus interactions may thus require the protruding D-regions on one filament to intercalate into the cavities between the subunits in an adjacent filament, and may even involve residues in the exposed segment of α1-N. In support of this idea, a Glu158→Leu mutation in TcpA, which profoundly disrupts pilus:pilus interactions, was rescued by three different suppressor mutations in α1-N (Kirn et al., 2000). Two of these mutations changed large hydrophobic residues in the exposed segment of α1-N to glycine (Val15→Gly, Val20→Gly). This size reduction may allow the cavity to more easily accommodate an intercalating D-region bulge that now possesses a bulky but uncharged residue at position 158 (colored orange in Fig. 4D). Interestingly, a Glu158→Ala mutation had little effect on pilus:pilus interactions suggesting that the negative charge at residue 158 is only helpful to this interaction if the side chain is bulky.
TcpA mutagenesis to test the refined TCP model
We tested the accuracy of the TCP model by generating missense mutations for select residues at the α2: α1-N/α3 interface. Arg26 is one of only 3 charged residues in α1-N and is oriented such that its side chain could form an intermolecular salt bridge with Glu83 on the αβ-loop of a neighboring subunit (Fig. 4F). To test the involvement of Arg26 and Glu83 at this subunit:subunit interface, we changed Arg26 to Glu (TCPR26E) and Glu83 to Arg (TCPE83R), both individually and as a double mutation (TCPR26E/E83R), and assessed the V. cholerae mutants for their ability to assemble TCP. V. cholerae TcpA mutants were tested for (i) their ability to produce pilin protein, assessed by immunoblot analysis of whole cell lysates using anti-TcpA antibody; and (ii) their ability to assemble TCP, assessed by TEM analysis and by the presence of TcpA protein in the supernatant of homogenized cells. Pilus morphology and bundling characteristics were also assessed by TEM. TCP functions were assessed by (i) observing aggregation or “autoagglutination” of overnight cell cultures, which indicates pilus:pilus interactions; and (ii) phage transduction assays, since TCP are the receptors for CTXΦ. Pilin protein was observed at wild type levels in whole cell cultures for both V. cholerae mutants, TCPR26E and TCPE83R, indicating that these mutations do not disrupt the pilin fold (Fig. 6A). This is in contrast to a folding mutation, Cys120→Ala, (TCPC120A), which disrupts the conserved disulfide bridge and abrogates pilin production. No pilin band was observed in the homogenized cell supernatant for TCPR26E, and only a very faint pilin band was seen for TCPE83R (Fig. 6A, B). Some TCPE83R filaments were also observed by TEM (Fig. 6C), confirming that low levels of pilus expression occurred for this strain. The dramatic reduction in pilus expression for the TCPR26E and TCPE83R mutants clearly demonstrates a role for Arg26 and Glu83 in pilus assembly. Importantly, the TCPR26E/E83R double mutant partially rescued pilus assembly, producing ~ half the level of wild type TCP (Fig. 6A), thus supporting a direct interaction between Arg26 and Glu83, as predicted by our model (Fig. 4F). Interestingly, although pilus morphology appeared normal for TCPE83R and TCPR26E/E83R by TEM (Fig. 6C, D), the typical rope-like bundles seen for wild type TCP were not observed: TCPE83R formed tangled masses, and the TCPR26E/E83R filaments twisted around each other and themselves to a greater degree than wild type. Furthermore, both strains showed reduced autoagglutination and phage transduction beyond what could be accounted for by reduced pilus expression (Fig. 6B). These observations suggest that in addition to affecting pilus assembly, these mutations have altered the surface characteristics of the pili affecting their function.
Figure 6. Analysis of pilin expression, pilus assembly and pilus functions in TcpA mutants. (A).
Immunoblot showing pilin subunit expression in whole cell lysates (top panel) and pilus assembly, indicated by the presence of pilin protein in sheared cell homogenates (bottom panel). Wild type refers to V. cholerae strain O395, classical biotype. (B) Pilin and pilus expression were also assessed by autoagglutination and phage transduction assays. (C) TEM of TCPE83R and (D) TCPR26E/E83R.
