Abstract
The activity of transcription factors is usually governed by allosteric physicochemical signals or metabolites, which are in turn produced in the cell or obtained from the environment by the activity of the products of effector genes. Previously we identified a collection of more than 110 transcription factors and their corresponding effector genes in Escherichia coli K-12. Here we introduce the notion of “triferog”, which relates to the identification of orthologous transcription factors and effector genes across genomes and show that transcriptional sensing systems known in E. coli are poorly conserved beyond Salmonella. We also find that enzymes that act as effector genes for the production of endogenous effector metabolites are more conserved than their corresponding effector genes encoding for transport and two-component systems for sensing exogenous signals. Finally we observe that on an evolutionary scale enzymes are more conserved than their respective TFs, suggesting a homogenous cellular metabolism across genomes and the conservation of transcriptional control of critical cellular processes like DNA replication by a common endogenous signal. We hypothesize that extensive variation in the domain architecture of TFs and changes in endogenous conditions at large phylogenetic distances could be the major contributing factors for the observed differential conservation of TFs and their corresponding effector genes encoding for enzymes, causing variations in transcriptional responses across organisms.
Keywords: transcriptional sensing systems, cellular signaling, conservation, transcriptional regulation, allosteric regulation, effector metabolites, prokaryotes
1. Introduction
Organisms constantly monitor environmental conditions in order to respond to changes. This is normally achieved by physicochemical signals, which are recognized by the cell as messengers of environmental composition or their own metabolic state [1-3]. The binding of transcription factors (TFs) to these specific signals determines their active or inactive conformation and affinities to interact with specific sequences on cis-regulatory regions of transcription units or with the rest of the transcriptional machinery [4]. In turn these signals are produced or delivered as a result of the activity of the protein products of the effector genes, which indirectly but effectively control the activity of the TFs, providing a concerted link between the genetic and metabolic components of a cell in the regulation of transcription. Recent studies demonstrated that the components of the network of transcriptional interactions (i.e a network where TFs and their regulated genes form the nodes and the directed regulatory interactions form the edges) in bacterial genomes evolve rapidly [5,6]. However it is unclear if the genetic repertoire forming the core of the transcriptional regulation in response to allosteric signals follows the same trend of poor conservation across organisms. In this work, we study the conservation of the genetic components for transcriptional sensing systems (TSSs), that is to say, the TFs and their corresponding effector genes, across a range of prokaryotic organisms and show that the TSSs identified in the gram negative γ-proteobacterium, E. coli K-12 [7-9], are poorly conserved across the phylogenetic spectrum, with closely related species sharing a higher proportion of the TF-effector pairs. We find that despite poor conservation of TSSs across genomes, certain TF-effector gene pairs sensing basic metabolites like ATP, biotin, amino acids and some metals are highly conserved. Our observations suggest that TSSs behave like functional modules as their component TF and signal genes were found to be significantly co-detected across a set of non-redundant genomes. Furthermore, we also demonstrate that there is variation in the extent of conservation of different transcriptional signal sensing categories as defined by the effector genes constituting them. The results reported here should enhance our understanding of the evolution of transcriptional sensing machinery in prokarya.
2. Materials and Methods
2.1 Dataset
Information about TFs and their corresponding effector genes with experimental evidence from literature was gathered from the RegulonDB database, which contains extensive information centered on transcriptional regulation in Escherichia coli strain K-12 [10]. Our initial dataset comprised 84 TFs and 291 corresponding effector genes as described in Martinez-Antonio et. al [7]. It should be noted that a TF to effector gene relation can be many to many, so a TF can have multiple effector genes and multiple TFs can be controlled by a single effector gene. In RegulonDB, however, information on their signal effectors is available for only a minor fraction of all known two-component systems which are experimentally characterized. Therefore we added to this list those cases for which there was indirect evidence (such as their experimental characterization in closely related species). Hence our final dataset consisted of 113 TFs covering around 38% of the roughly 297 predicted TFs in E. coli [7,11,12]. The complete classification of TFs and effector genes is available as Supplementary Material.
