Abstract
The phosphotransferase system (PTS) is the sugar transportation machinery that is widely distributed in prokaryotes and is critical for enhanced production of useful metabolites. To increase the glucose uptake rate, we propose a rational strategy for designing the molecular architecture of the Escherichia coli glucose PTS by using a computer-aided design (CAD) system and verified the simulated results with biological experiments. CAD supports construction of a biochemical map, mathematical modeling, simulation, and system analysis. Assuming that the PTS aims at controlling the glucose uptake rate, the PTS was decomposed into hierarchical modules, functional and flux modules, and the effect of changes in gene expression on the glucose uptake rate was simulated to make a rational strategy of how the gene regulatory network is engineered. Such design and analysis predicted that the mlc knockout mutant with ptsI gene overexpression would greatly increase the specific glucose uptake rate. By using biological experiments, we validated the prediction and the presented strategy, thereby enhancing the specific glucose uptake rate.
Keywords: Escherichia coli, glucose uptake, phosphotransferase system, regulation, system analysis
Introduction
The goals of systems biology are to understand the mechanism of how biochemical networks generate particular cellular functions in response to environmental stresses or genetic changes, and to rationally design the molecular processes to meet an engineering purpose. Robustness, which is an inherent property for cells to maintain homeostasis against a variety of environmental stresses, genetic changes, and stochastic fluctuations, is the key feature to analyze or design the molecular architecture that produces various cellular functions.
In the field of biotechnology, there have been many studies that aimed at producing useful metabolites using genetically engineered organisms, which often require a rational strategy of how the molecular processes of complex and robust cells are designed in order to achieve enhanced production. Since various mechanisms generating robustness, such as pathway redundancy and feedback (FB) regulation, often cancel the effect of genetic manipulations, it is important to understand the molecular mechanism of how a biochemical network generates robustness to genetic perturbations.
To rationally design biological systems at the molecular interaction level, it is essential to identify a biochemical network map, to build a dynamic model of the system, and to perform system analysis, thereby linking the molecular architecture to cellular functions. Computer-aided design (CAD) is now a key concept in building dynamic models and to mathematically simulate the molecular architecture of a genetically engineered cell (Kurata et al, 2005). Perturbation analysis is useful for identifying critical parameters that affect the system's performance. To modify the metabolism using biotechnology, the most common and convenient methodology for perturbation analysis is changing the copy number of metabolic pathway coding genes by using genetic recombination technology. Most of the previous metabolic engineering studies proposed qualitative alternations of the nature of enzymes or metabolite concentrations in cells.
Furthermore, module-based analysis has been presented that decomposes a biochemical network map into hierarchical modules, functional and flux modules, in a manner analogous to control engineering architectures (Kurata et al, 2006). If an engineering purpose is provided for a biochemical network, functional modules are readily determined and an engineering function can be assigned to each module. This greatly facilitates the rational design of a genetic manipulation responsible for achieving the desired outcome, and provides an intuitive understanding of how the biochemical network of interest is improved or modified.
In the field of industrial production of useful substances by microorganisms, glucose (Glc) is one of the most important substrates necessary for substance production or cell growth. Escherichia coli is a commonly used microbe in the industrial field, including the manufacture of amino acids (Ikeda, 2003) and organic acids (Wendisch et al, 2006). To enhance the ability to utilize substrates is of critical importance to increase the productivity of useful substances from the viewpoint of economical impact. Based on biological knowledge of the phosphotransferase system (PTS), several modeling and simulation studies have been performed (Rohwer et al, 2000; Thattai and Shraiman, 2003; Sauter and Gilles, 2004; Rodriguez et al, 2006). Rohwer et al (2000) measured the kinetic parameters of the E. coli glucose PTS and built the enzymatic reaction model. To study catabolite repression in E. coli, Sauter and Gilles (2004) developed a numerical model describing the transport and signal processing function by the sucrose PTS, validating their model with several experiments.
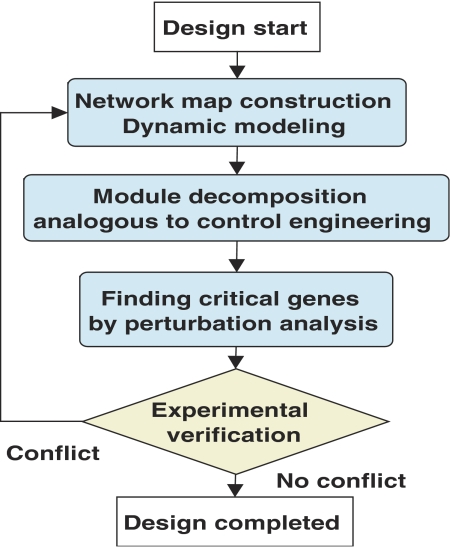
To perform perturbation analysis or to analyze the effect of the copy number of genes, we constructed an extended mathematical model of the E. coli glucose PTS model including the transcriptional regulation of the PTS proteins. The final objective was to increase the fermentation productivity through genetic improvement of glucose uptake efficiency in the PTS model. We propose a general strategy for computer-aided rational design of biochemical models as shown in Figure 1. CAD supports construction of a biochemical network map, mathematical modeling, dynamic simulation, and system analysis. Using the CADLIVE system we designed the molecular architecture of the glucose PTS that consists of the following genes: ptsG, ptsH, ptsI, crr, cyaA, crp, and mlc (Kurata et al, 2005). In order to make a rational plan of how the PTS could be genetically engineered for an enhanced glucose uptake rate, or to determine which genes should be deleted or overexpressed, the PTS was decomposed into hierarchical modules, functional and flux modules, in a manner analogous to control engineering architectures, and subsequently the effects of changes in gene expression on the glucose uptake rate were simulated to predict which genes are critical for enhanced uptake. Finally we validated the predicted performance of rationally engineered cells with experimental results, demonstrating the feasibility of the computer-aided rational design of biochemical networks.
Figure 1.
Algorithm chart for computer-aided rational design of biochemical networks.
Results and discussion
Modular architecture of the glucose PTS and mathematical modeling
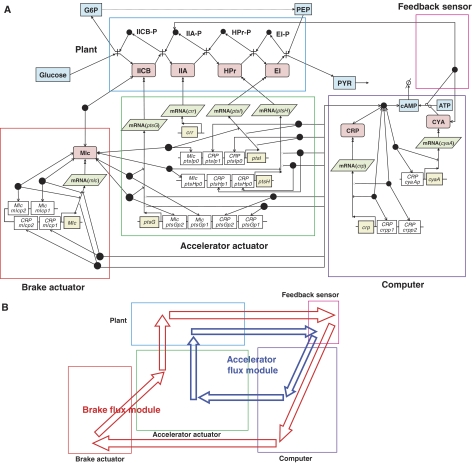
We present a biochemical map of the E. coli glucose PTS (Figure 2) and developed a dynamic model from this map (Supplementary Tables S1 and S2; see also Materials and methods) by using CADLIVE (Kurata et al, 2005). Details of the graphical notation are described elsewhere (Kurata et al, 2003). Generally, it is of great importance to increase the sugar uptake rate for enhanced substance production. We aimed at increasing the glucose uptake rate. To assign a particular function to each gene in terms of an enhanced uptake rate, the biochemical map of the glucose PTS was decomposed into functional modules (Figure 2A) and flux modules (Figure 2B) analogous to an engineering control systems block diagram (Kurata et al, 2006). Specifically, if we define the phosphotransfer cascade of IICBGlc (ptsG), IIAGlc (crr), HPr (ptsH), and EI (ptsI) as the plant to be controlled, then the entity that drives this plant is the signal from the accelerator actuator. The phosphorylation signal of IIAGlc-P is sensed by the FB sensor module and sent to the computer module that consists of adenylate cyclase (CYA), cyclic AMP (cAMP), and cAMP receptor protein (CRP), while it is also transmitted to the signal transduction pathways to enhance other carbon transport systems. The complex of IIAGlc-P:CYA, which is formed in the FB sensor module, produces the cAMP signal in the computer module, resulting in the formation of CRP:cAMP. CRP:cAMP is the output of the computer module, which modulates the accelerator and brake actuator modules. The accelerator actuator module comprises the synthesis process of the IICBGlc, HPr, and EI proteins, supplying these PTS proteins to the plant module; the brake actuator module of the Making Large Colonies Protein (Mlc) suppresses the synthesis of the same PTS proteins.