Two other residues were tested for their involvement in pilus assembly. Leu76, which is centrally located in the α2: α1-N/α3 interface at the C-terminal end of α2 (Fig. 4F), was changed to lysine. The TCPL76K mutant made pilin but not pili, as detected by immunoblotting, TEM and functional studies (Fig. 6A, B). These results implicate Leu76 in filament assembly, demonstrating that the α2: α1-N/α3 interface involves both hydrophobic and ionic interactions. Finally, Lys68 is in the middle of α2 (Fig. 4F) within a segment that was readily labeled in our DXMS studies (Fig. 3A), and is thus not expected to contribute to an interaction interface. Consistent with this, changing Lys68 to alanine had no effect on pilus assembly or pilus functions (Fig. 6A, B).
DISCUSSION
Our DXMS analyses were combined with structural data to generate a new molecular model for TCP, which is supported by mutational analyses. This structure is a solid helical filament devoid of any central channel. Subunits are anchored by hydrophobic interactions among the N-terminal α-helices and by both hydrophobic and polar interactions between the αβ-loops and α1-N/α3, leaving most of the globular domain solvent-accessible. The surface of the filament is decorated with bulges created by the protruding D-regions and by cavities created between the loosely-packed globular domains, which expose a segment of α1-N. This structure provides explanations for pilus features and functions, and has important implications for the design of antibacterial and anti-virulence therapies.
Several residues implicated in pilus bundling and cell autoagglutination (Kirn et al., 2000) localize to the protruding D-region, prompting us to propose a mechanism for pilus:pilus interactions whereby the D-region bulges of one filament intercalate into the cavities of adjacent filaments and vice versa. These intimate lateral interactions, that may even involve the exposed α1-N segment, extend over long lengths of the pilus filaments (see Fig. 1A, B) and would provide a strong cumulative force to cement the V. cholerae cells together in microcolonies on the intestinal epithelium. Such bulge-in-cavity interactions between pili may explain pilus-mediated aggregation for other bacteria. N. gonorrhoeae GC Type IV pili are involved in microcolony formation and also possess undulating surfaces, with protruding D-region hypervariable loops and grooves running between the globular domains (Craig et al., 2006). Interestingly, a pilin-like protein, PilX, was recently identified as being required for N. meningitidis pilus:pilus interactions (Helaine et al., 2005). PilX, which is incorporated throughout the N. meningitidis pilus, shares the conserved structural core of Type IV pilins, and its protruding D-region is necessary for pilus-mediated bacterial aggregation (Helaine et al., 2007). Although no such accessory protein has been identified for TcpA, this and other Type IVb pilins have a much longer D-region (~55 amino acids) than those of the Type IVa pilins (~22 residues), so the D-region bulge may be a characteristic feature of this pilus subgroup.
The exposed N-terminal α-helix and surface gap may also be present in other Type IVb pilins, which share an amphipathic, glycine-rich segment in α1-N. The gap created by the loose packing between the globular domains would provide a compression space to allow for the extreme flexibility observed for TCP. This flexibility would be further enhanced by the presence of three glycine residues in α1-N. A full-length crystal structure of a Type IVb pilin has not been solved, and it is possible that the N-terminal α-helical segment is distorted around the glycines. Interestingly, an analogous segment of α1-N is also partially exposed in a groove between subunits of the GC pilus, which belongs to the Type IVa class (Craig et al., 2006). However, this groove is much narrower and the side chains of the exposed section of α1-N are considerably more hydrophobic in GC pilin compared to TcpA. Still, there have been reports that the N-terminal α-helices of Type IVa pilins are partially exposed and contribute to pilus functions (Marceau et al., 1995; Watts et al., 1983). GC and other Type IVa pilins possess a single glycine, Gly14, and a proline at residue 22 (Fig. 5A), which induces a kink in α1-N that is likely important in filament flexibility for this pilus subclass. These structural features help to explain how Type IV pili can possess high tensile strength (> 100 pN (Maier et al., 2002)) yet be extremely flexible.