2.2 Identification of orthologous transcription factors and effector genes across genomes
Orthologs are defined as those genes in different species that evolved from a common ancestor by speciation [13] and usually have the same function. Our working definition of orthology consisted of BLASTP reciprocal best hits, along with additional rigorous parameters to take into account the multi-domain nature and extensive duplication in transcription factors [11] as described earlier [6]. The majority of TFs in prokarya are comprised of at least a DNA-binding domain and an effector domain. In order to identify and distinguish orthologous sequences from those arising due to lineage specific duplications and recombinations, which are known to be the common phenomena driving the evolution of TFs [11,14], it becomes important to consider domain organization and orientation to detect functionally equivalent orthologs. Therefore in addition to traditional bi-directional best hits it was required that Pfam domains [15] of query and target proteins match to consider them as orthologs. Using this approach we identified the orthologs of all protein coding genes in E. coli across a collection of 216 genomes shown in the Supplementary Material. More relaxed definitions of orthology which rely only on reciprocal best hits and typically gain on the coverage of orthology detection but loose on sensitivity [5] did not vary our conclusions (see Supplementary material).
2.3 Validation of detected triferogs
In order to assess how likely triferogs are to be detected in a genome due to chance alone and to measure the significance of their conservation, we compared the number of triferogs identified in a given genome for the complete set of TSSs in E. coli against the conservation of 1000 randomly constructed TSS collections. Each random collection was created by randomly assigning an effector gene to a TF by simply altering the label of the effector gene with any protein coding gene in E. coli except that of the TF itself, while the label of the TF was retained as such. In order to avoid over-scoring on the extent of conservation due to the over-representation of genomes which are evolutionarily very close, we filtered out strains and species of the same bacterial genus, keeping the strain or species with the maximum number of genes among a given genus of organisms to generate a non-redundant set of genomes as described earlier [6]. In addition, to avoid any affects due to the sample size of conserved pairs, we considered only those non-redundant genomes in which at least 20 triferogs were identified for presenting our results. P-values were calculated from Z-scores assuming a normal distribution of the random observations since the number of conserved pairs for the random TSS datasets followed a gaussian distribution.
3. Results and Discussion
3.1 Triferog definition
A transcriptional sensing system (TSS) is comprised of three elements: i) a transcription factor protein, ii) an effector gene and, iii) a corresponding signal effector. These systems may or may not constitute transcriptional sensing circuits, i.e, the effector gene may or may not be directly regulated by its corresponding TF (see Figure 1a, b). The effector gene may not be directly regulated by its corresponding TF when it is involved in a regulatory cascade of two or more TFs where an effector gene regulated by one TF could be producing an effector metabolite modulating the activity of a second TF (see Figure 1b). Apart from this, there could be a possibility that the first and second metabolites correspond to higher and lower parts of the same metabolic pathway. In this way the transcriptional sensing systems play important roles to link the transcriptional regulation of genes whose products are involved in different parts of metabolism or a regulatory cascade and shape the cell physiology to varying exogenous and endogenous conditions [2]. In the whole network of TF-effector gene pairs we found that only ∼36% of the links could be accounted for transcriptional interactions between a TF and its effector gene, suggesting that majority of the effector genes are not under the transcriptional control of their respective TFs but control the activity of the TFs in an indirect manner (see Supplementary material). In this work, we introduce the notion of “triferog”, which refers to the presence of both an orthologous transcription factor and its effector gene in different bacteria. If these two genetic components are present in distant bacteria it is probable that they constitute a TSS involving the same signal effector as seen in the reference or source genome (see Figure 1c). The process of identifying a putative triferog in an organism of interest involves the assignment of an orthologous effector gene to its corresponding TF which is likely to control or modulate its activity. This concept is important to understand the conservation of transcriptional sensing machinery across bacteria. It should be noted that the concept of triferog is different from that of regulog, while in the later the transfer of annotation is limited to a putative transcriptional regulatory interaction between a TF and its regulated gene, i.e. orthologous regulons, [5,6,8,16] while in the former, a TF and its effector gene constituting a regulatory interaction, is more of an exception than a rule as this happens in less than 33% of the regulons (see Supplementary material).