Figure 2.
The modular architecture of the glucose PTS in E. coli. (A) Functional modules. The symbol ‘-' indicates modification. The arrows indicate various reactions: binding ( ), protein synthesis or conversion (
), protein synthesis or conversion ( ), catalyzing a reaction (
), catalyzing a reaction ( ), suppressing protein synthesis (
), suppressing protein synthesis ( ), and activating protein synthesis (
), and activating protein synthesis ( ). (B) The two major flux modules. The inner arrows are the accelerator flux module that goes through the plant, FB sensor, computer, and accelerator actuator modules; the outer arrows are the brake flux module that goes through the plant, FB sensor, computer, brake actuator, and accelerator actuator modules.
). (B) The two major flux modules. The inner arrows are the accelerator flux module that goes through the plant, FB sensor, computer, and accelerator actuator modules; the outer arrows are the brake flux module that goes through the plant, FB sensor, computer, brake actuator, and accelerator actuator modules.
Superimposed on these functional modules, we identify two major flux modules, the accelerator flux module and the brake flux module. Although our description of these fluxes is qualitative, the components of the fluxes can be easily identified in Figure 2A. First, we identified the accelerator flux module where the signal of the IIAGlc-P increase goes through the FB sensor, computer, accelerator actuator, and plant modules to induce the synthesis of IICBGlc, HPr, and EI (Figure 2B). Second, we identified the brake flux module where the signal of the IIAGlc-P increase goes through the FB sensor, computer, brake actuator, accelerator actuator, and plant modules, which regulates the PTS component synthesis by suppressing the accelerator actuator.
We provide an example of the signal processing under glucose depletion conditions. Since the phosphorous of IIAGlc-P is not transferred to glucose, the IIAGlc-P concentration immediately increases. The sensor module then assesses an increase in the signal molecule, transmitting its information to the computer module. The computer module calculates the output signal necessary for an adequate control action, which is transmitted simultaneously to the accelerator actuator module and the brake actuator module. The phosphotransfer activity in the plant module is determined by the balance between the accelerator and brake actuator modules.
The mathematical equations can be connected to the functional modules for the full model (Supplementary Table S1). In this model, Equations (1.1–1.8) in Supplementary Table S1 describe the plant module, which corresponds to the phosphotransfer reactions in the PTS. Equations (2.1–2.3) in Supplementary Table S1 describe the FB sensor module, which senses the extracellular glucose concentration. The computer module corresponds to Equations (3.1–3.13), which contains transcription and translation of the crp and cyaA genes and cAMP production. The brake actuator module corresponds to Equations (4.1–4.13), which describes the role of the Mlc protein. Equations (5.1–5.26) show the accelerator actuator module, which contains the transcription and translation reactions of the ptsG, ptsH, ptsI, and crr genes.
Dynamic simulation
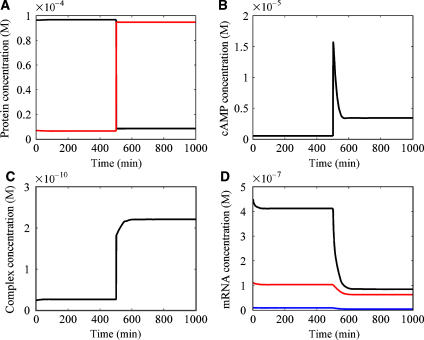
To demonstrate how the mathematical model reproduces the dynamic behaviors of the glucose PTS with respect to glucose, we simulated the model as shown in Figure 3. These dynamic behaviors were very consistent with the experimental data. As shown in Figure 3A, the concentration of IIAGlc-P rapidly increased just after glucose depletion, while that of IIAGlc decreased. Since phosphorus is not transferred to glucose due to its depletion, the phosphorylated proteins accumulate in the plant module. As shown in Figure 3B, the concentration of cAMP greatly increased in response to glucose depletion and gradually decreased after the peak, then reached a steady-state. The accumulated IIAGlc-P protein, which is assumed to be the signal for adenylate cyclase activation, binds to adenylate cyclase in the FB sensor module, thereby enhancing the synthesis of cAMP. As shown in Figure 3C, the concentration of the complex of Mlc and promoter 1 for the ptsG gene increased in response to glucose depletion, which resulted from the decrease in the IICBGlc concentration. Consequently, the complex of Mlc and promoter 1 of the ptsG gene suppresses the gene expression of the ptsG gene. In the same manner, ptsH and ptsI gene expression was suppressed by the complex of Mlc and promoter 0 of the ptsH and ptsI genes. If the concentration of IICBGlc is high, IICBGlc captures Mlc to prevent Mlc from binding to promoter 1 of the ptsG gene, which permits transcription to proceed. As shown in Figure 3D, the expression of the ptsG, ptsH, and ptsI genes diminished in response to glucose depletion. This suppression was caused mainly by the brake actuator module. Actually, since the effect of the brake actuator module is greater than that of the accelerator actuator module in the absence of glucose, the synthesized Mlc protein suppresses the transcription of the ptsG, ptsH, and ptsI genes. The phosphotransfer activity decreases in the plant module, thereby decreasing glucose uptake. In the PTS, to solve the problem of glucose depletion, the signal of glucose depletion, the accumulated IIAGlc-P, is transferred to the signal transduction pathways to enhance other carbon transport systems.
Figure 3.
Simulated time course for molecular concentrations in response to glucose depletion in the PTS. The extracellular glucose concentration was changed from 0.2 M to 0.2 nM at 510 min. (A) Time evolution for IIAGlc protein in the phosphate relay cascade. The black line shows the IIAGlc concentration and the red line shows the IIAGlc-P concentration. (B) Time evolution for the intracellular cAMP concentration. (C) Time evolution for the concentration of the complex of Mlc and promoter 1 of the ptsG gene. (D) Time evolution for the expression of the ptsG, ptsH, and ptsI genes. The black line shows ptsH mRNA, the red line ptsG mRNA, and the blue line ptsI mRNA.
Validation of the mathematical model by experimental data
To further validate the mathematical model, we compared the simulated results with experimental data that were not used for the mathematical modeling, as shown in Figure 4. Such comparison of the simulated results with experimental data demonstrated that the dynamic behavior of the mathematical model was consistent with that of the experimental data.