Indirect evidence suggests that the repeating cavities on the TCP surface may also provide binding sites for pilus assembly proteins and inner membrane anchoring proteins. The pili used for the DXMS study carry the H181A mutation, which causes them to fall off cells. In spite of this, V. cholerae H181A mutants possess wild type levels of pili on their surfaces (Lim and Craig, unpublished data), suggesting that TCPH181A shear off the cells because they grow too long rather than because they are less stable. His181 lies on the protruding segment of the D-region at the edge of the cavity (colored blue in Fig. 4D), and thus, may interact with ruler-like protein(s) that sense and control pilus length, or with an as-yet-unidentified retraction motor. The TCP cavities may also bind exogenous agents such as CTXΦ. In support of this idea, a point mutation at Asp113, which lies deep in the cavity (colored yellow in Fig. 4D), disrupts TCP-mediated CTXΦ transduction, although the pili are expressed at wild type levels (Lim and Craig, unpublished data). We are currently investigating the role of residues within the cavities with respect to pilus bundling, phage interactions and pilus assembly.
The surface cavity, and the exposed α-helix, may represent good targets for therapeutics for bacterial pathogens that utilize Type IV pili. Such agents would bind in the cavity, perhaps to accessible residues on α1-N, and block essential pilus functions such as bundling or retraction. This strategy could be particularly effective for organisms that infect the lungs and gastrointestinal tract, such as P. aeruginosa and V. cholerae respectively, since pilus-specific reagents could be inhaled or ingested for delivery to infectious bacterium without having to travel through the bloodstream, thus increasing their efficacy and minimizing their toxicity. In the case of retractile pili, bacteriocidal or bacteriostatic agents could be taken up by the bacteria selectively during pilus retraction. Although pilus retraction has not been observed for V. cholerae, it is a common feature of many Type IV pili that facilitates twitching motility and DNA uptake (Burrows, 2005).
Our mutagenic analysis of the α2: α1-N/α3 interface revealed that mutations in this region had a dramatic effect on pilus expression, confirming the involvement of this interface in pilus assembly. However, if this interface were essential for pilus stability, no TCP filaments should have been observed for the TCPE83R mutant. The α2: α1-N/α3 interface is relatively small compared to the extensive, multiple interfaces formed among the N-terminal α-helices, so it is remarkable that single mutations such as R26E, L76K, and E83R have such dramatic effects on pilus assembly. We propose that the α2: α1-N/α3 interface is not actually required for filament stability, but is instead involved in initiating the addition of each subunit to the growing pilus. During pilus biogenesis, pilin subunits are translocated across the inner membrane and likely remain anchored in this membrane via their hydrophobic α1-N helices prior to being incorporated into a growing pilus filament. Previously we proposed that pilus assembly was driven in part by charge complementarity between the conserved Glu5 side chain of the membrane-localized pilin and the positively-charged N-terminus in the terminal subunit of the nascent filament (Craig et al., 2006; Parge et al., 1995). Our mutational results suggest that charge complementarity between Arg26 and Glu83 is also important for initiating pilus assembly. These electrostatic attractions would provide a driving force for transferring a pilin subunit from the hydrophobic environment of the inner membrane to that of the pilus filament. In addition, since no pili were made in the L76K mutant, it is clear that overall chemical complementarity and not just electrostatic complementarity at this small interface is important for the assembly process. In the absence of complementarity between these two surfaces, pilus assembly may occur but is much less efficient, explaining the dramatic reduction in pilus expression for the α2: α1-N/α3 interface mutants. Once assembled, it is the extensive hydrophobic interactions among the N-terminal α-helices that firmly anchor the subunits in place to provide structural stability.
Our new TCP model helps to explain how Type IV pili can assemble via a conserved mechanism and architecture yet display extraordinary functional diversity. All Type IV pili are held together by hydrophobic interactions among their conserved N-terminal α-helices. The β-sheets of the globular domains provide a physical link between the α-helical core of the filament and the exposed subunit surface, which displays substantial sequence, length and structural variability. The arrangement of the globular domains on the filament surface introduces another level of structural variation, resulting in a filament landscape with unique chemical and shape properties that together define the diverse and often unique functions of Type IV pili in the bacterial life cycle and in pathogenesis. We expect that all Type IV pili have grooves and gaps around the globular domains to allow for filament flexibility and to provide interaction interfaces. Our data hint at a new and central role for the N-terminal α-helix in defining the filament surface and hence its functions, in addition to its well-established role in pilus assembly. The TCP model provides testable hypotheses to relate Type IV pilus structure to the assembly mechanism and its many functions, and offers the possibility of novel strategies for antibacterial therapeutic agents.