Figure 1.

a) An example showing the mechanism of action of a transcriptional sensing system (TSS) observed in E. coli. The effector gene cyaA encodes for the enzyme adenylate cyclase that catalyzes the formation from cyclic-AMP using ATP as substrate in the cytoplasm. The cAMP acts as signal metabolite (mainly in carbon source starvation) and when it is bound by the cAMP receptor protein (CRP), its transcriptional activity is affected by allosterism. This CRP-cAMP complex regulates the transcription of many transcription units [26-29]. b) Representation of two interconnected TSSs involved in the regulation of sulphur assimilation in E. coli K-12. CysB and Cbl are controlled by their corresponding signal metabolites produced by the effector genes for transport and synthesis. CysB acts depending on the availability of transportable metabolites containing sulfur (in this case thiosulphate is the signal metabolite) and Cbl responds to the signal metabolite (Adenosine 5-Phosphate) which is a product of the initial biochemical steps in the synthesis of L-cysteine. These TSSs in a) and b) show how the effector gene may or may not be directly regulated by its respective TF but can still respond to a functionally related TF. In this example CysB is the master regulator for sulfur metabolism in E. coli and it is possible that cbl, an accessory regulatory partner, is an ancient duplicate of cysB as these genes share 60% amino acid identity [30,31]. c) Representation of the Triferog concept using the TSS shown in a) for E. coli K12. As the genetic sensing components in the source organism are identified and validated as orthologs (dashed lines) in a different genome (target organism) it is highly probable that this genetic sensing system might be responding to the same signal effector. The black lines represent the signal effector availability by the effector(s) gene(s) and their allosteric effect on the respective transcription factor in the source organism. The break lines in the target organism represent the putative TF-effector interaction between the orthologs of genetic sensing components observed in the source organism. In this case the target organism is Vibrio cholerae where the TSS is experimentally shown to be conserved [32,33].
3.2 Transcriptional sensing systems identified in E. coli are poorly conserved across γ-proteobacterial genomes
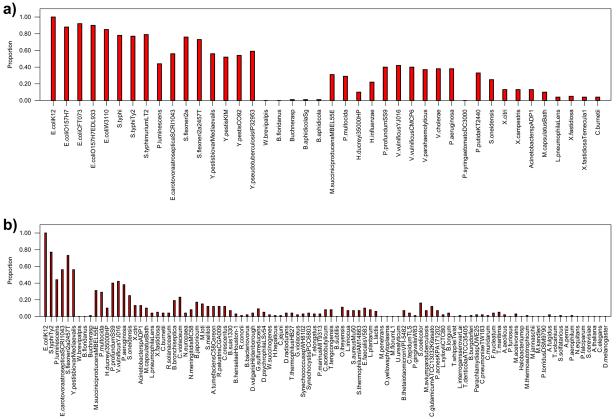
To study the conservation of TSSs in E. coli we identified their triferogs across a collection of 216 completely sequenced genomes tabulated in the Supplementary material. In Figure 2a, we show the proportion of TSSs conserved across all the γ-proteobacterial genomes from this set. As the phylogenetic distance with respect to E. coli K-12 increases, the proportion of triferogs identified across genomes decreases. All the 5 strains of E. coli share more than 80% of the TSSs known in E. coli K-12, while all the Shigella and Salmonella species share between 70-80%. All Yersinia strains and the Pectobacterium, Erwinia carotovora share between 50-60% of the TSSs known in E. coli. Vibrionaceae, Photorhabdus luminescens and Pseudomonas aeruginosa were found to share between 35-50% while Pasteurellaceae, which include Haemophilus influenzae, Pasteurella multocida and Shewanella species have around 20-35% of the TSSs conserved, indicating that the genetic components composing the TSSs in E. coli K-12 are poorly conserved beyond Salmonella and Shigella genomes.
Figure 2.