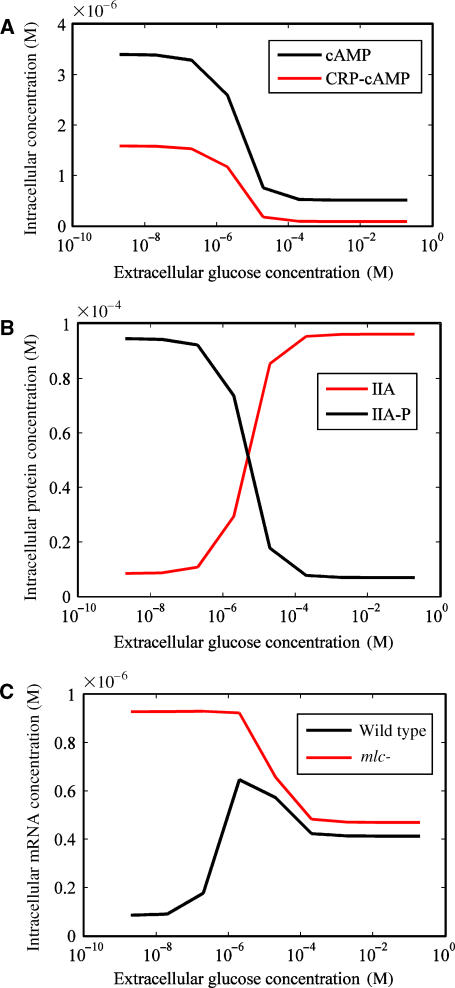
Figure 4.
Validation of the mathematical model. (A) The intracellular cAMP concentration and CRP:cAMP complex concentration were simulated with respect to the extracellular glucose concentration. The black line and red line show the cAMP concentration and the CRP:cAMP complex concentration, respectively. (B) IIAGlc protein phosphorylation was simulated with respect to the extracellular glucose concentration. The black line and red line show the phosphorylated IIAGlc protein concentration and the unphosphorylated IIAGlc protein concentration, respectively. (C) The ptsG mRNA concentration was simulated with respect to the extracellular glucose concentration. The black line and red line show the ptsG mRNA concentrations of wild type and of an mlc knockout mutant, respectively.
As shown in Figure 4A, the dynamic model reproduced the experimental behavior that the concentration of intracellular cAMP and CRP:cAMP varied with the extracellular glucose concentration in a dose-dependent manner (Notley-McRobb et al, 1997). The cAMP concentration in glucose-abundant medium was lower than that in glucose-deficient medium. As shown in Figure 4B, the IIAGlc protein was dephosphorylated at a high extracellular glucose concentration and the phosphorylation status of IIAGlc protein was responsible for the extracellular concentration (Hogema et al, 1998). These simulated results were consistent with the experimental data. Zeppenfeld et al (2000) measured the ptsG promoter activity in wild type and an mlc knockout strain (mlc−) using a lacZ fusion in minimum medium with glucose and glycerol. In wild-type cells, the ptsG promoter activity in glucose medium was higher than that in glycerol medium, which may result from the de-repression of Mlc. In both glycerol and glucose medium, the ptsG promoter activity in an mlc− knockout strain was higher than that in wild-type cells. As shown in Figure 4C, the expression of the ptsG gene was successfully reproduced by the simulation, where the glucose dose dependency for ptsG gene expression was observed both in wild type and an mlc knockout mutant and the gene expression level in the mlc− mutant was higher than that in wild-type cells regardless of the extracellular glucose concentration.
Extraction of critical genes for enhanced glucose uptake
The functional and flux module decomposition of the PTS model (Figure 2) readily provides a rational strategy for enhanced glucose uptake. The mlc and crp genes of the transcription regulation factors affect the expression of multiple genes. The mlc gene is responsible for the brake actuator, thus mlc knockout is expected to enhance the synthesis of the PTS proteins. The crp gene belongs to the computer module for the accelerator actuator, and thus overexpression of the crp gene has a potential to enhance the synthesis of the PTS proteins. Since the ptsG, ptsH, ptsI, and crr genes are the catalysts for the phosphorylation cascade in the plant module, it is reasonable to increase their expression for enhanced glucose uptake rates. It is critical for enhanced glucose uptake to increase the PTS proteins in the plant module (the ptsG, ptsH, ptsI, and crr genes), to delete the brake actuator module (the mlc gene), or to enhance the accelerator actuator module through the computer module (the crp gene). Regarding the crp gene manipulation it is necessary to consider the quantitative balance between the accelerator and brake flux modules, because CRP:cAMP not only enhances the accelerator actuator but also the brake actuator.
Simulation and perturbation analysis of the rationally designed PTS
To further explore target genes critically responsible for enhancing glucose uptake, we performed perturbation analysis with regard to crp, mlc, ptsG, ptsH, ptsI, and crr gene expression, as shown in Table I, where the specific glucose uptake rates were simulated with respect to a 10-fold change in the copy number of these genes. When the concentration of a target gene was varied by 10-fold from the default condition (wild type), the ratio of the specific glucose uptake rate for a mathematical mutant to that for the wild type was simulated. The ratio of the phosphoenolpyruvate (PEP) concentration to the pyruvate (Pyr) concentration ([PEP]/[Pyr]) was set to 1 and each concentration was set to 1 mM (Chassagnole et al, 2002). We calculated the effect of the mlc gene disruption and crp gene amplification (crp+) on the specific glucose uptake rate. The ptsI gene amplification showed the most enhanced specific glucose uptake rate for all the strains: wild type, the mlc knockout mutant, and the crp-overexpressing mutant. An mlc knockout mutant that overexpressed the crp and ptsI genes was predicted to enhance the specific glucose uptake rate by the greatest degree. Since the ratio of the intracellular PEP to the Pyr concentration affected the specific glucose uptake rate by the PTS (Hogema et al, 1998), we simulated the ptsI amplification effect on an enhanced specific glucose uptake rate using various values of the ratio of [PEP]/[Pyr], by changing each molecule concentration from 0.1 to 10 mM and confirmed that ptsI amplification was always effective in enhancing the glucose uptake (data not shown). Based on this result, the subsequent simulations were performed with 1:1 ratio at 1 mM of PEP and Pyr concentration.
Table 1.
Prediction of changes in the specific glucose uptake rate for mathematical mutants
| Genetic background |
|||
|---|---|---|---|
| Gene amplification | Wild | mlc− a | crp+ b |
| None | 1.00c | 1.02 | 2.91 |
| ptsG | 0.81 | 0.81 | 2.91 |
| ptsH | 7.95 | 8.05 | 8.23 |
| ptsI | 10.83 | 11.08 | 9.53 |
| crr | 3.48 | 3.51 | 3.51 |
The values are the ratios of the specific glucose uptake rate for a mutant to that for wild type.
aThe mlc− mutant indicates that the mlc gene is deleted.
bIn the crp+ mutant the crp gene concentration is 10-fold higher than the wild type.
cThe value was normalized by providing 1 to the ratio for wild type.