EXPERIMENTAL PROCEDURES
Plasmids and bacterial strains
V. cholerae strains RT4236, RT4225, RT4524, O395 and CL101, E. coli strain S17, plasmid pTK1 and anti-TcpA antibody were gifts from Ronald Taylor, Dartmouth Medical School. E. coli Origami DE3 cells (Novagen) transformed with the pET:15b vector (Novagen) containing the gene encoding N-terminally-truncated TcpA (ΔN-TcpA) were used for expression of the TcpA monomer containing an N-terminal his-tag/linker (Craig et al., 2003). Details of bacterial strains are described in Table 1.
Table 1.
Bacterial strains and plasmids
| Bacteria | Description | Reference/Source |
|---|---|---|
| V. cholerae O395* | Wild type | (Taylor et al., 1987) |
| V. cholerae RT4225 | TcpA:H181A:toxT | (Kirn et al., 2000) |
| V. cholerae RT4236 | pMT5:toxT | (DiRita et al., 1996) |
| V. cholerae RT4524 | tcpA::lacZ | R.K. Taylor |
| V. cholerae ML10 | TcpA C120A | This study |
| V. cholerae ML19 | TcpA K68A | This study |
| V. cholerae ML21 | TcpA R26E | This study |
| V. cholerae ML22 | TcpA L76K | This study |
| V. cholerae ML23 | TcpA E83R | This study |
| V. cholerae ML24 | TcpA R26E/E83R | This study |
| V. cholerae CL101 | pCTX-KmΦ | (Kirn et al., 2003) |
| E. coli S17 | λpir | (Skorupski and Taylor, 1996) |
| E. coli Origami(DE3)-pET:15b-TcpA | E. coli K12 gor-/trxB-, Δ1–28, hisTcpA | Novagen & (Craig et al., 2003) |
all V. cholerae TcpA mutant strains are derived from O395, classical biotype
Purification of TCP filaments
V. cholerae strains expressing TCPWT (RT4236) and TCPH181A (RT4225) cells were grown for 16 hours under TCP-expressing conditions (Luria broth [LB], pH 6.5, 30 °C) on a roller shaker in media supplemented with 100 μg/ml ampicillin and 0.4 mM isopropyl β-D-thiogalactopyranoside. For strain RT4225, cultures were centrifuged to remove cells and TCPH181A were precipitated from the supernatant using 30% ammonium sulfate, resuspended in phosphate-buffered saline (PBS) with 10 mM EDTA and dialyzed exhaustively. TCPWT were sheared from RT4236 cells by passing the bacteria through a 25-gauge needle followed by vigorous vortexing and homogenization using an Ultra-Turrax T8.01 disperser (Ika). The pili were concentrated by centrifugation at 5000 Xg, solubilized in PBS/EDTA and dialyzed exhaustively. Further details on TCP preparations are provided in Supplemental Data.
Purification of monomeric TcpA
N-terminally-truncated TcpA containing an N-terminal his-tag/linker was expressed in E. coli Origami DE3 cells (Novagen) transformed with ΔN-TcpA-pET-15b constructs as described previously (Craig et al., 2003). After purification, the pilin subunits were dialyzed in PBS/EDTA.
Chymotrypsin Digestion
Protein concentrations were determined by the Bradford method (ref). Chymotrypsin diluted in PBS/EDTA was added to pre-incubated samples at a chymotrypsin:protein mass ratio of 3:1 and incubated at 37 °C while shaking. Aliquots were removed at specific time points and quenched with phenylmethylsulfonyl fluoride (10 mM final concentration). The proteins were analyzed by SDS-PAGE followed by Coomassie staining or immunoblotting using a polycolonal antibody against residues 174–199 of TcpA (Sun et al., 1991).
Transmission electron microscopy
Five μl samples of whole cell culture were applied to glow-discharged carbon-coated copper grids (Electron Microscopy Sciences), stained with 1% phosphotungstic acid, and imaged on a FEI Tecnai F20 at 120 keV.