Conservation of TSSs across genomes as the phylogenetic distance with respect to E. coli increases. Calculation of phylogenetic distance and construction of non-redundant set of genomes was done as described earlier [6]. a) Conservation of proportion of transcriptional sensing systems (TSSs) known in E. coli across 42 γ-proteobacterial genomes. b) Conservation of the proportion of TSSs across 105 non-redundant genomes showing that the TSSs are poorly conserved across the phylogenetic spectrum.
3.3 Conservation of the genetic machinery for sensing drops rapidly as the phylogenetic distance with respect to E. coli increases
Figure 2b shows the conservation of TSSs across 101 non-redundant set of genomes (see methods and reference [6] e. g. only one E. coli genome of the 5 sequenced is considered) from three domains of life. We found that in more than 90% of the genomes less than 30% of the TSSs are conserved and in more than 95% of the genomes less than 50% are conserved suggesting that in majority of the genomes there are very few detected triferogs, which can be thought to be functionally equivalent to those observed in E. coli K-12. From the figure 2b it is also clear that as the phylogenetic distance with respect to E. coli increases, conservation drops off very rapidly although there are small abrupt jumps at certain distances corresponding to those of endosymbionts which usually have small genome sizes accompanied by substantial decrease in TF content suggesting their survival to limited conditions in their host environment. A comparison of the extent of conservation of TSSs against that of the complete protein coding genes in E. coli in all the genomes shown in Figure2b, suggests that TSS are less conserved than gene repertoire of E. coli in most genomes (see Supplementary material).
To assess the likelihood of identifying true triferogs in a genome based on conservation of the individual TF and effector genes we compared the conservation of the known TSSs against that of randomly constructed sets, as described in the methods section. We found that in 75% of the genomes where at least 20 triferogs were detected, the conservation was significantly higher (with P-values < 0.001) than compared to the conservation of randomly constructed TSSs (see methods) suggesting that the TSSs from E. coli K-12 have a strong tendency to occur together despite their poor conservation across genomes and the effector gene assigned to the TF on the basis of orthology is very likely to be functionally equivalent to those known in E. coli K-12 (see Supplementary Material for significance values seen in each genome).
3.4 TF-effector pairs constituting enzymes as effector genes are more conserved than those comprising transporters and sensor proteins
An obvious question which arises, given that TSSs identified in E. coli are poorly conserved across genomes with increasing phylogenetic distance is: are there TSSs or their components which are evolutionarily more conserved and hence are likely to be ancient and more stable, and if so, what are their functions? To address this, we analyzed the conservation of TSSs by taking into account the type of effector genes constituting them. We were able to identify only three major classes of TSSs, depending on the effector genes constituting them, namely enzymes, transporters and sensor proteins. However, there might be some poorly represented or unidentified class of effector genes in the complete regulatory repertoire of the cell which we can not take into consideration at the moment due to the incompleteness of our knowledge about transcriptional regulation in E. coli K12. The first kind of the three classes that we consider in this study includes TFs sensing signal metabolites synthesized by enzymes in the cell cytoplasm and therefore might correspond to the sensing of intracellular signals while the other two comprise of TFs for which transporters and sensor proteins (of two-component systems) act as their effector genes and sense mostly exogenous signals [7,8]. Figure 3 shows the conservation of TSSs based on this classification. There is a clear bias in the extent of conservation of TSSs constituting enzyme genes (i.e preponderance of triferogs of internal sensing systems identified in