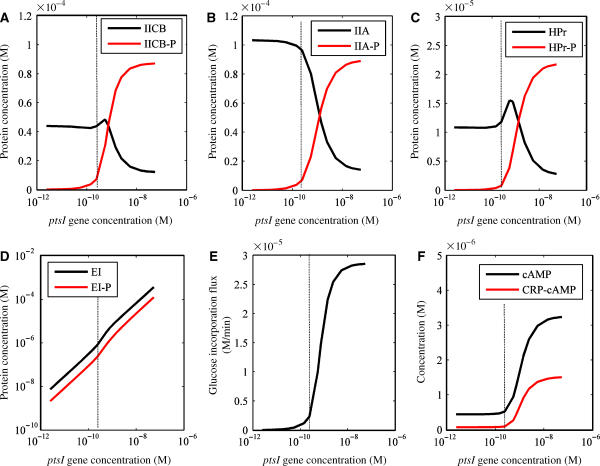
To analyze the mechanism of how the glucose uptake rate is increased by ptsI gene amplification, the PTS component concentrations were simulated with respect to a change in the ptsI gene dose, as shown in Figure 5. Since the specific glucose uptake rate is defined as the rate of the phosphotransfer reaction from phosphorylated IICBGlc to glucose per cell, that is, the production rate of glucose-6-phosphate (G6P), the glucose uptake rate is closely related to that for the phosphorylated status of IICBGlc protein. As shown in Figure 5A–D, the concentrations of phosphorylated proteins increased with an increase in the concentration of the ptsI gene. This simulated result suggests that the increase in the PTS flux is caused by an increase in the EI protein concentration. In the simulation, the phosphorylated IIAGlc protein becomes abundant when the ptsI gene is 10-fold amplified compared with the default condition. The phosphorylation status of the IIAGlc protein is known to depend on extracellular sugars and most of the IIAGlc proteins are unphosphorylated when glucose is used in the medium. The phosphorylation of the IIAGlc protein is suggested to be the signal for increasing the activity of adenylate cyclase that converts ATP into cAMP (Hogema et al, 1998). As shown in Figure 5E and F, the specific glucose uptake rate and the intracellular cAMP and CRP:cAMP complex concentrations increased with an increase in the EI concentration. The increase in the CRP:cAMP concentration activates the transcription of the ptsG gene and pts operon, subsequently increasing the specific glucose uptake rate.
Figure 5.
Effects of ptsI gene amplification on the PTS components in the dynamic simulation. The ptsI gene concentration in a cell was changed within a range from 2.43 pM to 54.3 nM. The default concentration was 0.24 nM (one gene per a cell), as shown by the vertical dashed line. (A) IICBGlc protein phosphorylation. The black line and red line show the unphosphorylated and phosphorylated IICBGlc concentrations, respectively. (B) IIAGlc protein phosphorylation. The black line and red line show the unphosphorylated and phosphorylated IIAGlc protein concentrations, respectively. (C) HPr protein phosphorylation. The black line and red line show the unphosphorylated and phosphorylated HPr protein concentrations, respectively. (D) EI protein phosphorylation. The black line and red line show the unphosphorylated and phosphorylated EI protein concentrations, respectively. (E) Glucose uptake rate. (F) The cAMP and CRP:cAMP complex concentrations. The black line and red line show the cAMP and CRP:cAMP complex concentrations, respectively.
Out of the ptsG, ptsH, crr, and ptsI genes, the ptsI gene was selected for detailed analysis because its amplification enhanced the specific glucose uptake rate most strongly. In addition, some background information supports this selection. The ptsI gene encodes the EI protein, which catalyzes the phosphate transfer reaction from PEP to the EI protein. This reaction is suggested to be a rate-limiting step for glucose uptake in the PTS (Weigel et al, 1982; Patel et al, 2006), and the enhanced EI protein concentration is expected to lead to an increase in the glucose PTS flux. The transcriptional attenuation of ptsH and ptsI occurs and the amount of ptsI transcripts is suggested to be one-tenth of that of the ptsH transcripts (De Reuse and Danchin, 1988). Supposedly, the attenuation of the ptsI transcript is one of the intrinsic points governing the glucose PTS flux. Therefore, the ptsI gene was a reasonable choice among the enzymes of the phosphorylation cascade in the plant module.
Experimental validation of rationally designed cells
The mlc knockout mutant that overexpresses both the crp and ptsI genes was predicted to enhance the specific glucose uptake rate most strongly. To evaluate the validity of the presented strategy, we carried out biological experiments. Since CRP is one of the global regulators whose functions are not fully clear, unexpected effects may be caused by crp gene amplification. Thus, we excluded crp-overexpressing mutants.
First, we investigated the effect of ptsI gene amplification. The simulation results suggest that more than 10 copies of the ptsI gene are required for the enhancement of specific glucose uptake; thus, a high-copy number plasmid was used for ptsI gene expression. The enhancement of EI protein expression was confirmed by SDS–polyacrylamide gel electrophoresis (data not shown). To test our prediction on the ptsI-amplified strain, MG1655/pUC118-ptsI, and a control strain, MG1655/pUC118, were cultivated in 20 ml of M9 glucose medium in a shaking incubator for 18 h. Growth, glucose uptake, specific glucose uptake, and extracellular cAMP concentration were measured (Table II). Regardless of the similar growth of both strains, the specific glucose uptake rate of the ptsI-amplified strain was significantly higher than that of the control strain. These experimental findings supported the prediction that an increased EI protein concentration enhances the specific glucose uptake rate under normal cell growth conditions.
Table 2.
Experimental results of growth, glucose uptake, specific glucose uptake, and cAMP concentration in growing cells
| Strains | OD600 | Glucose uptake |
Specific glucose uptakea |
cAMP concentration |
|---|---|---|---|---|
| M | M/OD | fM | ||
| E. coli MG1655 pUC118 | 0.81±0.17 | 0.012±0.003 | 0.015±0.001 | 10.88±0.28 |
| E. coli MG1655 pUC118-ptsI | 0.94±0.34 | 0.017±0.004 | 0.018±0.004 | 7.54±3.71 |
| E. coli MG1655M pUC118 | 0.67±0.17 | 0.011±0.003 | 0.016±0.001 | n.t. |
| E. coli MG1655M pUC118-ptsI | 0.11±0.02 | 0.0029±0.001 | 0.026±0.008 | n.t. |
cAMP, cyclic AMP; OD600, optical density at 600 nm.
aThe statistical significance was tested by the Bonferroni t-test. Except for the difference between MG1655 pUC118-ptsI and MG1655 M pUC118, all of the possible combination showed significant difference at least 5% level.
n.t.: not tested.
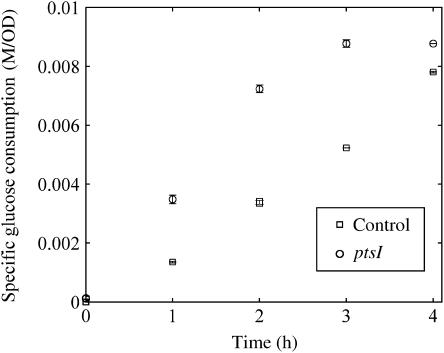
Second, we compared the specific glucose uptake rate between the ptsI-overexpressing strain and a control, using non-growing cells in minimum medium without thiamine required for JM109 growth (Figure 6). The specific glucose uptake rate of the ptsI-amplified strain, JM109/pUC118-ptsI, was higher than that of the control strain, JM109/pUC118. The observed growth (ΔOD600) was less than 0.6 in both strains. Mainly, the incorporated glucose was converted into acetic acid or other organic acids (data not shown). This result indicates that the ptsI gene amplification enhances the specific glucose uptake rate. This is also supported by previous kinetic studies that the EI protein concentration is critical for increasing the phosphate flux in the PTS (Weigel et al, 1982; Patel et al, 2006).