DXMS analysis
Detailed protocols are provided in Supplemental Data and summarized below. Protein solutions were diluted to 2 mg/ml in regular or deuteration buffer (25 mM Tris-HCl, 150 mM NaCl in D2O, pH 7.1) for varying intervals (10 to 3000 s) prior to addition of ice-cold quench solution (1.0 M Tris-2-carboxyethyl phosphine [TCEP], 7.4 M guanidinium hydrochloride, pH 2.9) After a 5 minute incubation at 0 °C samples were mixed with 140 μl of ice-cold 0.8% formic acid and immediately frozen and stored at −80 °C. Stored samples were transferred to dry ice thenindividually melted at 5 °C and pumped over a protease column containing immobilized porcine pepsin (Sigma), coupled to 20AL support from PerSeptive Biosystems (30 mg/ml, 66 μl column bed volume) flowing in 0.05% trifluoroacetic acid (TFA) at 100 ml/min. Proteolytic fragments were contemporaneously collected on a C18 HPLC column (Vydac) then eluted by a linear acetonitrile gradient (5–45% solvent B in 20 min, 50 ml/min: solvent A, 0.05% TFA; solvent B, 80% acetonitrile, 0.01% TFA). Column effluent was directed to the mass spectrometer (ESI LCQ Classic, Thermo Finnigan Inc.) operated with capillary temperature at 200 °C and spray voltage of 5000 V, with data acquisition in either MS1 profile mode or data-dependent MS1:MS2 mode (Black et al., 2004). Peptides were identified and analyzed as described in Supplemental Data.
Refinement of the TCP model
The new TCP model was generated by the method described in (Craig et al., 2003) using our original published model as a starting point. Three rotation angles, x, y and z, for the TcpA subunit, as well as its radial distance from the filament axis, were varied. Filament models were generated by systematically applying an axial translation (rise) and rotation (twist) to the newly-positioned TcpA subunit using the values determined for the original model (7.5 Å and 140°, respectively for a left-handed 1-start helix). Unlike the original modeling procedure, which oriented the TcpA subunit about a pivot point at Asp29-Cαfor the x, y and z rotations, our current models were generated by pivoting about Leu76-Cα. This allowed us to expose the D-region without forcing the αβ-loop into the center of the filament. Models were evaluated computationally, eliminating those with Cα-Cαclashes or unacceptably large (>10 nm) diameters, and selecting for models having close distances between residues Leu76-Cαand Arg26-Cα, and ≥ 10 Å between Ile179 -Cαand Lys68-Cα. A model that conformed to the DXMS data was then refined by hand, iteratively adjusting the radial distance and x, y and z rotations of the TcpA subunit to optimize the subunit packing as dictated by the DXMS results. Further details are provided in Supplemental Data.
Introduction of mutations into the tcpA gene
tcpA mutations were generated by the allelic exchange procedure of Skorupski and Taylor (Skorupski and Taylor, 1996). QuikChange mutagenesis (Stratagene) was used to generate missense mutations in the tcpA gene in pTK1, which was derived from the pKAS32 suicide vector (Skorupski and Taylor, 1996). E. coli S17 cells carrying pTK1-tcpA* (i.e. the mutated tcpA gene) were mated with V. cholerae strain RT4524, and transconjugants were selected with ampicillin and gentamycin. Transconjugants were then screened for a second homologous recombination event whereby pTK1 is excised out of Chromosome I, taking with it the endogenous tcpA gene and leaving behind tcpA*. In RT4524, the endogenous tcpA gene has a segment replaced with a lacZ gene, allowing positive strains to be identified as light blue colonies on agar plates containing 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside and gentamycin. Strains that retained the endogenous tcpA-lacZ hybrid gene appear as dark blue colonies. All mutations were confirmed by sequencing the entire tcpA gene.