E. coli) in comparison to those comprising transporter and sensor proteins (see Figure 3a, b and c respectively, the X axis shows the conservation of TF and effector gene pairs across bacterial genomes). This finding makes biological sense because signals sensed by well conserved TSSs of the enzyme class correspond to important building block metabolites involved in cell structure, intermediate metabolic pathways or as cellular fuel. The most conserved TSS with its triferog detected in 130 genomes is that constituted by DnaA (DNA-replication initiator protein) and some of the enzymes involved in the ATP (adenosine triphosphate) biosynthesis, followed by those corresponding to the synthesis of arginine (ArgR), biotin (BirA), glycerol-3-phosphate (GlpR) and leucine (Lrp), ranking from 71 to 58 genomes in their extent of conservation (see Figure 4 and Figure 3a). It is important to note that arginine, leucine and glycerol-3-phosphate are effector signals of hybrid TSSs in E. coli as they can be produced both endogenously (using enzymes as effector genes) and exogenously (using transporters as effector genes) [7]. The cell, in addition to having enzymes necessary for synthesizing these metabolites internally, can also import them from the exterior of the cell using transport systems. This means that certain TFs have effector genes encoding for both enzymes and transporters and therefore are capable of sensing both exogenous and endogenous conditions. The transport systems used for obtaining these metabolites from the milieu are less conserved across bacteria than their corresponding enzymes (see Figure 4) and this bias is observed in other hybrid systems (see Supplementary data of sensing systems and Figure 4). Among the TSSs involving the transport of exogenous metabolites the most conserved are those for internalizing metals like zinc and ferric ion (in 70 and 51 genomes respectively) while in two-component systems highly conserved pairs occur in less than 50 genomes (Figures 3b and c). Taken together, these observations allow us to conclude that TSSs for internal signals in E. coli K-12 are more conserved in bacteria in comparison to those for sensing exogenous signals. This becomes very evident from those cases where the cell, despite having the machinery for transporting the metabolites from the milieu in addition to synthesizing them internally, prefers to conserve biosynthesis systems over transport systems. This might be due to the fact that certain internal signals are also important metabolites for the cellular metabolism and hence can be expected to be more homogeneously distributed across different bacterial kingdoms in comparison to the external signals, which change depending on the composition of each biological niche. As a consequence the transcriptional machinery to detect the external signals might be niche-dependent.
Figure 3.

Conservation of TSSs in prokaryotes. X axis shows the number of genomes where the TF and effector gene pairs from E. coli K12 are detected, Y axis shows the number of genomes where the effector gene and transcription factor are conserved individually. a) Genomic conservation of TSSs where the effector genes encode for enzymes (pink dots), b) TSSs where the effector genes encode for transporters (green dots) and c) where the effector genes encode for a sensor protein of two-component systems (light blue dots). Correlations (R2) of the best fit are shown in broken lines for TF and in continuous lines for effector genes.
Figure 4.

Conservation of TSSs in prokaryotes from the perspective of Escherichia coli K12. Dark blue nodes represent the TFs; different types of effector genes are represented in different colors: pink for enzymes, green for transporters and light blue for the sensor proteins of two-component systems. The effector genes whose products might be sensing external signals are represented in the external circle (transport and sensor proteins). The enzymes and the TFs sensing internal signals are represented in the inner circle. Thickness of the edges represents the extent of conservation: thick edges, putative TSS conservation in more than 100 genomes; edges with medium thickness, TSS conservation in between 50 and 99 genomes; thin lines, conservation in less of 49 genomes.