Figure 6.
Comparison of the specific glucose uptake between wild type and the ptsI-overexpressing mutant. The glucose uptake and cell growth of JM109/pUC118-ptsI (circle) and JM109/pUC118 (square) were measured under non-growing conditions. Three replicate experiments were performed and the averaged data are shown.
Third, we examined the effect of how ptsI gene amplification in the mlc knockout mutant derived from MG1655 (MG1655M) enhances the specific glucose uptake rate. The ptsI amplification strain, MG1655M/pUC118-ptsI, and a control strain, MG1655M/pUC118, were cultured in 20 ml of M9 glucose medium in a shaking incubator for 18 h. We compared the specific glucose uptake among the four strains (Table II). The MG1655M/pUC118-ptsI strain showed the highest specific glucose uptake even though it had the lowest growth. The next highest was the MG1655/pUC118-ptsI strain. The predictions for specific glucose uptake rates were validated by the experiments (Table I), demonstrating that amplification of the ptsI gene increases the specific glucose uptake rate and further enhances it in an mlc knockout mutant.
Discrepancies between simulation and experiment
In the simulation, the intracellular cAMP concentration in the ptsI-amplified strain was higher than that in wild type, but this was not consistent with the experimental data, as shown in Table II. We measured the cAMP concentration in the medium. The cAMP concentration in the ptsI gene amplification strain was lower than that in the control strain. In vivo, the ATP level may decrease or adenylate cyclase may not be activated by the ptsI gene amplification. Thus, we investigated the discrepancy in cAMP concentration. Inada et al (1996) showed that the dephosphorylation activity for the phosphorylated IIAGlc protein was observed when E. coli was cultured with glucose. Recently, not only the phosphorylated IIAGlc protein but also other factors have been reported to be involved in the activation of adenylate cyclase (Park et al, 2006). These mechanisms may explain the reason why the cAMP concentration shows the discrepancy between the mathematical and biological models.
Subsequently, we improved the glucose PTS model that the activation of adenylate cyclase does not occur in the presence of glucose and simulated the model as shown in Table III. Q[2] in Equation (3.4) was set to zero when glucose was present in the medium (Supplementary Table S1). In the improved model the cAMP concentration hardly decreased (data not shown), which was more consistent with experimental data than the previous model (Table I). Furthermore, the model improvement solved another discrepancy regarding the relationship between glucose uptake rates and the IICBGlc enhancement experiment. The glucose uptake rate was suggested to be increased by IICBGlc enhancement (Rohwer et al, 2000), while the glucose uptake rate was simulated to decrease with an increase in the copy number of the ptsG gene (Table I). By model improvement, ptsG gene amplification increased the glucose uptake, which was consistent with the experimental result (Table III).
Table 3.
Prediction of changes in the specific glucose uptake rates in the improved dynamic model that excludes adenylate cyclase activation by IIAGlc-P
| Genetic background |
|||
|---|---|---|---|
| Gene amplification | Wild | mlc− a | crp+ b |
| None | 1.00c | 1.21 | 3.85 |
| ptsG | 1.25 | 1.27 | 4.55 |
| ptsH | 3.49 | 4.55 | 9.86 |
| ptsI | 3.87 | 5.70 | 10.87 |
| crr | 0.86 | 1.04 | 3.25 |
The values are the ratios of the specific glucose uptake rate for a mutant to that of the wild type.
aThe mlc− mutant indicates that the mlc gene is deleted.
bIn the crp+ mutant the crp gene concentration is 10-fold higher than the wild type.
cThe value was normalized by providing 1 to the ratio for wild type.
The simulation of the improved model suggests that the effect of the phosphorylated IIAGlc-activated adenylate cyclase is very limited in the presence of glucose; in other words, unknown factors other than the phosphorylated IIAGlc play an important role in controlling the computer module.
Severe growth suppression
The specific glucose uptake rate was suppressed by Mlc in the wild type, while it was retained in an mlc− knockout mutant. Mlc is reported to suppress the expression of the ptsG gene and the pts operon (Plumbridge, 1998; Kim et al, 1999; Tanaka et al, 1999), thereby governing glucose uptake. As the dynamic simulation predicted, the mlc knockout enhanced the specific glucose uptake rate through synthesis of the PTS components, but our experimental results showed unexpected severe growth suppression with the combination of mlc gene deletion with ptsI gene amplification.
It is known that the excess accumulation of G6P inhibits cellular growth in E. coli (Kadner et al, 1992). The growth suppression in MG1655M/pUC118-ptsI may be caused by the excess accumulation of sugar phosphate due to the rapid glucose uptake (Table II). Recently, small RNAs in E. coli have been shown to respond to an intracellular sugar concentration and the transcripts of the ptsG gene are degraded by RNaseE (Morita et al, 2003). We speculate that E. coli has some unknown severe control systems for the pool size of sugar phosphate and no activation of adenylate cyclase in glucose medium can be due to such control systems. A further refined and extended model including G6P metabolism will clarify the control mechanism of glucose uptake.
Power of computer-aided rational design
We have presented a general strategy for the rational design of biochemical networks for an engineering purpose. The strategy consists of constructing a biochemical network, dynamic simulation, module-based analysis, perturbation analysis, and experimental validation. CAD supports construction of a biochemical network map, building its dynamic model, and system analysis. Module-based analysis decomposes a biochemical network map into hierarchical modules, functional and flux modules, in a manner analogous to control engineering architectures. If an engineering purpose is determined, functional modules are readily determined and an engineering function is assigned to each module. This facilitates rational design of a genetic manipulation responsible for achieving the purpose and an intuitive understanding of how the biochemical network of interest can be improved or modified. Perturbation analysis further explores critical genes responsible for achieving the engineering purpose.
Using CADLIVE we performed mathematical modeling, simulation, and system analysis to make a strategy of how the glucose PTS can be genetically engineered for enhanced glucose uptake. We found the critical factors responsible for an enhanced specific glucose uptake rate, for example, ptsI amplification and mlc knockout. Mathematical simulation and subsequent perturbation analysis predicted that the combination of amplification of ptsI and deletion of mlc would enhance the specific glucose uptake rate. Finally, experimental data validated this prediction.
Rational design of a robust system
Rational design requires an understanding of the mechanism of how a biochemical network provides a robust property to a target performance, or identifying the genes that show fragility of the performance to their genetic change. Manipulation for a particular flux module has a great potential to change the network performance of interest, for example, deletion of a negative FB loop or enhancement of a positive FB loop readily alters the cellular performance. In the glucose PTS, the brake actuator flux module has a negative FB loop. An increase in ptsI gene expression enhances phosphorylation of the PTS proteins (Figure 5), IIAGlc-P:CYA binding, CRP:cAMP synthesis, and Mlc production in turn, thereby suppressing ptsI gene expression. By contrast, the accelerator actuator flux module has a positive FB loop. An increase in ptsI gene expression enhances phosphorylation of the PTS proteins, IIAGlc-P:CYA binding, and CRP:cAMP synthesis in turn, which further enhances ptsI gene expression. Genetic manipulation for such a negative or positive FB loop shows a great potential to enhance the specific glucose uptake or activation of the PTS proteins. Knockout of the mlc gene is reasonable for ptsI gene overexpression, because it removes the negative FB loop that suppresses EI protein expression. Perturbation analysis in both the mathematical model and biological experiment support this design strategy.