Analysis of pilin and pilus expression in TcpA mutants
V. cholerae cells were grown overnight in 2-ml cultures under pilus-inducing conditions (LB, pH 6.5, 30 °C) and examined visually for autoagglutination. Strains that express functional pili will cause the V. cholerae cells to aggregate (autoagglutinate) and fall to the bottom of the culture tube. Autoagglutination was scored relative to wild type classical V. cholerae, strain O395. Cell densities were normalized based on optical density measurements and pilin production was assessed by running 24 μl of normalized whole cell cultures on sodium dodecyl sulfate (SDS) polyacrylamide gels and Coomassie-staining or immunoblotting. Pilus expression was assessed by physically shearing the pili from the cells using an Ultra-Turrax T8.01 disperser and analyzing the cell homogenate by SDS-PAGE and immunoblotting.
CTXΦ transduction assays
V. cholerae strain CL101 was grown overnight under pilus expressing conditions. CL101 cells produce CTXΦ-kan, in which the ctxA gene is replaced with the marker for kanamycin resistance (Waldor and Mekalanos, 1996). Cells were centrifuged and residual cells were removed on a 2 μm pore bottletop filter (Nalgene). Equal volumes of culture supernatant containing CTXΦ and overnight cultures of mutant or wild type V. cholerae were mixed and incubated at room temperature for 30 min, serially diluted and plated on LB-kanamycin agar plates, grown overnight at 37 °C, then counted for colony-forming units.
Figure preparation
Fig. 3B, 4 and Movie S1 were prepared using PyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Ronald Taylor for insightful discussions, guidance in making tcpA mutations and comments on the manuscript. This work is supported by National Institutes of Health grants CA099835 and CA118595 to VLW, GM070996 to MP and AI061051 to LC.
Abbreviations
- DXMS
hydrogen/deuterium exchange mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- TCP
toxin-coregulated pili
- TEM
transmission electron microscopy
References
- Audette GF, Irvin RT, Hazes B. Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor-binding architecture. Biochemistry. 2004;43:11427–11435. doi: 10.1021/bi048957s. [DOI] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV Pilus Structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- DiRita VJ, Neely M, Taylor RK, Bruss PM. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci U S A. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron JA, Gomez-Duarte OG, Jarvis KG, Kaper JB. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili - a minireview. Gene. 1997;192:39–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J Mol Biol. 2000;299:1005–1017. doi: 10.1006/jmbi.2000.3801. [DOI] [PubMed] [Google Scholar]
- Hegge FT, Hitchen PG, Aas FE, Kristiansen H, Lovold C, Egge-Jacobsen W, Panico M, Leong WY, Bull V, Virji M, et al. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc Natl Acad Sci U S A. 2004;101:10798–10803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol. 2005;55:65–77. doi: 10.1111/j.1365-2958.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0707581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Jr, Bakaletz LO. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- Keizer DW, Slupsky CM, Kalisiak M, Campbell AP, Crump MP, Sastry PA, Hazes B, Irvin RT, Sykes BD. Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili. J Biol Chem. 2001;276:24186–24193. doi: 10.1074/jbc.M100659200. [DOI] [PubMed] [Google Scholar]
- Kirn TJ, Bose N, Taylor RK. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol. 2003;49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- Maier B, Potter L, So M, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Beretti JL, Nassif X. High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol. 1995;17:855–863. doi: 10.1111/j.1365-2958.1995.mmi_17050855.x. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- Ramboarina S, Fernandes PJ, Daniell S, Islam S, Simpson P, Frankel G, Booy F, Donnenberg MS, Matthews S. Structure of the bundle-forming pilus from enteropathogenic Escherichia coli. J Biol Chem. 2005;48:40252–40260. doi: 10.1074/jbc.M508099200. [DOI] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Sun D, Seyer JM, Kovari I, Sumrada RA, Taylor RK. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–118. doi: 10.1128/iai.59.1.114-118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga JJ, Nguyen V, O’Brien DK, Rodgers K, Walker RA, Melville SB. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol. 2006;62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- Watts TH, Sastry PA, Hodges RS, Paranchych W. Mapping of the antigenic determinants of Pseudomonas aeruginosa PAK polar pili. Infect Immun. 1983;42:113–121. doi: 10.1128/iai.42.1.113-121.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XF, Tan YW, Lam L, Hackett J, Zhang M, Mok YK. NMR structure of a type IVb pilin from Salmonella typhi and its assembly into pilus. J Biol Chem. 2004;279:31599–31605. doi: 10.1074/jbc.M404727200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.