3.5 Transcription factors and their effector genes are conserved across genomes to unequal extents when compared against the conservation of their TSSs
It is well known that different cellular components are differentially conserved across the phylogenetic tree. In the case of transcriptional regulatory networks it has been shown recently that the transcription factors evolve faster than their target genes [5,6]. In a similar way it is possible to analyze separately the genomic conservation of transcription factor and effector genes. Thus in the Y axis of Figure 3 conservation of the components of the TSSs in bacterial genomes is shown. It is easy to note that enzymes are more conserved than their TFs while transporters and their respective TFs seem to be conserved to the same extent. On the other hand, sensor components are less conserved than their corresponding response regulators (Figure 3a, b and c respectively). It is possible that enzymes are more conserved than other kind of effector genes with respect to their TFs as they form part of the basal cellular metabolism producing important intracellular metabolites as discussed in the previous section. For example Eno (enolase, conserved in 177 genomes) is the enzyme involved in the interconversion of 2-phosphoglycerate to phosphoenolpyruvate. Following are some well conserved enzymes: GlyA (serine aldolase subunit, 172 genomes) involved in the interconversion of glycine and serine, some components of ATP synthesis (like AtpA, 162 genomes), GpsA catalyzing the conversion of dihydroxyacetone-phosphate to produce glycerol-3-phosphate (157 genomes) and MetK (S-adenosylmethionine synthetase, 148 genomes). All these enzymes are more conserved than the most conserved TFs, DnaA and BirA (146 and 86 genomes respectively). It is interesting to note that in the case of TSSs where the effector genes encode for transporters, TFs and their respective effector genes are conserved to equal extents (Figure 3b), except in the case of Fur (ferric uptake regulator), where the TF is far more conserved than its effector gene for transport. A possible explanation for this observation could be that most of these TSSs are either encoded in the same operon or in close chromosomal proximity which can aid them to evolve as chromosomal modules [9]. On the other hand lower conservation of sensor genes with respect to their response regulators in two-component systems is difficult to explain given that these systems also tend to be encoded proximal on the E. coli chromosome and are found to be interacting even at the level of protein products [9]. One possible reason for this differential conservation of TFs and their cognate sensor genes could be due to the extreme flexibility in the genetic components of two-component systems as a result of horizontal gene transfer events and lineage-specific expansions in bacteria [17]. In addition it is well known that the signal input domain in the histidine kinases is highly variable [18] and therefore detection of orthologs might be effected, causing imbalance in the detection of two-component systems. Although in general there is a tendency for an equilibrium in the extent of conservation of response regulators and histidine kinases in bacteria, there are some groups where there are clearly more histidine kinases than response regulators as seen in cyanobacteria and green sulfur bacteria and on the contrary beta and epsilon-proteobacteria have less histidine kinases than response regulators [19,20]. Thus it might be that sensor components of the external sensing machinery vary more quickly in bacteria given that some redundancy and cross talk in some of their components is known to exist [21,22]. Thus it seems that much of the variation in transcriptional sensing machinery across bacteria can be accounted for the changes in the effector genes used for sensing external conditions, which are poorly conserved, than those for sensing the changes in the endogenous conditions.
In this work we show that TSSs identified in E. coli, which can be interpreted to comprise of TF-effector gene pairs, are poorly conserved across genomes and the conservation falls off rapidly with increasing phylogenetic distance. We also find that TSSs from different categories are conserved to varying extents in complete genomes, with those comprising enzymes as their effector genes conserved the most. Our results suggest that transcriptional sensing machinery involved in the sensing of signals mostly synthesized by enzymes in the cytoplasm is well conserved across organisms, while the one sensing exogenous conditions is poorly shared with phylogenetically distant organisms. We note that some of the TSSs shown in Figure 4 (mainly those constituted by BirA, Lrp, GlpR, ArgR and Fur) in E. coli form transcriptional sensing circuits, where the TF regulates its effector gene (note that less than 35% of the TFs show this property). However, due to the lack of information about transcriptional regulation in phylogentically distant genomes it would be premature to conclude if these kind of circuits would tend to be more conserved than those TSSs not forming circuits, across bacterial genomes. The differential conservation of TF and effector gene (enzyme) pairs that we observe in this work can be explained by the following factors a) although the cellular metabolism might be conserved, each group of bacteria depending on their life-history could be using different set of endogenous metabolites to control their gene expression and consequently use different sets of effector genes to affect the activity of non-orthologous or different TFs. In fact, even if they use the same endogenous metabolite and hence same effector gene, transcriptional responses could be very different across organisms due to a plethora of possibilities in the domain combinations of TFs. b) post-transcriptional mechanisms like regulation by riboswitches might be substituting regulation at the level of transcription played by TFs, by responding to identical regulatory signals, as seen in the biosynthesis of the amino acids, methionine and tryptophan, where in the regulation in B. subtilis and E. coli is operated by different means [23-25]. It is possible to hypothesize based on our observations that at large phylogenetic distances there could be extensive variations in the domain architecture of the repertoire of TFs to accommodate and suffice the variations in the endogenous conditions of the cell, despite keeping some of the core enzymatic roles a constant. It would be interesting to explore in greater detail, on a case by case basis across genomes, where the metabolic pathway is known to be conserved but no corresponding TF is detected as this will enhance our knowledge about novel mechanisms linking metabolic and transcriptional networks.