In the improved model, positive and negative FBs were cancelled by disabling the function that IIAGlc-P:CYA synthesizes cAMP, where the glucose PTS system forms a more straight forward network. The brake flux module without the negative FB shows less suppression of PTS proteins with respect to EI or HPr overexpression, and the accelerator flux module without the positive FB weakens the expression of them. Actually, the specific glucose rate would be determined through the quantitative balance between the brake and accelerator modules. In the improved model, the decrease in the specific glucose uptake rate for ptsI or ptsH overexpression for wild type and an mlc knockout mutant (Tables I and III) would be due to deletion of the positive FB loop, while the increased uptake rates for other mutants would be caused by removal of the negative FB loop. It is of critical importance for rational design to understand the quantitative mechanism of how a flux module provides robustness to genetic changes.
Toward a perfect design
The simulation results did not necessarily explain the experimental results. As mentioned above, there was a discrepancy in the change in the cAMP concentration between the mathematical and experimental models. The combination of ptsI amplification and mlc deficiency was predicted to be a better strategy for enhanced specific glucose uptake, but it led to severe growth inhibition. From the mathematical simulation of specific glucose uptake, the ptsI gene-overexpressing strain was predicted to have a ratio of 3.87 compared with control, but experimentally at best a ratio of 1.2 (0.018/0.015) was observed (Tables II and III). Likewise, the ratio for the mlc− background was 5.70, but experimentally 1.7 (0.026/0.015) (Tables II and III). These differences seem to be due to the insufficient model size or lack of various FB regulations of enzyme and gene expressions that provide robustness to genetic modifications in the actual cells. In addition the other proteins, mannose PTS and non-PTS transporter, GalP, which are involved in permeation of glucose (Curtis and Epstein, 1975; Hernandez-Montalvo et al, 2003), may affect the experimental results.
The PEP/Pyr ratio is determined as a result of many enzymatic reactions including glycolysis, pentose-phosphate pathway, and anaplerotic pathways. Since growth is the output of complex interactions of a variety of molecules, it is still hard to accurately predict cell growth at the molecular interaction level. A large and complex model would be necessary that includes various regulations of enzyme and gene expressions in the glycolysis, pentose-phosphate pathway, TCA cycle, and anaplerotic pathway. An extended model is expected to clearly describe the experimental results.
If some discrepancies are found between the mathematical and experimental models, we need to feed this information back into model improvement. Ideally we should iterate the process consisting of mathematical analysis and experimental validation until prediction agrees with experimental results completely. This iteration cycle is key to the rational design of biochemical systems.
In conclusion, a computer-aided rational design approach was successfully applied to microbe engineering or breeding and verified by biological experiments. This methodology will lead to rapid development for not only applied biology that designs biochemical networks within a cell, but also fundamental research that reveals the mechanism of how biochemical networks generate particular cellular functions.
Materials and methods
Computer-aided rational design of biochemical networks
CAD is widely defined as computer-based tools that assist engineers, architects, and other design professionals in their design activities. CAD has been employed to conduct basic and applied research in drawing, modeling, simulation, and optimization of various engineering systems. The concept of CAD can be applied to biochemical systems, as shown in Figure 1. CAD supports not only constructing biochemical network maps but also mathematical modeling, simulation, and system analysis. To rationally design a biochemical network, it is important to present an engineering purpose and to decompose it into hierarchical functional and flux modules according to the purpose. In a manner analogous to control engineering architecture, a particular function is assigned to each functional module and mathematical simulation with perturbation analysis explores the genes critically responsible for achieving the purpose. These algorithms present a promising strategy for rational design of biochemical networks, providing instruction or prediction of how genes should be modified or manipulated. To validate the prediction, biological experiments are required. If there is any difference between the prediction and the experiment results, biochemical networks need to be redesigned through an understanding of the mechanisms that cause such discrepancies.
Biological model
The PTS transports sugars from the periplasm to the cytoplasm, coupled with phosphorylation (Postma et al, 1993). The source of phosphate is PEP in the cytoplasm. The PTS relays the phosphate from PEP to sugars (for a review see Postma et al, 1996). The glucose PTS in E. coli has been studied intensively. It consists of IICBGlc, IIAGlc, HPr, and EI proteins, which are encoded by the ptsG, crr, ptsH, and ptsI genes, respectively (schematically shown in Figure 2; for a review see Postma et al, 1996). Rohwer et al (2000) measured the kinetic constants of the phosphotransfer reactions and made a kinetic model of the glucose PTS.
E. coli prefers glucose rather than other sugars for its growth and glucose represses the catabolism of other sugars (Hogema et al, 1997). The glucose PTS is related to catabolite repression, chemotaxis regulation, glycogen phosphorylase, and multiple regulatory interactions (Lux et al, 1995; Postma et al, 1996; Seok et al, 1997; Saier, 1998; Eppler et al, 2002). Adenylate cyclase is activated when glucose in the medium is depleted. Although the detailed mechanisms of this activation are still unclear, we assume the complex of adenylate cyclase and phosphorylated IIAGlc protein is the active form of adenylate cyclase (Reddy and Kamireddi, 1998).
The gene expression regulation of the glucose PTS has been investigated (for a review see Postma et al, 1996). The ptsH, ptsI, and crr genes form an operon; however, the crr gene possesses its own promoter (Fox et al, 1992; De Reuse and Danchin, 1988). The global transcriptional regulators, CRP, Mlc, and Fis, play important roles in gene expression of the glucose PTS (Plumbridge, 2002). Mlc is able to bind both IICBGlc protein and DNA, and regulates expression of the ptsG, ptsH, and ptsI genes and its own gene (Tanaka et al, 1999). The mlc gene possesses two promoters and they are both positively and negatively regulated by Mlc and CRP:cAMP (Decker et al, 1998; Shin et al, 2001). The regulation of fis gene expression is complex and is related to the growth of E. coli (Ball et al, 1992; Ninnemann et al, 1992; Pratt et al, 1997; Walker et al, 1999; Nasser et al, 2001; Mallik et al, 2004). The fis gene is excluded from the mathematical model for simplification. Basically, CRP:cAMP positively regulates the ptsG gene and pts operon expression, and Mlc negatively regulates them (Fox et al, 1992; De Reuse and Danchin, 1988; Plumbridge, 1998; Kim et al, 1999; Tanaka et al, 1999). The expression of the crp gene, which encodes the CRP protein, is regulated both positively and negatively by CRP:cAMP (Hanamura and Aiba, 1991, 1992; Ishizuka et al, 1994).
Adenylate cyclase catalyzes the reaction from ATP to cAMP and it is encoded by the cyaA gene. cyaA gene expression is negatively regulated by CRP:cAMP (Aiba, 1985; Inada et al, 1996). The adenylate cyclase reaction is considered to follow the Michaelis–Menten equation (Yang and Epstein, 1983). Intracellular cAMP is degraded to AMP by phosphodiesterase, which is encoded by the cpdA gene (Imamura et al, 1996), or is excreted into medium (Goldenbaum and Hall, 1979). The degradation and excretion of cAMP is assumed to follow a linear equation (Equation (3.4) in Supplementary Table S1). For simplification, only cAMP+CRP → CRP:cAMP is considered for the pathway of CRP:cAMP complex formation. The numbers of binding sites for CRP:cAMP on the whole genome were estimated using EcoCyc (Keseler et al, 2005).
Mathematical model
Based on the molecular interactions described above, we built a detailed mathematical model of the E. coli glucose PTS system (Supplementary Tables S1 and S2) using CADLIVE (Kurata et al, 2005). The CADLIVE system possesses a graphical user interface for inputting the biological process, building up mathematical equations, and system analysis. In the model, the mathematical formula consisted of 44 algebraic equations and 19 differential equations with 131 parameters. The model uses first-order mass action kinetics to describe the synthesis, proteolysis, and binding of proteins. We make the common assumption that binding reactions occur on a faster time scale than production and degradation of proteins. Therefore, we replace the differential equations describing these fast binding reactions by algebraic equations. The kinetic parameters for the PTS model were picked or estimated from existing PTS-related literature. The intracellular metabolite concentrations were set to a constant based on the data from Chassagnole et al (2002).
Module decomposition analogous to control engineering
A module has been characterized as a subsystem that possesses a function that is separable from that of other modules, in the sense that it is capable of maintaining most of its identity (Kurata et al, 2003, 2006). To provide a useful characterization of modularity, a multi-resolution scheme can then be used to assess different aspects of the modular decomposition, zooming out from the molecular description (molecular modules) to a block diagram-like picture (functional modules). At a lower level of resolution, the components of a system can be divided into functional groups that we refer to as functional modules. We sought analogies with the modules that are traditionally identified in control engineering schemes. The process to be controlled is identified and the rest of the network is classified in terms of the function that it accomplishes to facilitate this regulation. A block represents each of these modules and the interconnection of such blocks is frequently referred to as a block diagram. We define a flux module as a pathway that traces the mechanisms of interaction of a group of molecules involved together in the performance of a certain function. A flux module ideally connects functional modules, but forms an entity that possesses its own functionality. Therefore, a flux is essentially the flow of information in the network.
Strains and plasmids
The strains and plasmids used in this study are summarized in Table IV. E. coli MG1655 mlc− was constructed according to the method of Datsenko and Wanner (2000).
Table 4.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or gene | Source or reference |
|---|---|---|
| Strain | ||
| MG1655 | Wild type | |
| MG1655M | MG1655 mlc∷Cmr | This study |
| JM109 | Takara Bio | |
| Plasmid | ||
| pUC118 | Plasmid vector, Ampr | Takara Bio |
| pUC118-ptsI | pUC118-carrying promoter of phoC from Morganella morganii and ptsI gene from E. coli. | This study |
Briefly, 5′-AGAACCGTTATACATCGCGTCTTTTACCAGTGCAGCTGAAGCCTGCTTTTTTAT-3′ and 5′-TCTGCGCATTAGTCGCGGGGAGATTTTCCTTGCTCTCGCTCAAGTTAGTATAAA-3′ were used as PCR primers. The amplified fragment was used to replace the cat gene. Since the ptsI gene is located in the middle of the pts operon, it does not possess its own promoter (Fox et al, 1992; De Reuse and Danchin, 1988). We used the promoter sequence of the acid phosphatase gene, phoC from Morganella morganii, for stable ptsI gene expression in E. coli, with glucose in the medium (Mihara et al, 2000). Plasmid pUC118 and E. coli strain JM109 were used for cloning the ptsI gene.
For ptsI gene amplification, 5′-GCCTGCAGCCGTAAGGAGGAATGTAGATGATTTCGAGCATT-3′ and 5′-GCAAGCTTCTCGTGGATTAGCAG-3′ were used as PCR primers. As a promoter sequence, 5′-GCGGATCCATTTTTTCAATGTGATTTTAACTTTTACTTACAGATGAC TATAATGTGACTAAAAACAAAACCATTGTTCTGGACATCTGCAGCG-3′ was used. PCR products were treated with PstI and HindIII and the promoter sequence was treated with BamHI and PstI. After the ligation reaction using pUC118 digested with BamHI and HindIII and these fragments, the products were used to transform E. coli JM109.
Culture condition
Cells were grown in L-broth, M9 medium with 20 mM glucose (Sambrook et al, 1989), or EI medium containing 20 mM NH4Cl, 2 mM MgSO4·7H2O, 40 mM NaHPO4, 30 mM KH2PO4, 0.01 mM CaCl2, 0.01 mM FeSO4·7H2O, 0.01 mM MnSO4·5H2O, 5 mM citrate, 50 mM MES–NaOH (pH 6.8), and 20 mM glucose. Cultivation was carried out at 37°C using a shaking incubator.
Analysis
For glucose uptake assay with non-growing cells, cells were collected from 50 ml of overnight culture grown in L-broth, and then washed and inoculated into 50 ml of M9–glucose medium and shaken at 37°C, aerobically. For the glucose uptake assay with growing cells, cells were collected from 1 ml of overnight culture grown in L-broth, and then washed and inoculated into 20 ml of M9–glucose medium and shaken at 37°C for 18 h under aerobic conditions. Into each culture, ampicillin (100 μg/ml) was added as required.
The cAMP concentration was measured using a cAMP competitive ELISA kit (ENDOGEN, USA).
Supplementary Material
Supplementary Table S1
Supplementary Table S2
Supplementary Information
Supplementary Information
Acknowledgments
We thank Akira Imaizumi, Shintaro Iwatani, Yohei Yamada, and Takayuki Tanaka for invaluable discussions. We also thank Hiroyuki Aoyagi and Fumiko Yamamoto for excellent technical support.