Supplementary Material
Supplementary information - Additional figures and tables related to this work can be obtained from the webpage below.
http://www.ccg.unam.mx/Computational_Genomics/TRNS/Triferog/
The regulatory sub-systems classified according to the location of signal metabolites, used for the entire analysis and additional figures can be obtained from http://www.ccg.unam.mx/Computational_Genomics/regulondb/CellSensing/
Acknowledgements
AMA was supported by an INSERM fellowship for Etránger Chercheur in the unit U511 at CHU-Pitié Salpetrière in Paris, France. SCJ has been supported by grants given to Julio Collado-Vides. We thank Arthur Wuster for critically reading a previous version of this manuscript and Irma Lozada-Chavez for discussions in the preliminary stages of this work.
Abbreviations
- TF
transcription factor
- TSS
transcriptional sensing system, includes a TF, an effector gene and its corresponding effector signal
- Triferog
orthologous pair of transcription factor and effector gene, the effector gene may or may not be directly regulated by the corresponding TF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–9. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Shi Y. Metabolic enzymes and coenzymes in transcription--a direct link between metabolism and transcription? Trends Genet. 2004;20:445–52. doi: 10.1016/j.tig.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 5.Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–33. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–45. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Seshasayee AS, Bertone P, Fraser GM, Luscombe NM. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr Opin Microbiol. 2006;9:511–9. doi: 10.1016/j.mib.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Janga SC, Salgado H, Collado-Vides J, Martinez-Antonio A. Internal Versus External Effector and Transcription Factor Gene Pairs Differ in Their Relative Chromosomal Position in Escherichia coli. J Mol Biol. 2007;368:263–72. doi: 10.1016/j.jmb.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Salgado H, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–7. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madan Babu M, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–44. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–47. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 14.Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–6. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, et al. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–18. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mascher T, Helmann JD, Unden G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiology and Molecular Biology Reviews. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–82. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11:359–66. doi: 10.1016/s0966-842x(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 22.Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–67. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gollnick P, Babitzke P, Antson A, Yanofsky C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- 24.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding Sadenosylmethionine. Nat Struct Biol. 2003;10:701–7. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 25.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–53. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci U S A. 1970;66:104–10. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmer M, deCrombrugghe B, Pastan I, Perlman R. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc Natl Acad Sci U S A. 1970;66:480–7. doi: 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botsford JL, Harman JG. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–22. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol. 2003;6:482–9. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Bykowski T, van der Ploeg JR, Iwanicka-Nowicka R, Hryniewicz MM. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5'-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol Microbiol. 2002;43:1347–58. doi: 10.1046/j.1365-2958.2002.02846.x. [DOI] [PubMed] [Google Scholar]
- 31.van der Ploeg JR, Eichhorn E, Leisinger T. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch Microbiol. 2001;176:1–8. doi: 10.1007/s002030100298. [DOI] [PubMed] [Google Scholar]
- 32.Skorupski K, Taylor RK. Sequence and functional analysis of the gene encoding Vibrio cholerae cAMP receptor protein. Gene. 1997;198:297–303. doi: 10.1016/s0378-1119(97)00331-4. [DOI] [PubMed] [Google Scholar]
- 33.Chattopadhyay R, Parrack P. Cyclic AMP-dependent functional forms of cyclic AMP receptor protein from Vibrio cholerae. Arch Biochem Biophys. 2006;447:80–6. doi: 10.1016/j.abb.2006.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information - Additional figures and tables related to this work can be obtained from the webpage below.
http://www.ccg.unam.mx/Computational_Genomics/TRNS/Triferog/
The regulatory sub-systems classified according to the location of signal metabolites, used for the entire analysis and additional figures can be obtained from http://www.ccg.unam.mx/Computational_Genomics/regulondb/CellSensing/