References
- Aiba H (1985) Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP–cAMP receptor protein. J Biol Chem 260: 3063–3070 [PubMed] [Google Scholar]
- Ball CA, Osuna R, Ferguson KC, Johnson RC (1992) Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol 174: 8043–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M (2002) Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol Bioeng 79: 53–73 [DOI] [PubMed] [Google Scholar]
- Curtis SJ, Epstein W (1975) Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol 122: 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Reuse H, Danchin A (1988) The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J Bacteriol 170: 3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K, Plumbridge J, Boos W (1998) Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol 27: 381–390 [DOI] [PubMed] [Google Scholar]
- Eppler T, Postma P, Schutz A, Volker U, Boos W (2002) Glycerol-3-phosphate-induced catabolite repression in Escherichia coli. J Bacteriol 184: 3044–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DK, Presper KA, Adhya S, Roseman S, Garges S (1992) Evidence for two promoters upstream of the pts operon: regulation by the cAMP receptor protein regulatory complex. Proc Natl Acad Sci USA 89: 7056–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenbaum PE, Hall G (1979) Transport of cyclic adenosine 3′,5′-monophosphate across Escherichia coli vesicle membranes. J Bacteriol 140: 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Aiba H (1991) Molecular mechanism of negative autoregulation of Escherichia coli crp gene. Nuceic Acids Res 19: 4413–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Aiba H (1992) A new aspect of transcriptional control of the Escherichia coli crp gene: positive autoregulation. Mol Microbiol 6: 2489–2497 [DOI] [PubMed] [Google Scholar]
- Hernandez-Montalvo V, Martinez A, Hernandez-Chavez G, Bolivar F, Valle F, Gosset G (2003) Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol Bioeng 83: 687–694 [DOI] [PubMed] [Google Scholar]
- Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, Takahashi H, Aiba H, Postma PW (1998) Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol 30: 487–498 [DOI] [PubMed] [Google Scholar]
- Hogema BM, Arents JC, Inada T, Aiba H, van Dam K, Postma PW (1997) Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol Microbiol 24: 857–867 [DOI] [PubMed] [Google Scholar]
- Ikeda M (2003) Amino acid production processes. Adv Biochem Eng Biotechnol 79: 1–35 [DOI] [PubMed] [Google Scholar]
- Imamura R, Yamanaka K, Ogura T, Hiraga S, Fujita N, Ishihama A, Niki H (1996) Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J Biol Chem 271: 25423–25429 [DOI] [PubMed] [Google Scholar]
- Inada T, Takahashi H, Mizuno T, Aiba H (1996) Downregulation of cAMP production by cAMP receptor protein in Escherichia coli: an assessment of the contributions of transcriptional and posttranscriptional control of adenylate cyclase. Mol Gen Genet 253: 198–204 [DOI] [PubMed] [Google Scholar]
- Ishizuka H, Hanamura A, Inada T, Aiba H (1994) Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J 13: 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadner RJ, Murphy GP, Stephens CM (1992) Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J Gen Microbiol 138: 2007–2014 [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD (2005) EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res 33: D334–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Nam TW, Shin D, Koo BM, Seok YJ, Ryu S (1999) Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J Biol Chem 274: 25398–25402 [DOI] [PubMed] [Google Scholar]
- Kurata H, El Samad H, Iwasaki R, Othake H, Doyle JC, Grigorova I, Gross C, Khammash M (2006) Module-based analysis of robustness tradeoffs in the heat shock response system. PLoS Comput Biol 2: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata H, Masaki K, Sumida Y, Iwasaki R (2005) CADLIVE dynamic simulator: direct link of biochemical networks to dynamic models. Genome Res 15: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata H, Matoba N, Shimizu N (2003) CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle. Nucleic Acids Res 31: 4071–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW (1995) Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA 92: 11583–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik P, Pratt TS, Beach MB, Bradley MD, Undamatla J, Osuna R (2004) Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J Bacteriol 186: 122–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara Y, Utagawa T, Yamada H, Asano Y (2000) Phosphorylation of nucleosides by the mutated acid phosphatase from Morganella morganii. Appl Environ Microbiol 66: 2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H (2003) Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem 278: 15608–15614 [DOI] [PubMed] [Google Scholar]
- Nasser W, Schneider R, Travers A, Muskhelishvili G (2001) CRP modulates fis transcription by alternate formation of activating and repressing nucleoprotein complexes. J Biol Chem 276: 17878–17886 [DOI] [PubMed] [Google Scholar]
- Ninnemann O, Koch C, Kahmann R (1992) The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J 11: 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Death A, Ferenci T (1997) The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 143: 1909–1918 [DOI] [PubMed] [Google Scholar]
- Park YH, Lee BR, Seok YJ, Peterkofsky A (2006) In vitro reconstitution of catabolite repression in Escherichia coli. J Biol Chem 281: 6448–6454 [DOI] [PubMed] [Google Scholar]
- Patel HV, Vyas KA, Savtchenko R, Roseman S (2006) The monomer/dimer transition of enzyme I of the Escherichia coli phosphotransferase system. J Biol Chem 281: 17570–17578 [DOI] [PubMed] [Google Scholar]
- Plumbridge J (1998) Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol Microbiol 29: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Plumbridge J (2002) Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR (1993) Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57: 543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR (1996) Phosphoenolpyruvate; carbohydrate phosphotransferase systems. In Escherichia Coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC (ed), pp 1149–1174. Washington, DC: ASM Press [Google Scholar]
- Pratt TS, Steiner T, Feldman LS, Walker KA, Osuna R (1997) Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J Bacteriol 179: 6367–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Kamireddi M (1998) Modulation of Escherichia coli adenylyl cyclase activity by catalytic-site mutants of protein IIA(Glc) of the phosphoenolpyruvate: sugar phosphotransferase system. J Bacteriol 180: 732–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JV, Kaandorp JA, Dobrzynski M, Blom JG (2006) Spatial stochastic modeling of the phosphoenolpyruvate-dependent phosphotransferase (PTS) pathway in Escherichia coli. Bioinformatics 22: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Rohwer JM, Meadow ND, Roseman S, Westerhoff HV, Postma PW (2000) Understanding glucose transport by the bacterial phosphoenolpyruvate:glycose phosphotransferase system on the basis of kinetic measurements in vitro. J Biol Chem 275: 34909–34921 [DOI] [PubMed] [Google Scholar]
- Saier MH Jr (1998) Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol Bioeng 58: 170–174 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a Laboratory Manual, 2nd edn, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sauter T, Gilles ED (2004) Modeling and experimental validation of the signal transduction via the Escherichia coli sucrose phosphotransferase system. J Biotechnol 110: 181–199 [DOI] [PubMed] [Google Scholar]
- Seok YJ, Sondej M, Badawi P, Lewis MS, Briggs MC, Jaffe H, Peterkofsky A (1997) High affinity binding and allosteric regulation of Escherichia coli glycogen phosphorylase by the histidine phosphocarrier protein, HPr. J Biol Chem 272: 26511–26521 [DOI] [PubMed] [Google Scholar]
- Shin D, Lim S, Seok YJ, Ryu S (2001) Heat shock RNA polymerase (E sigma(32)) is involved in the transcription of mlc and crucial for induction of the Mlc regulon by glucose in Escherichia coli. J Biol Chem 276: 25871–25875 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kimata K, Inada T, Tagami H, Aiba H (1999) Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli. Genes Cells 4: 391–399 [DOI] [PubMed] [Google Scholar]
- Thattai M, Shraiman BI (2003) Metabolic switching in the sugar phosphotransferase system of Escherichia coli. Biophys J 85: 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KA, Atkins CL, Osuna R (1999) Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol 181: 1269–12680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel N, Kukuruzinska MA, Nakazawa A, Waygood EB, Roseman S (1982) Sugar transport by the bacterial phosphotransferase system. Phosphoryl transfer reactions catalyzed by enzyme I of Salmonella typhimurium. J Biol Chem 257: 14477–144791 [PubMed] [Google Scholar]
- Wendisch VF, Bott M, Eikmanns BJ (2006) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol 9: 268–274 [DOI] [PubMed] [Google Scholar]
- Yang JK, Epstein W (1983) Purification and characterization of adenylate cyclase from Escherichia coli K12. J Biol Chem 258: 3750–3758 [PubMed] [Google Scholar]
- Zeppenfeld T, Larisch C, Lengeler JW, Jahreis K (2000) Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICB(Glc) and induction behavior of the ptsG gene. J Bacteriol 182: 4443–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Table S2
Supplementary Information
Supplementary Information